CRISPR/Cas9-mediated knockout of SLC15A4 gene involved in the immune response in bovine rumen epithelial cells
JIANG Mao-cheng ,HU Zi-xuan ,WANG Ke-xin ,YANG Tian-yu ,LIN Miao, ,ZHAN Kang,#,ZHAO Guo-qi,#
1 Institute of Animal Culture Collection and Application,College of Animal Science and Technology,Yangzhou University,Yangzhou 225009,P.R.China
2 Faculty of Health Sciences,University of Macau,Macau 999078,P.R.China
3 Institutes of Agricultural Science and Technology Development,Yangzhou University,Yangzhou 225009,P.R.China
4 Joint International Research Laboratory of Agriculture and Agri-Product Safety of Ministry of Education,Yangzhou University,Yangzhou 225009,P.R.China
Abstract The objective of this study was to determine the role of SLC15A4 in the muramyl dipeptide (MDP)-mediated inflammatory response of bovine rumen epithelial cells (BRECs). First,changes in the mRNA expression of proinflammatory factor genes in BRECs following 10 µg mL–1 MDP treatments were examined. RT-qPCR results showed that the expression of pro-inflammatory factor (IL-1β,IL-6,and TNF-α) mRNAs were significantly increased under MDP stimulation (P<0.001). Moreover,SLC15A4-Knockout (SLC15A4-KO) cells were obtained through lentivirus packaging,transfection,screening,and cell monoclonal culture. In order to gain further insight into the potential function of SLC15A4,we utilized transcriptome data,which revealed a change in the genes between WT-BRECs and SLC15A4-KO. Five down-regulated pro-inflammatory genes and 13 down-regulated chemokine genes related to the inflammatory response were identified. Meanwhile,the down-regulated genes were mostly enriched in the nuclear factor κB (NF-κB) and mitogen-activated protein kinase (MAPK) signaling pathways. The results of RTqPCR also verified these detected changes. To further determine the mechanism of how WT and SLC15A4-KO BRECs are involved in inflammatory responses,we investigated the inflammatory responses of cells exposed to MDP. WT-BRECs and SLC15A4-KO were treated with a culture medium containing 10 µg mL–1 MDP,in comparison to a control without MDP. Our results show that SLC15A4-KO BRECs had reduced the expression of genes (IL-6,TNF-α,CXCL2,CXCL3,CXCL9,and CCL2) and proteins (p-p65 and p-p44/42) from the MDP-mediated inflammatory response compared to WT-BRECs (P<0.05). In this experiment,CRISPR-Cas9 was used to KO the di/tripeptide transporter SLC15A4,and its role was confirmed via the MDP-induced inflammatory response in BRECs. This work will provide a theoretical basis for studying the pro-inflammatory mechanism of MDP and its application in the prevention and treatment of subacute rumen acidosis in dairy cows.
Keywords: SLC15A4,CRISPR/Cas9,immune response,proton-coupled oligopeptide transporter (POT) families,MDP
1.Introduction
The bovine rumen epithelium is not only a major absorption and transport site for nutrients but also the main site for rumen microbial attachment and growth (Trosviket al.2018). The rumen epithelium,in addition to its nutrient absorptive functions,is a physical barrier between the rumen microbes and the underlying host immune system(Steeleet al.2016). According to previous research,cows on a high grain diet experience a fast rise in short chain fatty acids (SCFAs) in the rumen,which will cause subacute rumen acidosis and the death of the majority of rumen bacteria. At the same time,the death and disintegration of bacteria will release a large amount of lipopolysaccharide (LPS) and muramyl dipeptide (MDP),resulting in damage to the rumen epithelium (Coulombeet al.2009;Andradeet al.2021). MDP is a conserved peptidoglycan structure found in the majority of Grampositive and -negative bacteria (Liet al.2016). Moreover,a recent study demonstrated that proton-coupled oligopeptide transporter (POT) families mediate the transport of MDP (Huet al.2018). The PEPT1 (SLC15A1),PEPT2(SLC15A2),PHT1 (SLC15A4),and PHT2 (SLC15A3)subfamilies make up the POT family (Solcanet al.2012).The POT family can transport dipeptides/tripeptides (Bissaet al.2016). Currently,important transport substrates include antibiotics,antiviral,and anticancer drugs (Smithet al.2013). There is mounting evidence that the POT family can transport chemotactic peptides produced from bacteria (Girardinet al.2003). According to the research of Hitotsumatsuet al.(2008),MDP can cause inflammation in mice (Hitotsumatsuet al.2008). Thus,substrates with physiological activity may be involved in the regulation of the innate immune response through the POT family.
