Identification and characterization of the chalkiness endosperm gene CHALK-H in rice (Oryza sativa L.)
PlAO Ri-hua ,CHEN Mo-jun ,MENG Fan-mei ,Ql Chun-yan ,KOH Hee-jong ,GAO Meng-meng ,SONG An-qi,JlN Yong-mei,YAN Yong-feng#
1 Rice Research Institute,Jilin Academy of Agricultural Sciences,Gongzhuling 136100,P.R.China
2 Research Institute of Agricultural Biotechnology,Jilin Academy of Agricultural Sciences,Changchun 130033,P.R.China
3 Department of Agriculture,Forestry and Bioresources,Plant Genomics and Breeding Institute,Seoul National University,Seoul 08826,Republic of Korea
4 College of Agricultural Sciences,Yanbian University,Yanji 133000,P.R.China
Abstract Chalkiness is one of the most important agronomic traits in rice breeding,which directly affects the quality of rice seed.In this study,we identified a chalkiness endosperm mutant,chalk-h,from N-methyl-N-nitrosourea (MNU)-induced japonica rice cultivar Hwacheong (HC). Compared with wild type (WT)-HC,chalk-h showed severe chalkiness in the endosperm,yellowish green leaves,as well as reduced plant height. Scanning electron microscopy (SEM)analysis showed that starch grains in the chalk-h mutant were irregular in size and loosely arranged,with large gaps between granules,forming ovoid or orbicular shapes. MutMap analysis revealed that the phenotype of chalk-h is controlled by a single recessive gene LOC_Os11g39670 encoding seryl-tRNA synthetase,which is renamed as CHALK-H. A point mutation occurs in chalk-h on the sixth exon (at nucleotide 791) of CHALK-H,in which adenine(A) is replaced by thymidine (T),resulting in an amino acid codon change from glutamine (Glu) to valine (Val). The chalk-h mutant exhibited a heat-sensitive phenotype from the 3-leaf stage,including yellow-green leaves and reduced pigment content. The transcriptional expression of starch synthesis-related genes was down-regulated in the chalk-h mutants compared to WT-HC at different grain-filling stages. With an increase in temperature,the expression of photosynthesis-related genes was down-regulated in the chalk-h mutant compared to WT-HC. Overexpression of CHALK-H rescued the phenotype of chalk-h,with endosperm and leaf color similar to those of WT-HC. Our findings reveal that CHALK-H is a causative gene controlling chalkiness and leaf color of the chalk-h mutant. CHALK-H is the same gene locus as TSCD11,which was reported to be involved in chloroplast development under high temperature. We suggest that CHALK-H/TSCD11 plays important roles not only in chloroplast development,but also in photosynthesis and starch synthesis during rice growth and development,so it has great application potential in rice breeding for high quality and yield.
Keywords: CHALK-H,Seryl-tRNA synthetase,chalkiness,rice
1.Introduction
Rice (OryzasativaL.) is one of the most important food crops in the world,and starch is the main component of rice seeds,accounting for about 80% of the total dry matter in the endosperm of rice (Fanget al.2014).Therefore,starch accumulation has an extremely important economic value for the formation of rice yield and quality. The formation of starch is a complex enzymatic process,and the starch is stored as powder in the form of amylose and amylopectin (Nakamura 2002).The formation of storage starch is accompanied by the proliferation and development of powder and starch granules in endosperm cells,which requires the synergistic action of various enzymes and regulatory factors (Kuboet al.1999;Hirose and Terao 2004;Kawagoeet al.2005;Wooet al.2008). The mutation of genes related to starch synthesis and metabolism will lead to changes in starch content or structure.
Chalkiness is the white opaque part formed due to insufficient grain filling,a loose arrangement of starch and protein particles in the endosperm,and the gaps between them affect the transmission of light (Guoet al.2011).According to its position on the endosperm,it can be divided into white-belly,white-back,and white-core (Peng Bet al.2014). Chalkiness is one of the most important factors affecting rice quality,including appearance quality,processing quality,cooking quality and nutritional quality,which is directly related to the marketability and market value of the rice (Guoet al.2011;Bianet al.2013). Grain chalkiness is generally evaluated by the area of chalky endosperm and the percentage of chalky grains. Rice grains with a chalkiness rate of more than 20% are generally not accepted in the market. Therefore,reducing chalkiness cannot only improve the appearance quality and palatability of rice,but also enhance its commercial value.
Exploring chalkiness related genes and analyzing their molecular mechanism can greatly promote the breeding of high-quality rice varieties. Many genes related to chalkiness have been cloned and analyzed in rice,such asosbt1,flo4,flo5,flo7,gpa1,ms-handWCR1(Kanget al.2005;Ryooet al.2007;Wooet al.2008;Wanget al.2010;Liet al.2014;Cakiret al.2016;Zhanget al.2016;Caiet al.2018;Wuet al.2022).Cakiret al.(2016) found that the mutantosbt1shows a heart-white phenotype,and theOsBT1gene encodes the ADPG transporter,which is involved in the regulation of starch synthesis and the formation of complex starch particles during the development of rice seeds.flo4is a T-DNA insertion mutant with a white-core endosperm phenotype,reduced grain weight,and increased protein and fat contents.OsPPDKBencodes pyruvate phosphate double kinase,which plays an important role in starch metabolism and the structure of the rice endosperm (Kanget al.2005). Theflo5mutant shows a white-core phenotype,reduced grain weight,and significantly reduced amylopectin long chains.OSSIIIaencodes starch synthase,which is mainly involved in the extension of the amylopectin B2–B4 chain(Ryooet al.2007). Wanget al.(2010) found that the mutantgpa1shows a white-core endosperm,and the complex powder granules in the endosperm are round and loosely wrapped.OsRab5aencodes a GTPase(GTPase),which is involved in protein transport in the vacuoles of endosperm cells.chalk5was the first cloned major QTL for controlling chalkiness,and it encodes a vacuolar H+-translocating pyrophosphatase (V-PPase).Overexpression ofChalk5can increase endosperm chalkiness,which may be caused by interference with the pH homeostasis of the developing seed intimal transport system. This process affects proteome formation and is coupled with a significant increase in vesicle-like structures,thus forming gas space in the endosperm storage material and leading to grain chalkiness (Liet al.2014). Although many chalkiness related genes have been cloned,there are still many unknown events in starch synthesis. Therefore,it is important to identify new chalkiness related mutants and clone their regulating genes in order to further clarify the starch synthesis and metabolic pathways for rice quality improvement.
In this study,we identified a novel chalkiness endosperm mutant,chalk-h,from the HC-MNU population.We characterized the phenotype ofchalk-hand cloned its regulating geneCHALK-Hthrough a MutMap strategy.CHALK-His the same locus ofTSCD11,which is considered to be the determinant of the albino leaves of mutanttscd11under high temperature (Fanget al.2020).Our results provide new evidence for the pleiotropic effect ofCHALK-H/TSCD11on the leaf albino and chalkiness traits.
2.Materials and methods
2.1.Plant materials and growth conditions
A chalkiness endosperm mutant line was induced by the chemical mutagenesis of ajaponicarice cultivar,wild type(WT)-Hwacheong (HC),using N-methyl-N-nitrosourea(MNU). The seeds of the chalkiness endosperm mutant(chalk-h) used in this study were taken from the M11generation. An F2population derived from a cross between thechalk-hand the WT-HC was used for whole genome sequencing. For genetic mapping,Milyang 23,a Korean Tongil-type rice cultivar,was crossed withchalk-hto obtain the mapping population. The parents and mapping populations were cultivated using conventional cultural practices on experimental farms at Seoul National University (Suwon,South Korea) and the Rice Research Institute of Jilin Academy of Agricultural Sciences(Changchun,China),respectively.
For the temperature treatment experiments,WT andchalk-hmutant seeds were disinfected with 3%hypochlorite for 20 min and placed in 1/2 MS medium in a 96-well plant hydroponic box. The seeds were grown in growth chambers (light and dark for 16 and 8 h,respectively) at different temperatures (25,30 and 35°C).
2.2.Measurement of pigment contents
The pigments were determined according to the method of Chenet al.(2023) with slight modifications. Samples of 0.2 g of the fresh leaves of WT andchalk-hmutants at different growth stages in the growth chamber were cut into small pieces. The leaves were completely soaked in 50 mL of 95% ethanol and kept in the dark at 26°C for 24–36 h. The contents of chlorophylla(Chla),Chlband carotenoids (Car) were measured at 663,645 and 475 nm,respectively,by an ultraviolet spectrophotometer.
2.3.Scanning electron microscopy analysis
Scanning electron microscopy (SEM) analysis was carried out according to Qiaoet al.(2010). Cut mature seeds were coated with gold and the mounted specimens were observed under a Stereoscan Leica Model 440 Scanning Electron Microscope (Leica Cambridge,UK) at an accelerating voltage of 10–20 kV.