The host’s first line of defense against invading microorganisms is innate immunity (Jinet al.2014).Epithelial cell pattern recognition receptors,such as the nucleotide-binding oligomerization domain (NOD)proteinsNOD1andNOD2,identify the pathogens that contribute significantly to innate immunity (Stroberet al.2006;Chenet al.2009). Several studies have shown thatSLC15A4is critical for the inflammatory response triggered by NOD-like receptors (Sasawatariet al.2011).Blasiuset al.(2010) and Sasawatariet al.(2011) inhibited the production of IFN-β by destroyingSLC15A4gene in mice,thereby improving intestinal inflammation in mice.Recently,studies have shown thatSLC15A4is linked to the development of many inflammatory diseases,including systemic lupus erythematosus (Hanet al.2009;Heet al.2010;Wanget al.2013) and inflammatory bowel disease (Leeet al.2008). Based on these observations,SLC15A4might play a significant role as a regulator in autoimmune diseases. However,the mechanism of howSLC15A4participates in the regulation of the inflammatory response in bovine rumen epithelium cells (BRECs) is still unclear. We hypothesized that MDP regulates the immune response in BRECsviaSLC15A4.
SLC15A4mediates the transport of di/tripeptides derived from bacteria,enhancing the nucleotide-binding oligomerization domain-dependent immune response in mouse bone marrow-derived macrophages,according to previous research (Huet al.2018). With this in mind,this study addresses two primary objectives: 1) To identify the regulatory effect of MDP on the immune response in BRECs,we first found the difference between mRNA expression of pro-inflammatory cytokines in BRECs with or without MDP using RT-qPCR analysis;and 2) to evaluate the effect of MDP on the production of inflammatory cytokines in wild-typeSLC15A4,we knocked out BRECs.Our findings demonstrated for the first time that MDPmediatedSLC15A4signaling pathways are essential for the rumen inflammatory response in ruminants.
2.Materials and methods
2.1.Cell culture
BRECs were provided by the Institute of Animal Culture Collection and Application (Yangzhou University,China).The establishment was according to Zhanet al.(2019).They successfully established immortalized BRECs from primary BREC by lentivirus transfection. In addition,they identified immortalized BRECs by Western blot and RT-qPCR. The BRECs were seeded in DMEM/F12 medium with 10% fetal bovine serum (Gibco,Grand Island,NY,USA),100 U mL–1penicillin and 100 µg mL–1streptomycin (Sangon Biotech,Shanghai,China).Under 5% CO2and 95% humidity,the culture was incubated at 37°C.
2.2.MDP stimulation of BRECs
The BRECs were seeded in 6-well plates (1×106cells well–1) and grown at 37°C in 5% CO2. Cells were divided into two experimental groups: (1) Control,DMEM/F12 medium;and (2) DMEM/F12 medium+10 µg mL–1MDP for 6 h. BREC were then tested for the mRNA expression of pro-inflammatory factors (IL-1β,IL-6,andTNF-α).
2.3.Design and cloning of the lentiviral vector for CRISPR knockout
Single guide RNA targeting exon 2 ofSLC15A4(Gene ID:510499 from National Center for Biotechnology Information)inBos taurus(cattle) was designed using online tools(http://chopchop.cbu.uib.no/) (Edicket al.2021). The four control sgRNA were selected and designed according to the method of Edicket al.(2021). The sequences of the sgRNA used in this study are shown in Table 1(synthesized by GENEWIZ Bioscience Co.,Ltd.,Suzhou,China). We performed vector construction to conduct CRISPR/Cas9 gene editing as described by Edicket al.(2021). For sgRNA construction,a pair of synthesized oligos was annealed,then ligated into the lentiCRISPR_V2 vector (Addgene,60954,USA). The lentiCRISPR_V2-SLC15A4 vector construction was performed by the Hengyi Biotechnology Company (Yangzhou,China). Briefly,the single guided RNA (sgRNA) sequence was generated by the CRISPR design tool (http://crispr.mit.edu) and the sgRNA targeted DNA sequence was then cloned into a lentiCRISPR/Cas9 v2 plasmid (Hanet al.2022). For lentivirus production,lentiviral vectors were packed using pMDLg/pRRE (Addgene,12251),pRSV-Rev (Addgene,12253),and pMD2.G (12259,Addgene) plasmids.Lentiviral plasmids were transfected into HEK293T cells(Q401,GenHunter,Nashville,TN) with lentivirus packaging plasmids,and packaging was performed as described previously (van der Gootet al.2018). The medium was replaced after overnight incubation and viral particles were harvested at 24 h,filtered through a 0.45-µm filter to remove any cell and debris,and stored at–80°C.
2.4.Cell infection
The BRECs were seeded into a 100-mm cell culture dish and grown to 50–70% confluence for infection. The culture medium was changed after the cells were infected with the lentiviruses for 24 h in the presence of 8 µg mL–1polybrene (TR-1003-G,Sigma-Aldrich,USA). Three days after infection,half of the cells were harvested for genome extraction,and the other half were cultured for the serial infection.