2.4.X-ray diffraction of starch
The X-ray diffraction patterns were obtained as described previously by Qiaoet al.(2010). The crystalline structure of starch was obtained using an X-ray powder diffractometer (D8 ADVANCE with DAVINCI,BRUKER,German) operating at 40 kV and 40 mA. The scanning region of the two-theta angle (2θ) ranged from 4.0 to 40.0° with a scan speed of 1 deg min–1.
2.5.Measurement of starch,lipid,and total protein contents
To measure the starch,amylose,lipid and total protein contents,we husked the rice grains and ground them to a flour consistency in a mill. Starch and amylose contents were determined according to the National industry standards of China (NY/T 11-1985,1985;NY/T 55-1987,1987),respectively. Lipid and protein contents were determined using the method described by Kanget al.(2005).
2.6.Genetic analysis and fine mapping of the CHALK-H gene
A total of 192 F2progeny from a cross betweenchalk-hand Milyang 23 was used for segregation analysis and gene mapping. For the initial mapping of thechalk-hgene,we performed recessive class analysis (RCA) using 136 polymorphic STS markers designed at the Crop Molecular Breeding Laboratory,Seoul National University,which were equally dispersed over the whole set of rice chromosomes. After BSA,fine mapping was performed using three additional STS markers designed against two rice databases (http://www.gramene.org;https://rapdb.dna.affrc.go.jp). PCR was performed as described by Piaoet al.(2009) with slight modifications. The primer sequences are listed in Table 1.
2.7.MutMap analysis
DNAs for the MutMap analysis were extracted from rice leaves with the CTAB method and treatment with RNaseI. HC and a bulked DNA pool were used for the MutMap analysis. The bulked DNA was prepared by mixing DNA equally from 32 F2individuals displaying the chalkiness endosperm phenotype. Sequencing libraries were generated using the Truseq Nano DNA HT Sample preparation Kit (Illumina,USA) and sequenced by the Illumina HiSeq4000 platform (Illumina,CA,USA). To generate the HC reference sequence,28.1 Gb of WT HC sequence reads was aligned to the Nipponbare reference genome using BWA (Burrows-Wheeler Aligner) Software.Alignment files were converted to SAM/BAM files through SAM tools,and the SNP index was calculated as described by Abeet al.(2012).
2.8.RNA isolation and quantitative real-time RTPCR
Total RNA was isolated from the mutants and WT using the OminiPlant RNA Kit (Jiangsu CoWin Biotech,China).Reverse transcription was performed with the HiFiScript gDNA Removal RT MasterMix (Jiangsu CoWin Biotech,China). The real time PCR primers related to starch synthesis and photosynthesis were designed according to Ohdanet al.(2005) and Fanget al.(2020),respectively,and theAction1gene was used as an internal control.qRT-PCR was performed using SYBR PremixEx Taq(TaKaRa,Japan) on an ABI Q3 real-time system (Thermo Fisher,USA).
2.9.Vector construction and rice transformation
For complementation testing,a PCR-amplified full-length ORF ofCHALK-H(LOC_os11g39670) was digested withSmaI andSpeI,and inserted into the pCAMBIA3300-ubi overexpression vector,which is a pCambia3300-modified vector containing the maize ubiquitin promoter. The Agrobacterium strain EHA105 harboring overexpression construct was used to transformation rice calli induced from mature embryos of thechalk-hmutants,according to the method described by Nishimuraet al.(2006),with slight modifications. The primers used for vector construction and genotyping are listed in Table 1.
3.Results
3.1.Characterization of the chalk-h mutant phenotype
We identified a chalkiness endosperm mutant,chalk-h,from the HC-MNU population,which was constructed by mutagenesis ofOryzasativassp.japonicacultivar HC with MNU. Thechalk-hmutants can be easily distinguished from WT plants by their general agronomic traits,such as leaf color,plant height,1 000-grain weight,seedsetting rate and spikelet fertility. Thechalk-hmutants exhibited a yellowish green phenotype at the 3-leaf stage,and the stem length was short at the seedling stage,leading to a significant reduction in plant height,and a low seed-setting rate and 1 000-grain weight (Fig.1-A–C;Table 2). Compared with the WT,the endosperm of grains inchalk-hmutants exhibited opaque white core endosperm (Fig.1-D and E). SEM analysis showed that the starch grains in WT endosperm were normal in size,tightly assembled and squeezed together to form regular polyhedrons,while those inchalk-hwere irregular in size and loosely arranged,with large gaps between granules,forming ovoid or orbicular shapes (Fig.2-A–D). X-ray diffraction analysis of seed endosperm starch showed a typical A-type crystal pattern in both WT and thechalk-hmutant (Fig.2-E). However,the peak intensity of thechalk-hmutant was lower than that of the WT-HC,indicating that the amount of crystallization was reduced in thechalk-hmutant without changing the crystallization type (Fig.2-E).
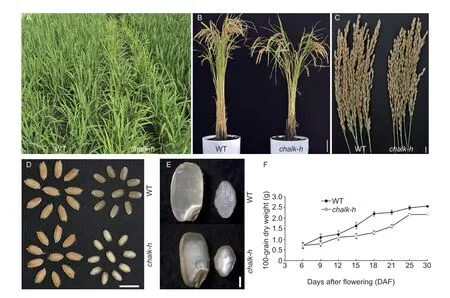
Fig.1 Phenotype comparisons of the wild type (WT) and chalk-h mutant. A,tillering stage on the experimental farm. B,whole plant at mature stage. C,mature panicles. D,de-husked seeds. E,vertical and transverse sections of polished seed endosperms.F,grain-filling process. Scale bars,20 cm in B;1 cm in C and D;1 mm in E. Bars in F mean SD (n=3).
During the whole grain-filling process,the dry matter accumulation level of thechalk-hmutant was significantly lower than that of the WT (Fig.1-F),indicating that the grain fullness ofchalk-hwas reduced,leading to the gaps between the starch grains. In thechalk-hmutant,the seed starch and amylose contents were lower,while the protein and lipid contents were higher than WT (Fig.2-F–I).These results suggest that thechalk-hmutation changed the biosynthesis of stored substances in the endosperm,especially starch synthesis.
3.2.Genetic analysis and candidate gene identification
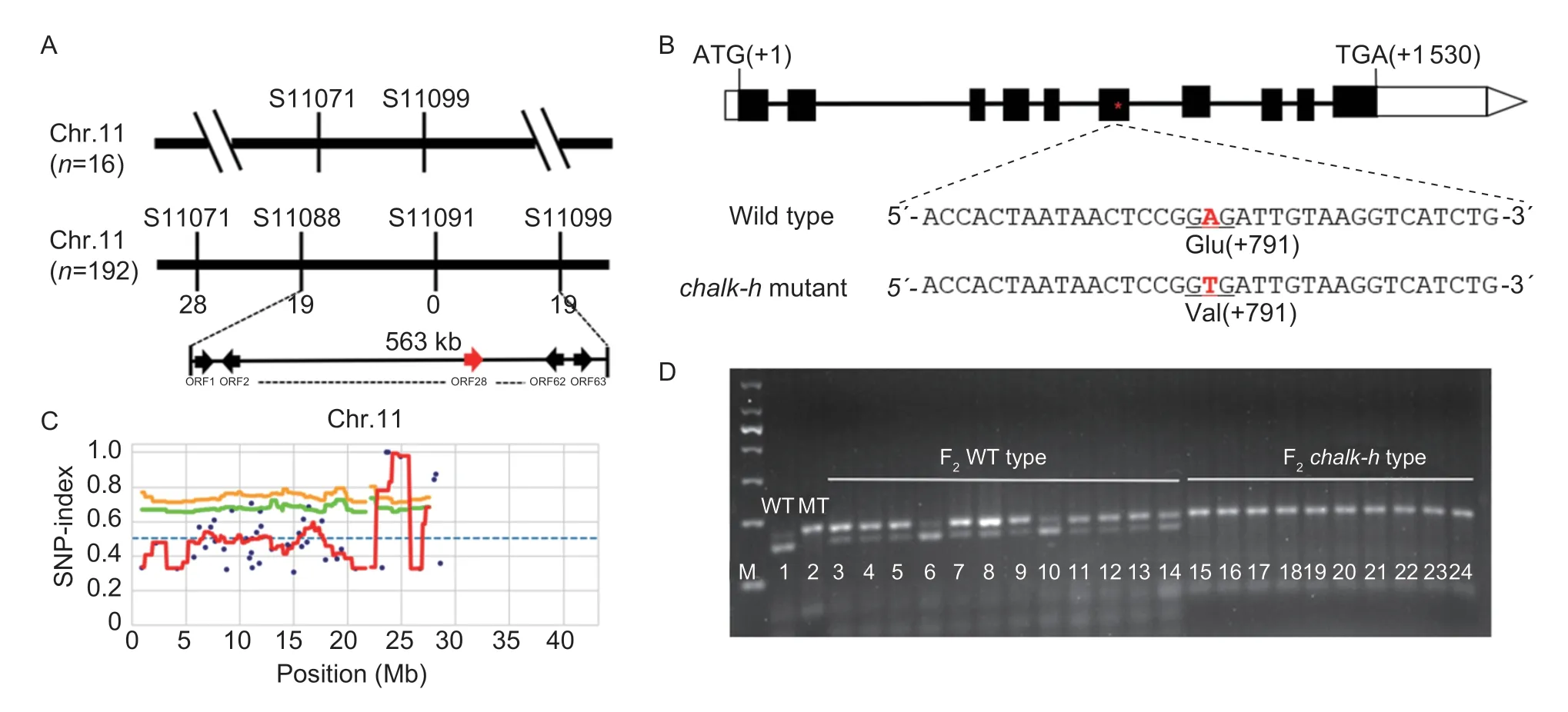
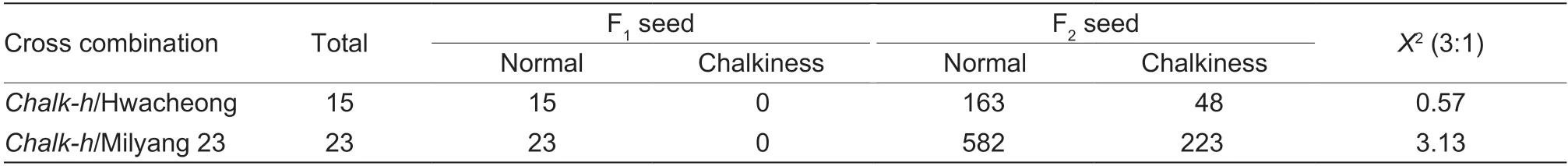
In order to generate a segregation population,chalk-h(japonica) was used as the female parent to cross withtwo WT plants,Milyang 23 (indica) and Hwacheongbyeo(japonica). All F1seed from these crosses had normal seed phenotypes,indicating that the chalkiness phenotype is recessive. In the F2seeds,some grains showed chalkiness similar to thechalk-hmutant. The results showed that the segregation ratio between normal endosperm and chalkiness endosperm was 3:1(X2 Table 2 Agronomic traits of the chalk-h mutant and wild-type (WT) cultivar Hwaheong (HC)1) To determine the physical location of theCHALK-Hgene,BSA analysis was performed on F2seeds derived from the cross between Milyang 23 andchalk-husing 118 STS (sequence-tagged site) polymorphic makers distributed on 12 chromosomes. The two markers(S11071 and S11099) on the end of the long arm of chromosome 11 appeared to be linked to the mutant phenotype and were used to genotype 192 F2chalkiness individuals. Linkage analysis showed that theCHALK-Hgene between the STS markers S11088 and S11099 was co-segregated with the STS marker S11091,and flanked markers with a physical distance of 563 kb including 63 open read frames (ORF) (Fig.3-A). Fig.3 Positional cloning of the chalk-h gene. A,genetic mapping of the chalk-h locus with sequence-tagged site (STS) markers.The candidate gene was mapped to a 563-kb region on chromosome 11. B,SNP index plot of chr.11 for the identification of genomic regions harboring the causal mutation of the chalk-h gene. C,schematic diagram of the chalk-h gene. Black rectangles represent exons and the red star represents the mutation site. D,co-segregation analysis of F2 plants using dCAPs markers. Lane 1,Hwacheong;lane 2,chalk-h mutant;lanes 3–14,normal phenotype;lanes 15–24,chalkiness phenotype. Table 3 Genetic segregation of the chalkiness phenotype in rice (=3.84) Table 3 Genetic segregation of the chalkiness phenotype in rice (=3.84) In order to locate the mutations and the gene responsible for the chalkiness phenotype,a bulk population (32 plants) of DNA from F2plants derived from a cross betweenchalk-hmutant and WT-HC were used for MutMap analysis. A total of 19.41 Gb of clean data was used in the MutMap analysis,with Q30 reaching 93.51%.The average ratio of sample to reference genome was 98.255%,the average coverage depth was 39.50X,and the genome coverage was 88.61%. The sequence reads were aligned to the HC genomic sequence. Based on the average SNP index peak,the most probable candidate region was detected in the 157 kb region of chromosome 11,which is located in the flanked STS marker region(Fig.4-B). Only two SNPs with an SNP index of 1 