2.5.Genomic DNA extraction and PCR amplification
The processes of DNA extraction and PCR amplification were described in Edicket al.(2021). Genomic DNA was harvested from cells using the Cell Genomic DNA Extraction Kit (TIANGEN,Beijing,China). DNA concentrations were determined using an OD1000 Instrument (Thermo Fisher Scientific,Waltham,MA,USA).
PCR amplification was performed to obtain a 386-bp region inSLC15A4exon 2 using PCR SuperMix (Thermo Fisher Scientific). The PCR primers for amplification of exon 2 of theSLC15A4-CX genome were as follows:5´-AGGTTAAGGATCGGGGTCCA-3´ (forward);and 5´-AGCACAGGACAGACAGAAGC-3´ (reverse). PCR was then performed by using 2×Hieff®PCR Master Mix(Yeasen,Shanghai). After PCR amplification,the PCR products were sequenced by Genewiz Bioscience Co.,Ltd.(Suzhou,China).
2.6.TA cloning and knockout efficiency
The PCR products were subsequently transferred into pMD 19-T vector (TaKaRa,Japan) and sequenced. At least 15 positive clones of each gene were picked for sequencing.Sequence analyses and genomic sequence alignments were performed using the DNASTAR Software package(DNASTAR,Madison,WI). The knockout efficiency predictive value was calculated according to the following formula: Knockout efficiency (%)=Number of positive clones/Number of genes knocked out×100 (Niuet al.2015).

Table 1 Single guide RNA (sgRNA) of SLC15A4 in bovine
2.7.Acquisition of SLC15A4-KO-BRECs
The limited dilution approach,as previously described (Tianet al.2018),was used to extract specific cell clones from the positive cell pool. Positive cells were diluted to one cell per 100 µL of media after being transfected with Cas9/sgRNASLC15A4expression plasmids as previously described.They were then added to 96-well cell culture plates and grown for 10–14 days to produce single clone colonies.The genomic DNA of individual clones was extracted,and theSLC15A4-CX primer was used in PCR. After the PCR products were purified,they were sequenced.
2.8.Illumina HiSeq mRNA sequencing
The transcriptomes of wild-type (WT) BRECs andSLC15A4-Knockout (KO) BRECs were compared using RNA-seq analysis. Using Trizol reagent,total RNA was extracted from cell samples (Invitrogen,Shanghai,China). A total of 1 µg of total RNA from samples with an RNA integrity number values of less than 7 were used to create the libraries.Following fragmentation and reverse transcription of the mRNA,GENEWIZ processed and examined the sequences(Suzhou,China). Transcriptome sequencing (RNA-seq)was performed on an Illumina HiSeq 4000 platform (Illumina,United Kingdom). The selection of the library quality inspection kit was either the DNA 1000 Assay Kit (Agilent Technologies Santa Clara,CA) or the High Sensitivity DNA Assay Kit (Invitrogen,Shanghai,China). The manufacturer’s instructions for onboard clustering were followed when creating the clusters (Illumina,San Diego,CA). The significant pathways that are connected to the genes and found to be expressed differentially were identified through the use of Kyoto Encyclopedia of Genes Genomes (KEGG)pathway analysis.
2.9.Cell treatments
WT BRECs andSLC15A4-KO BRECs were added to 6-well culture plates and adjusted to 2×105cells mL–1.The cells were cultured for 12 h,and new medium was replaced after they were divided into the following 3 groups: (1) WT BRECs for the control group;(2) WT BRECs+10 µg mL–1MDP for the WT+MDP group;and (3)SLC15A4-KO BRECs+10 µg mL–1MDP for the KO+MDP group. Cells were collected after 6 h of culture.
2.10.RNA extraction and quantitative real-time qPCR
TRIzol reagent (TaKaRa,Code No.RR036A,China) was used to extract the RNA from BRECs in accordance with the manufacturer’s instructions,and an OD1000 instrument was used to measure RNA concentrations. Then,in accordance with the instructions,RNA was reverse transcribed into cDNA using a typical reverse transcription kit (TaKaRa,Tokyo,Japan). The PCR reaction was carried out with the help of the SYBR®Premix Ex TaqTM II Kit(TaKaRa,Dalian,China). The GAPDH can be computed using the 2–ΔΔCTmethod,which is already known to be appropriate for BRECs (Kent-Denniset al.2020). All the primers used are listed in Appendix A (synthesized by GENEWIZ Bioscience Co.,Ltd.,Suzhou,China).