were found in the 156 kb candidate region,one of these two SNPs was located in the–452 bp promoter region ofLOC_Os11g39910,and the other was located in the sixth exon ofLOC_Os11g39670. The dCAPs makers were designed to analyze these two candidate genes,and the results showed thatLOC_Os11g39670has the mutation site. A point mutation occurred at 791 bp on the ORF ofLOC_Os11g39670(on the sixth exon),in which adenine (A)was substituted by thymidine (T),leading to the change of glutamine acid (Glu) to valine (Val) in thechalk-hmutant(Fig.4-A–C). A co-segregation analysis was performed using the derived split expanded polymorphic sequence(dCAPs) markers (Table 1). The results showed that the genotypes and phenotypes were completely co-segregated in the F2population (Fig.4-D). These results suggest that the chalkiness phenotype ofchalk-hwas derived from the point mutation ofLOC_Os11g39670. Therefore,we designatedLOC_Os11g39670asCHALK-H. In a previous study,LOC_Os11g39670was designated asTSCD11(temperature-sensitive chlorophyll-deficient 11)(Fanget al.2020). In order to test whether thechalk-hmutants have the same heat-sensitive phenotype,WT andchalk-hmutants were grown at 25,30 and 35°C. The WT andchalk-hmutants are cold regionjaponicarice varieties that are sensitive to high temperature. The phenotypes of all WT andchalk-hmutants were almost identical at the different temperatures until the 2-leaf stage (Fig.4-A–C),however,interestingly,they changed at 3-leaf stage.Thechalk-hmutants showed a yellow-green phenotype at 25°C and a white stripe phenotype at 30°C (Fig.4-A,B,D and E);and when the temperature reached 35°C,the number of white leaves increased compared with the 30°C treatment. Under the condition of 35°C,the newly grown 4th leaves showed the albino phenotype (Fig.4-C arrows),but the already grown 1st to 3rd leaves did not turn albino. With the increase in treatment temperature,the pigment contents of both WT andchalk-hmutants decreased to some extent (Fig.4-G–I). The contents of Chla,Chlband Car in thechalk-hmutants decreased the most significantly at 35°C,by 37.1,50.39 and 42.06%,respectively,compared with the WT (Fig.4-C,F and I). In order to better understand the role ofLOC_Os11g39670in rice development,the expression pattern ofLOC_Os11g39670in different organs and tissues of WT-HC at the reproductive stage was examined by quantitative RT-PCR (Fig.5). The transcriptional expression level ofLOC_Os11g39670was the highest in leaves,followed by panicle and stem.LOC_Os11g39670was mainly expressed in green tissues,and slightly expressed in endosperm during the grain-filling stage (Fig.5-A). Under different temperature treatments,the transcriptional expression pattern ofLOC_Os11g39670was significantly different in WT and thechalk-hmutant. At 25°C,the transcriptional expression level ofLOC_Os11g39670increased with increasing leaf age in both WT-HC and thechalk-hmutant;at both 30 and 35°C,it increased at first and then decreased with increasing leaf age in WT-HC,however,it decreased dramatically in thechalk-hmutant until almost no expression was observed (Fig.5-B and C). Fig.5 Transcriptional expression of LOC_Os11g39670. A,qRT-PCR analysis of LOC_Os11g39670 in various tissues (panicles,roots,leaves,stems,seeds at 6,9 and 12 days after flowering of wild type (WT)-Hwahenong (HC) plants,and the expression levels in the roots were set as the control. B and C,comparison of LOC_Os11g39670 expression in WT-HC and the chalk-h mutant under different temperatures (25,30 and 35°C),and the expression levels in the WT-HC at the 2-leaf stage under 25°C were set as the control. Bars mean SD (n=3). The contents of dry matter in the grain-filling stage and amylose