2.11.lmmunocytofluorescence
BRECs was added and adjusted to 2×103cells mL–1in an 8-well chamber slide (Thermo Scientific,Lab-TekTMII,Code No.154534,NY,USA). The cells underwent MDP treatment and multiplied by 80–90%. The slides were then fixed with 4% paraformaldehyde for 30 min at room temperature,washed three times with PBS (3 min),and then rinsed again before being submitted to antigen retrieval with EDTA-Na2(95°C,5 min). The slides were coated with PBS containing 3% horse serum and blocked for 1 h at room temperature.After removing the blocking solution,the phosphorylation-ERK1/2 (p-p44/42) rabbit mAb (1:1 000;Cell Signaling Technology,Shanghai,China) and phosphorylation-nuclear factor κB (NF-κB) (p-p65) rabbit mAb (1:500;Cell Signaling Technology,Shanghai,China) were added and incubated at 4°C for the duration of the next day. Anti-mouse IgG conjugated to fluorescein isothiocyanate (Cy3) (1:2 000;Santa Cruze,Shanghai,China) was then added,and the mixture was incubated for 1 h at room temperature without exposure to light. Before DAPI staining,cells were washed with PBS 3 times,and 8 min later,the cells were washed three times with PBS and observed with a confocal laser scanning microscope (Olympus,Tokyo,Japan).
2.12.Statistical analysis
Data are displayed as mean±standard error. Data were subjected to a one-way ANOVA and two-way ANOVA at a test significance level ofP<0.05 utilizing the SPSS Statistical Software Package (v.20.0,IBM Corp.,Armonk,NY,USA). Tukey’s postdoc multiple comparisons test was carried out at a significance level ofP<0.05. Values sharing the same letter are not significantly different(P>0.05),whereas those having different letters are significantly different (P<0.05).
The Student’s two-tailedt-test was used to determine whether differences in the mean±SEM of the cytokines discharged by cells from different groups were statistically significant. Differences were considered significant forP-values under 0.05.
3.Results
3.1.Effect of MDP on pro-inflammation factors of BRECs
To investigate the regulatory effect of MDP on mRNA expression in pro-inflammation cytokines,the levels were measured by RT-qPCR. As shown in Fig.1,we observed an increase in mRNA levels of proinflammatory cytokines (IL-1β,TNF-α,andIL-6) in 10 µg mL–1MDP-treated BRECs compared with the control group (P<0.001).
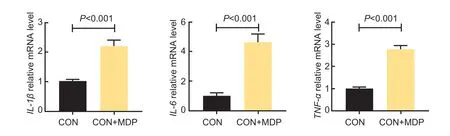
Fig.1 Effect of muramyl dipeptide (MDP) on the pro-inflammation factors of bovine rumen epithelial cells (BRECs). BRECs were treated either with 10 µg mL–1 MDP or without MDP (CON,control) for 6 h. The mRNA levels of the IL-1β,IL-6,and TNF-α genes in the MDP-treated BRECs were measured by RT-qPCR. Mean±SE (n=6) data are presented. Each treatment group’s data were normalized using data from the control group. An independent-sample t-test was used to calculate group-to-group comparisons.
3.2.CRISPR-Cas9-mediated knockout of SLC15A4 in BRECs
To characterize the role ofSLC15A4in the inflammatory response in MDP-treated BRECs,we used CRISPRCas9 KO technology to ablate the SLC15A4 protein.To this end,we designed four independent sgRNAs targeting exon 2 ofSLC15A4(Fig.2-A),and found that the four independent targets were successfully constructed (Fig.2-B). Cloning and sequencing of the PCR product revealed the predicted TA footprint left by the excisedSLC15A4(Fig.2-C). Knockout efficiency forSLC15A4was assessed by TA cloning sequencing of the knockout site. TheSLC15A4had a knockout efficiency in this assay of about 20%. Lentivirus infected cells were cloned to monoclonal lines by serial dilution screening.SLC15A4-KO cell lines were established and confirmed by genomic sequencing (Fig.2-D),consistent with the results of the TA sequencing. Although we have tested several commercially available antibodies againstSLC15A4,we failed to validate one suitable for immunoblot analysis. Therefore,the genomic mutations were confirmed by monoclonal genome sequencing and TA clone sequencing.
3.3.Genes differentially expressed between wildtype and SLC15A4-KO cells
To understand the gene expression differences between WT BRECs andSLC15A4-KO BRECs in regulating cellular responses,transcriptome analysis was performed. Transcriptome analysis revealed a marked difference in the gene expression profiles between the WT-BRECs andSLC15A4-KO BRECs. Knockout ofSLC15A4significantly reduced the expression ofIL-6andIL-8(Table 2;P<0.001). Compared with WT BRECs,the expression of mostCCLandCXCLchemokine-related genes was significantly reduced(Table 3;P<0.05). Interestingly,most of the related genes in the NF-κB signal pathway and MAPK signal pathway were also significantly down-regulated (Tables 4 and 5). Additionally,RT-qPCR was used to confirm the expression of chemokines and pro-inflammatory cytokines,which was compatible with the study of gene expression profiling (Fig.3). These findings suggest thatSLC15A4may regulate the gene expression of chemokines and pro-inflammatory cytokines in BRECs.
3.4.SLC15A4 mediated MDP-regulated pro-inflammation factors in BRECs
Compared with the control group,adding MDP significantly increased the expression ofIL-6andTNF-αmRNA (Fig.4;P<0.05). Interestingly,after knocking outSLC15A4,adding MDP reversed this result.
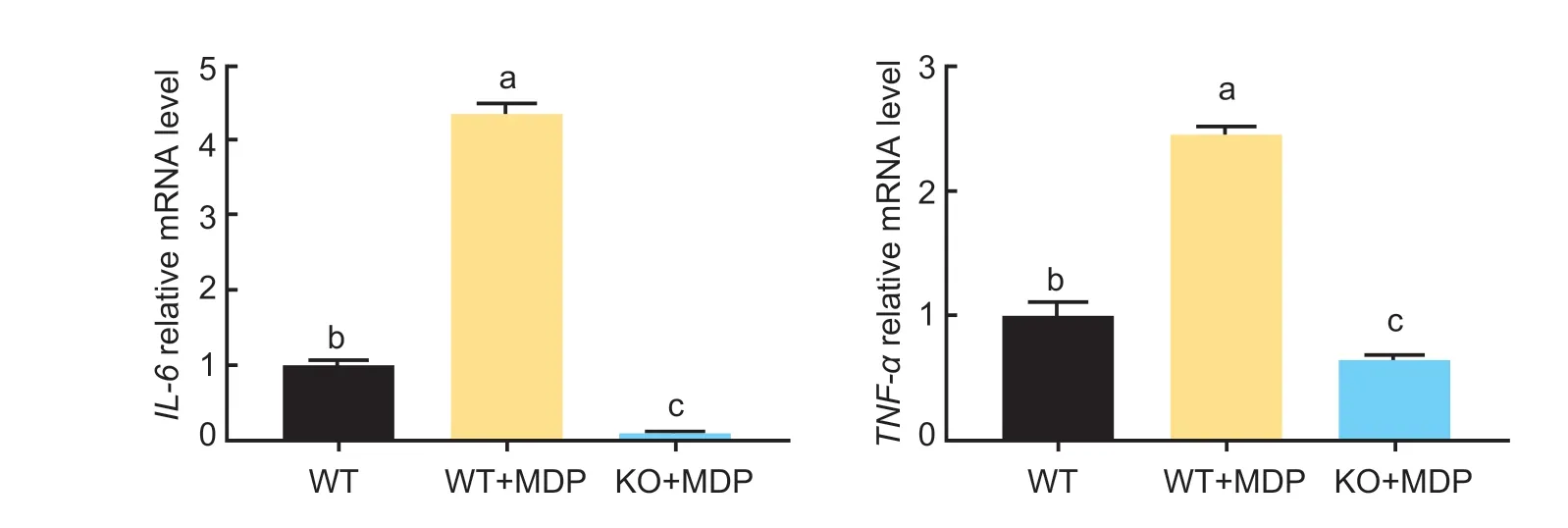
Fig.4 Effect of muramyl dipeptide (MDP) on the pro-inflammation factors of wild-type (WT) and SLC15A4-Knockout bovine rumen epithelial cells (BRECs) (KO). The mRNA levels of IL-6 and TNF-α genes in the SLC15A4-KO BRECs were measured by RT-qPCR. Mean±SE (n=3) data are presented. A one-way ANOVA was used to calculate group-to-group comparisons. The various lowercase letters in the bar charts indicate significant differences (P<0.05).
3.5.SLC15A4 mediated MDP-regulated chemokines in BRECs
Compared with the control group,adding MDP significantly increased the mRNA expression ofCXCL2andCCL2(Fig.5;P<0.05). Conversely,the addition of MDP significantly reduced the mRNA expression ofCXCL2,CXCL3,CXCL9,andCCL2inSLC15A4-KO BRECs.