in the mature stage in thechalk-hmutants were significantly different from those of the WT. Therefore,the expression of starch synthesis-related genes may be affected to some extent. The expression of starch synthesis-related genes in the endosperm of thechalk-hmutant and WT-HC was analyzed at 6,9 and 12 d after flowering (Fig.6-A). The results showed that most of the starch synthesis-related genes were down-regulated in thechalk-hmutants at different grain-filling stages. In particular,the expression levels of theOsAGPS2b,OsAGPL1,OsAGPL4,OsSSIIaandOsBEIIbgenes inchalk-hmutants were significantly down-regulated during the whole grouting period. In addition,the expression levels ofOsAGPS1,OsAGPS2a,OsISA1,OsISA3andOsPULgenes fluctuated between WT-HC and thechalk-hmutant. The leaf color change and reduction in pigment content in thechalk-hmutants at high growth temperature (Fig.4)may affect photosynthetic capacity. The expression levels of the photosynthesis-related genes Photosystem I Subunit A (PsaA),Photosystem II Subunit of A (PsbA),Rice Chlorophylla/bBinding Protein (CAB2R),and Rubisco Large Subunit (RbcL) were analyzed in thechalk-hmutants and compared to WT-HC under different temperature treatments (Fig.6-B–E). The results showed that the expression of most photosynthesisrelated genes inchalk-hmutants was down-regulated at 25°C from the 1-to the 3-leaf stage;but when the temperature was increased to 30 or 35°C,the expression of photosynthesis-related genes in thechalk-hmutants at each leaf stage was down-regulated,and the amplitude of down-regulation increased with an increase in the treatment temperature. To confirm whetherCHALK-His related to the chalkiness phenotype and leaf color change,we performed a complementation assay. A vector containing theCHALK-Hcoding sequence under the control of the maize-ubiquitin promoter (ubq:CHALK-H) (Fig.7-A)was introduced into thechalk-hmutant,and transgenic lines were identified by Test cards for GMO (PAT/bar-Bt Cry1Ab/Ac) (Genesino,China) and PCR analysis. A total of seven independent transgenic lines were obtained,among which five lines (OE-1,OE-2,OE-5,OE-6,and OE-39) were completely recovered to normal endosperm and their leaves returned to green. Fig.7 Overexpression transgenic plant analysis. A,schematic of the overexpression vector construct. LB,left board;RB,right board;Bar,bialaphos resistance gene;ubi,rice ubiquitin promoter. B,CHALK-H gene expression in the overexpression transgenic plants. C,SPAD value of the flag leaf of overexpression transgenic plants. Bars mean SD (n=3). *,0.01 The expression level ofCHALK-Hwas quantified in the transgenic lines by real-time PCR analysis. The results showed that the expression levels ofCHALK-Hin the two unrestored lines (OE-3 and OE-4) were significantly lower than in the restored lines (Fig.7-B). These results revealed thatCHALK-His the causal gene for thechalk-hmutant phenotype. With the improvement of living standards,people now have higher requirements for the quality of rice.Chalkiness is one of the most important indexes for evaluating rice appearance quality,but it also affects the taste quality,processing quality and nutritional quality of rice,as well as grain weight and rice yield (Liuet al.2011;Zhenget al.2012). In this study,we identified a chalkiness mutant,chalk-h,whose plant height,ear length,grain width,1 000-grain weight and other important agronomic traits were significantly reduced compared with the WT (Table 2),which was different from the findings of other studies on chalkiness-related mutants (Qiaoet al.2010;Chenet al.2020). We mapped the gene of thechalk-hmutant and named itCHALK-H. Our results suggest that a single point mutation