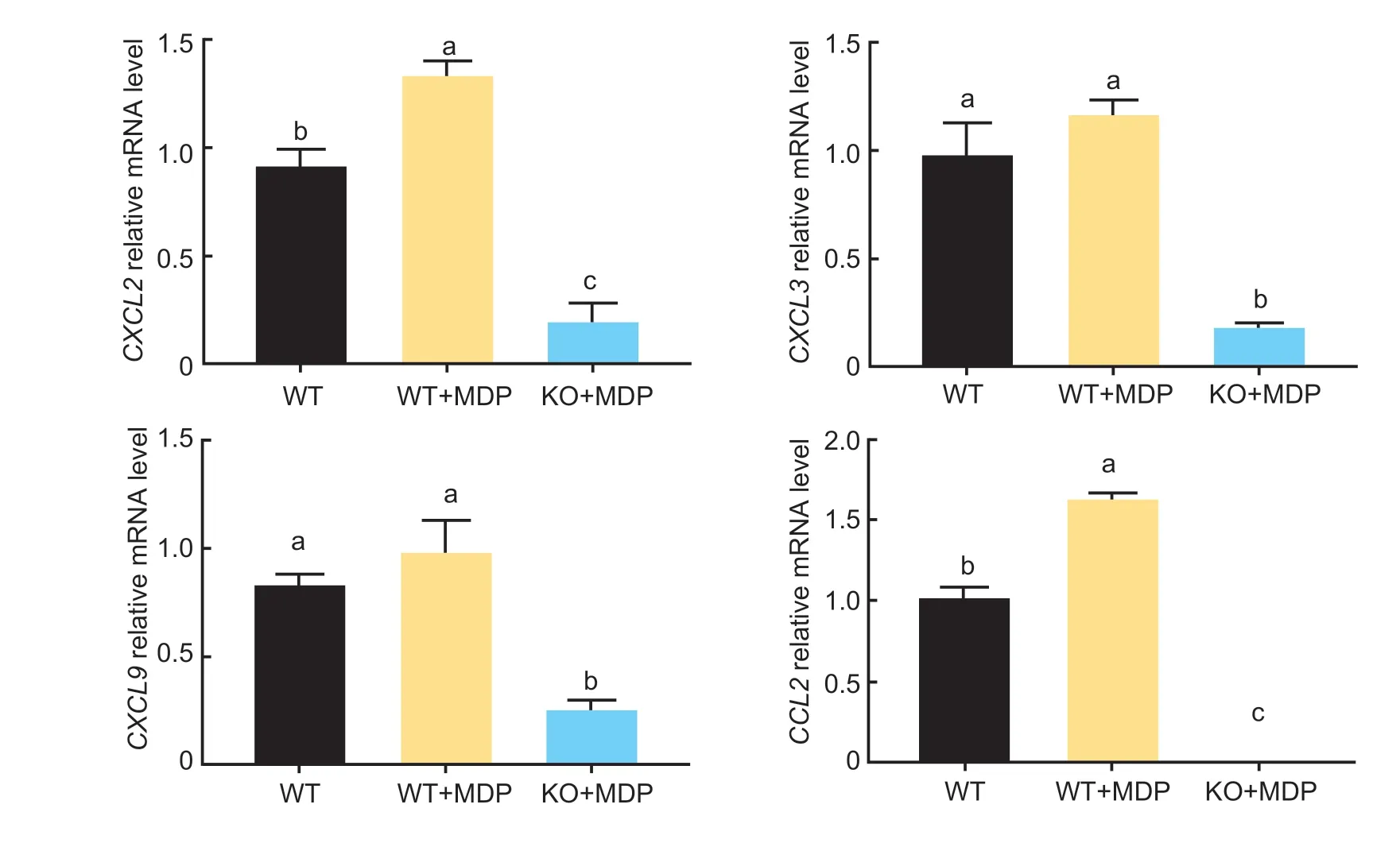
Fig.5 Effect of muramyl dipeptide (MDP) on chemokines of wild-type (WT) and SLC15A4-Knockout bovine rumen epithelial cells(BRECs) (KO). The mRNA levels of the CXCL2,CXCL3,CXCL9 and CCL2 genes in the SLC15A4-KO BRECs were measured by RT-qPCR. Mean±SE (n=3) data are presented. A one-way ANOVA was used to calculate group-to-group comparisons. The various lowercase letters in the bar charts indicate significant differences (P<0.05).
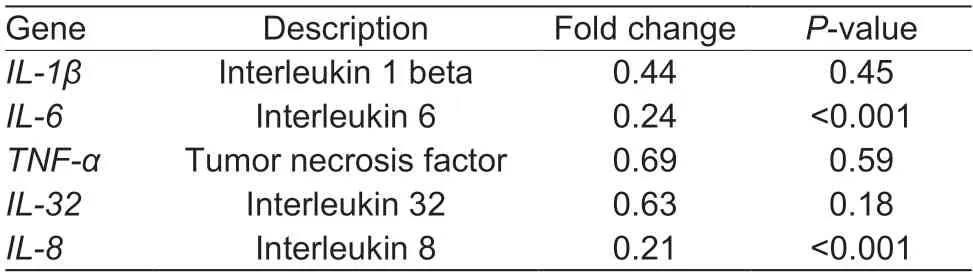
Table 2 Differential expression of pro-inflammatory genes in wild-type and SLC15A4-Knockout bovine rumen epithelial cells (BRECs)
3.6.SLC15A4 mediated MDP-regulated NF-κB and MAPK signaling pathways in BRECs
The results of immunofluorescence indicated that MDPactivates the NF-κB and MAPK signaling pathways in BRECs leading to increased levels of p-p65 and p-p44 proteins (Fig.6). Interestingly,the protein levels of p-p65 and p-p44 were significantly lower after knockdown of theSLC15A4gene,indicating that the p65 protein activation and translocation into the nucleus were inhibited. This analysis suggests thatSLC15A4plays important roles in regulating the immune response.
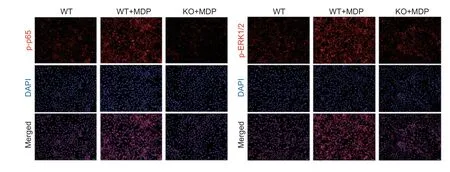
Fig.6 Effect of muramyl dipeptide (MDP) on NF-κB and MAPK signaling pathways in wild-type (WT) and SLC15A4-Knockout bovine rumen epithelial cells (BRECs) (KO). The BRECs were treated with 10 µg mL–1 MDP for 6 h. The immunofluorescence for phosphorylation-NF-κB (p-p65) and phosphorylation-ERK1/2 (p-p44) (red) was measured using the nuclear dye 4´,6-diamidino-2-phenylindole (DAPI;blue). Scale bar=100 µm.
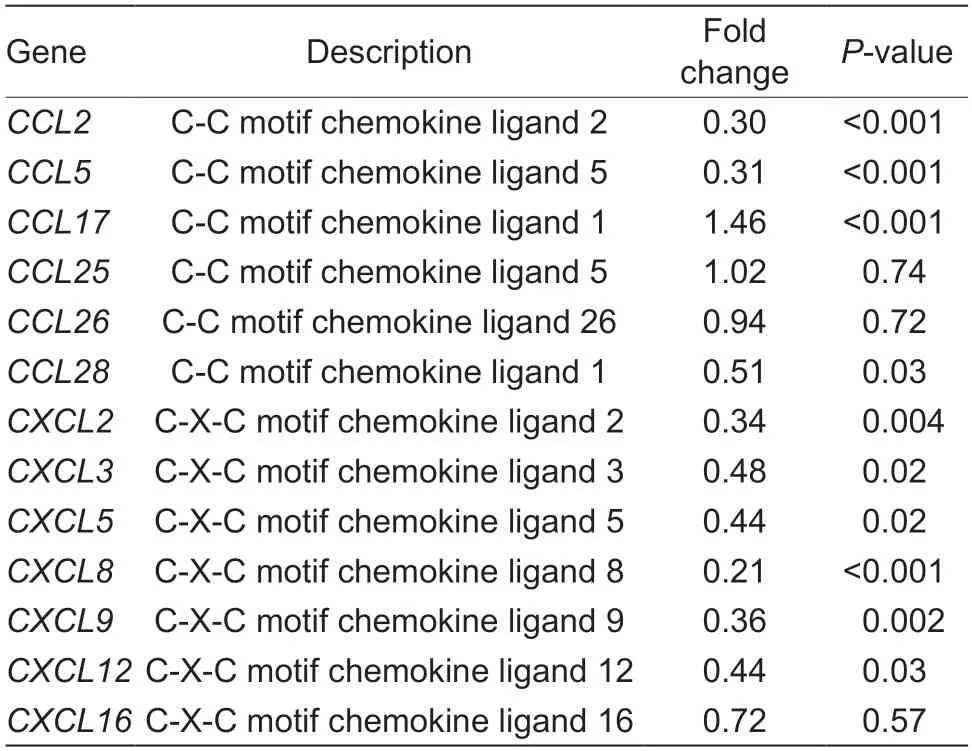
Table 3 Differential expression of chemokine genes in wild-type and SLC15A4-Knockout bovine rumen epithelial cells (BRECs)
4.Discussion
There is an increasing evidence that proton-coupled oligopeptide transporters (POTs) facilitate NODlike receptor signaling pathways and the subsequent production of pro-inflammatory cytokines,both of which play an important role in innate immunity regulation.Small peptides from the meal are transportedviathe di/tripeptide transporterSLC15A4in gastrointestinal epithelial cells,greatly aiding nutrient absorption (Huet al.2018).SLC15A4is capable of transporting oligopeptides produced by bacteria,such as fMLP,MDP,and Tri-DAP,which can harm the rumen epithelium (Buyseet al.2002;Vavrickaet al.2004;Dalmassoet al.2010). As a result,SLC15A4has important pathological significance because it plays a major role in the transport of bacterial MDP,which causes neutrophil aggregation and rumen epithelium injury(Buyseet al.2002). For bacterial lysate,MDP can be pumped into the cytosol of BRECs bySLC15A4(Huet al.2018). The expression ofSLC15A4and the transfer of MDP in aberrant rumen epithelia also cause inflammation and injury (Marina-Garciaet al.2009;Kieser Kagan 2017).We found that MDP treatment significantly increased the mRNA expression of pro-inflammatory factors (TNF-α,IL-6,andIL-1β) in BRECs,which is consistent with previously reported findings from Daiet al.(2022).