impedes normal plant development in thechalk-hmutant,and its growth restriction may be the main reason for the formation of chalkiness in the grains. Chalkiness is closely related to the coordination of“sourcing–sink flow” in rice plants,grain-filling dynamics and the formation and accumulation of starch in the endosperm,and is also easily affected by environmental conditions (Liuet al.2007;Zhouet al.2009). Compared with the WT-HC,thechalk-hmutant had an opaque white core endosperm (Fig.1-D and E) with loosely arranged starch grains that varied in size (Fig.2-A–D). In addition,the level of dry matter accumulation inchalk-hwas significantly lower than in WT-HC throughout the whole grain filling process (Fig.1-F). These results indicate that the chalkiness phenotype ofchalk-his caused by the gaps between starch grains in the endosperm due to reduced grain fullness. The main components of rice endosperm are starch and protein. The composition and structure of starch and protein in the chalkiness mutant is changed,and the expression of genes related to starch synthesis is also seriously affected (Fanget al.2014). Thechalk-hmutants had changes in the biosynthesis of stored substances in endosperm,in that the seed starch and amylose contents were lower,while the protein and lipid contents were higher than WT (Fig.2-F–I). In many chalkiness mutants,the expression levels of starch synthesis-related genes were much lower than in WT (Fanget al.2014;Peng Bet al.2014;Peng Cet al.2014;Wuet al.2015;Chenet al.2020). In this study,most of the starch synthesisrelated genes (OsAGPS2b,OsAGPL1,OsAGPL4,OsSSIIaandOsBEIIb) inchalk-hwere down-regulated as in other chalkiness mutants,however,interestingly,the repression amplitude was small (Fig.6-A). Therefore,we speculated that the chalkiness phenotype in thechalk-hmutant may not be caused by mutations in the genes associated with starch synthesis (Peng Cet al.2014;Wuet al.2015). Despite the many reports on chalkiness mutants,the formation mechanism of chalkiness is very complex and the molecular mechanisms of its formation and regulation has not been well studied,therefore,more cloning and functional studies on the related genes are needed. In the present study,we narrowed thechalk-hlocus betweenLOC_Os11g39910andLOC_Os11g39670on chromosome 11 using the MutMap strategy. A point mutation (A to T) occurred at locus 791 bp ofLOC_Os11g39670,in which Glu is substituted to Val (Fig.3).We designated it asCHALK-Hand found that it encodes a seryl-tRNA synthetase. Sertyl-tRNA synthetase is one member of the aminoacyl-tRNA synthetase family (aaRSs),which consists of 20 kinds of synthetases. aaRSs are the earliest proteins in the process of the evolution of life,which play an important role in the body by catalyzing the acylation of amino acids and corresponding tRNAs to ensure the accuracy of genetic information translation(Paket al.2018). Sertyl-tRNA synthase has been frequently reported in microorganisms and animals,but rarely in plants. Using virus-induced gene silencing(VIGS) techniques,Kimet al.(2005) found that reducing the expression of sertyl-tRNA synthase in tobacco led to severe leaf yellowing. A new heat-sensitive mutant identified in rice,tscd11,has a point mutation in sertyltRNA synthase,resulting in an albino leaf phenotype at the 3-leaf stage at 35°C. In this study,thechalk-hmutants showed different heat-sensitive phenotypes at the three leaf stages under different temperature treatments,with yellow-green at 25°C,white stripe at 30°C and white stripe and albino at 35°C (Fig.4). These results are different from those of thetscd11mutants,in that the leaf color change occurred at 35°C. In addition,the chalkiness