In BRECs,SLC15A4knockdown revealed that this protein plays a crucial role not only in MDP transport but also in the immune system. The CRISPR-Cas9 gene editing system was used in the creation of theSLC15A4knockout cells. The CRISPR-Cas9 knockout system proved to be more effective and specific compared with traditional knockout methods (Cobbet al.2015;Hanet al.2020). For this study,we used the 293T cells that were transfected with 3rd-generation lentiviral packaging plasmid and the lentiCRISPR v2 vector expression plasmid (Sanjanaet al.2014). Subsequently,the single cell masses were randomly selected and further screened by sequencing. DNA sequencing the TA clone confirmed a 1-bp insert mutation in theSLC15A4exon following the selection of a single clone (Fig.2-C and D). These results indicate that the CRISPR/Cas9 System successfully knocked out theSLC15A4gene.
To understand the gene expression differences between WT andSLC15A4-KO BRECs in regulating cellular responses,transcriptome analysis was performed.Transcriptome analysis revealed a marked difference in the gene expression profiles between the WT-BRECs andSLC15A4-KO BRECs. Notably,after knocking out theSLC15A4gene,the inflammatory cytokines (IL-6andIL-8)and mostCCLandCXCLchemokine-related genes were significantly downregulated. We also looked at several other genes that are involved in major signaling pathways,particularly the NF-κB and MAPK pathways. The results of transcriptome sequencing revealed down-regulation of the MAPK NF-κB pathways after knocking outSLC15A4.Therefore,we speculate thatSLC15A4may mediate the regulation of immune responses mainly through the NFκB and MAPK signaling pathways. Due primarily to their capacity to transfer NOD ligands from extracellular to cytosolic receptors,an increasing number of studies have demonstrated thatSLC15A4is an important immune response modulator (Leeet al.2009). Previous research has shown that the cytokines were reduced in bone marrow dendritic cells ofSLC15A4knockout mice,and theseSLC15A4deficient mice were more resistant to DSS-induced colitis (Sasawatariet al.2011). At the same time,qRT-PCR was performed to verify mRNA differential gene expression. We found that the RT-qPCR results were generally in line with those of the transcriptome sequencing results. These results confirmed that theSLC15A4might participate in regulating the immune response.
It is possible for MDP to activate NF-κB and other immune effector pathways as a NOD ligand,thereby enhancing the production of pro-inflammatory cytokines(Huet al.2018). Previous studies have shown that MDP is a substrate for the POT family of transporters(Wu and Smith 2013;Nakamuraet al.2014). Therefore,we speculate thatSLC15A4may regulate the immune response by transporting MDP to cytosol and activating the NOD-dependent innate immune response(Tyshkovskiyet al.2019). TheSLC15A4interacting with bacterially derived products and regulated the production of pro-inflammatory cytokines (such as,IL-6andTNF-α) in BRECs. Our results showed that knocking outSLC15A4reduced MDP-induced pro-inflammatory cytokines and chemokine mRNA expression in WT-BRECs.Immunostaining analysis showed that the expression levels of phosphorylated-ERK1/2 and phosphorylated-p65 were significantly reduced after knockout ofSLC15A4.Many studies have shown that MDP enters the cytoplasm and can cause the release of downstream inflammatory factors. One interesting phenomenon was that knocking outSLC15A4individually had an effect on the release of inflammatory factors and the protein expression levels of phosphorylated-ERK1/2 and phosphorylated-p65. Our studies have shown that the bacterial peptide MDPmediatedSLC15A4signaling pathways are essential for the rumen inflammatory response in ruminants.
5.Conclusion
In this experiment,we used the CRISPR/Cas9 System to knockout the di/tripeptide transporterSLC15A4,and confirmed its role in the MDP-induced inflammatory response in BRECs. This work will provide a theoretical basis for the study of the pro-inflammatory mechanism of MDP and its application in work on the prevention and treatment of subacute rumen acidosis in dairy cows.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (31972589),the earmarked fund for China Agriculture Research System (CARS-36) and the Postgraduate Research &Practice Innovation Program of Jiangsu Province,China (KYCX21-3283).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
This experiment was performed under protocols approved by the Animal Care and Use Committee of Yangzhou University,China (ethic code is DWLL-202011-201). The authors confirm that they have followed European Union standards for the protection of animals used for scientific purposes.
Appendixassociated with this paper is available on https://doi.org/10.1016/j.jia.2023.06.016
 Journal of Integrative Agriculture2023年10期
Journal of Integrative Agriculture2023年10期
- Journal of Integrative Agriculture的其它文章
- The association between the risk of diabetes and white rice consumption in China: Existing knowledge and new research directions from the crop perspective
- Linking atmospheric emission and deposition to accumulation of soil cadmium in the Middle-Lower Yangtze Plain,China
- Genome-wide association study for numbers of vertebrae in Dezhou donkey population reveals new candidate genes
- Are vulnerable farmers more easily influenced? Heterogeneous effects of lnternet use on the adoption of integrated pest management
- lnfluences of large-scale farming on carbon emissions from cropping:Evidence from China
- Spatio-temporal variations in trends of vegetation and drought changes in relation to climate variability from 1982 to 2019 based on remote sensing data from East Asia