phenotype is not mentioned in relation totscd11(Figs.1-A and 5-A–F). We suggest that the reason for the phenotype difference betweentscd11andchalk-hmutants is as follows. Thechalk-hmutation is in cold regionjaponicarice varieties that are more sensitive thantscd11to high temperature. Also,chalk-handtscd11have differences in their nucleotide mutation sites and amino acid codon changes in which the protein activity varies according to the mutation site ofCHALK-H/TSCD11,which causes thechalk-hmutant to exhibit a different phenotype fromtscd11;that is,chalkiness endosperm and yellow-green leaves occurred at the normal temperature in thechalk-hmutant. Complementation experiments confirmed that the mutation in theCHALK-Hgene was a causative factor for the chalkiness phenotype of thechalk-hmutant. A total of five independent overexpressing lines completely recovered from the chalkiness phenotype to normal endosperm,except for two lines (OE-3 and OE-4) in which theCHALK-Hexpression level and chlorophyll content were too low to recover the normal endosperm (Fig.5-B and C). The transcription expression of starch synthesisrelated genes showed that although there were expression changes in thechalk-hmutants (Fig.6-A),the amplitudes of those changes were obviously different from those in other endosperm mutants generated by changes in the expression of starch synthesis-related genes (Fanget al.2014;Chenet al.2020). In contrast,the photosynthesisrelated genes were significantly down-regulated in thechalk-hmutant (Fig.6-B–E). In particular,the expression levels of photosynthesis-related genes inchalk-hmutants were significantly lower than in the WT-HC at both 30 and 35°C. Therefore,we speculated that the root cause of chalkiness in thechalk-hmutant might be the reduction in its photosynthetic capacity,which leads to insufficient assimilation material,i.e.,the “source” is small while the “sink” is large. The chain reaction of the chlorophyll reduction,diminished photosynthetic capacity and insufficient assimilation material is ultimately responsible for producing the chalkiness grains. The molecular mechanism ofCHALK-Hin photosynthesis and starch synthesis will be clarified in future studies. In this study,we identified an MNU-induced chalkiness mutant,chalk-h,which is accompanied by yellowishgreen leaves,as well as reduced culm height,seedsetting rate,fertility and 1 000-grain weight. Inchalk-h,a single nucleotide substitution mutation from A to T occurred on the sixth exon (at nucleotide 791) ofCHALK-H,resulting in an amino acid codon change from Glu to Val. Complementation analysis further confirmed thatCHALK-His the causative gene for thechalk-hmutant.CHALK-Hencodes seryl-tRNA synthetase,which has been reported asTSCD11,conferring chloroplast development under high temperature. Although the typical phenotypes ofchalk-handtscd11are different,both are caused by reduced chlorophyll content. This study provides new insights into the role ofCHALK-Hin leaf color and grain development. This research was supported by the Administration of Central Funds Guiding the Local Science and Technology Development,China (202002069JC) and the earmarked fund for the China Agriculture Research System (CARS-01-10). The authors declare that they have no conflict of interest.
3.3.Phenotypes of the chalk-h mutant at different temperatures
3.4.Expression pattern of LOC_Os11g39670 in the different organs

3.5.Expression analysis of starch synthesis-and photosynthesis-related genes
3.6.Complementation of the chalk-h mutant

4.Discussion
5.Conclusion
Acknowledgements
Declaration of competing interest
 Journal of Integrative Agriculture2023年10期
Journal of Integrative Agriculture2023年10期
