Phylogenic and Functional Profile of Ser/Thr Kinases in Synechocystis sp. PCC 6803
WANG Xiaoting, ZHANG Pengpeng
(Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China)
Abstract:Signal perception and transduction for organisms are tightly associated with their adaptability to environments. For signal transduction systems, two-component kinase systems or Ser/Thr kinase systems both in eukaryotes and prokaryotes have been studied and reviewed. As a prokaryotic photosynthetic model organism, except for the typical two-component His-Rep kinase systems, Synechocystis sp. PCC 6803 also possesses 12 genes encoding putative eukaryotic Ser/Thr kinases, which can be divided into 2 subfamilies according to their structural features, PKN2 subfamily and ABC1 subfamily. Genes encoding Ser/Thr kinases belonging to the PKN2 subfamily have been investigated mostly, while the rest belonging to the ABC1 subfamily mostly are functionally unknown. In this paper, the prediction of conserved domains and motifs, as well as protein cellular localizations of these 12 Ser/Thr kinases in Synechocystis sp. PCC 6803 was investigated. As the representative monocotyledon and dicotyledon, rice and Arabidopsis were chosen as reference objects, and by sequence analysis together with 15 rice ABC1Ks and 17 Arabidopsis ABC1Ks, the potential functions and evolutionary relationship of Ser/Thr kinases in Synechocystis sp. PCC 6803 were discussed, which would provide new insights into a more comprehensive understanding of Ser/Thr kinase signaling process in Synechocystis sp. PCC 6803.
Key words:Synechocystis; signal transduction; Ser/Thr protein kinase; photosynthesis
Signaling is an indispensable mechanism for organisms to deal with environmental disorders,which mainly includes two-component kinase systems and Ser/Thr kinase systems. Generally, bacteria mostly possess two-component kinase systems for signal transduction, which spread not only in nearly all prokaryotes[1-3], but also in eukaryotes like plants,yeasts, and fungi[4]. Unlike prokaryotes, eukaryotes mostly use serine/threonine and tyrosine kinases to accomplish signal transduction[5-6]. Ser/Thr kinases had been thought to exist exclusively in eukaryotes and two-component kinase systems were the only way available for signal transduction in prokaryotes for quite a long time. Nevertheless, since the discovery of Ser/Thr kinase and Tyr kinase in a bacterium[7], genes encoding eukaryotic-like Ser/Thr kinases have been identified increasingly from different bacterial strains[8-9].
Cyanobacteria, the first oxygenic photosynthesis organism on earth, possesses a very strong ability to survive in different environments, such as extremes of temperature, salinity, nutrient inadequacy, and fluctuating light intensities. Therefore, they cover a large scope of terrestrial and aquatic habitats[10]. It’s presumed that the adaptability of cyanobacteria to various environments was probably due to their highly efficient and flexible signal sensing and transducing systems. In cyanobacteria, the first report about Ser/Thr modification was in 1994 by radioactive labeling[11].Subsequently, about 286 genes encoding Ser/Thr kinases were discovered from 21 cyanobacterial species,and the vast majority of them haven’t been characterized[12]. The phosphoproteomic studies in cyanobacteria have identified hundreds of Ser/Thr/Tyr phosphoproteins, and a comprehensive functional analysis also revealed that Ser/Thr/Tyr kinases participated in the regulation of many critical processes including photosynthesis, nitrogen metabolism, and other vital life activities[13-17]. Therefore, cyanobacteria not only possess typical two-component kinase systems but also eukaryotic Ser/Thr kinase systems, despite twocomponent systems are dominant in quantity.
1 Ser/Thr kinases in Synechocystis sp.PCC 6803
1.1 About Synechocystis sp. PCC 6803
Synechocystissp. PCC 6803, an unicellular,mesophilic, non-nitrogen-fixing cyanobacterium, was the first photosynthetic organism whose genome was fully sequenced[18]. Because of some kinds of unknown self-mutagenesis, the strain can grow either autotrophically or heterotrophically[19], depending on the availability of light and exogenous organic carbon sources. It has become a model organism for photosynthesis and biofuel researches due to its relatively short culturing time, small genome size, and easy genetic manipulation[20]. Moreover,Synechocystissp. PCC 6803 possesses a strong adaptability to diverse environmental stresses, such as high temperature, cold, nutrient deficiency, high light,therefore it may have evolved flexible and sophisticated signal perception and transduction systems, like two-component kinase systems and Ser/Thr kinase systems. Evolutionary analysis indicated that Ser/Thr kinases inSynechocystissp. PCC 6803 were present before the divergence between prokaryotes and eukaryotes during evolution or were laterally transferred into prokaryotes at the early stage of bacterial evolution[21]. Although two-component kinase systems are predominant inSynechocystissp.PCC 6803, here Ser/Thr kinase systems were mainly discussed in this review.
1.2 Category and Structure of Ser/Thr kinases in Synechocystis sp. PCC 6803
A total of 12 genes encoding putative Ser/Thr kinases (STKs) inSynechocystissp. PCC 6803, 7 of them encode proteins belonging to the PKN2 subfamily (spkA-spkG), and the rest 5 belonging to the ABC1 (activity of bc1 complex) subfamily (spkHspkL) (Table 1)[22]. The atypical kinase ABC1 family was characterized by a highly conserved ABC1 domain and one or more protein kinase domains[23].This ancient family was highly conserved among bacteria, archaea, and eukaryotes, and had undergone substantial expansion in photosynthetic organisms[24], which suggested that they might play fundamental biological roles[25].

Table 1 12 Ser/Thr kinases in Synechocystis sp. PCC 6803
Sequence analysis of the first 7 protein kinases(SpkA-SpkG) revealed that they all contained subdomain Ⅰ~Ⅺ[26], and most of the catalytic domains were located at the N-terminal[27]. Sequence analysis also showed that 5 of the 12 protein kinases have one (SpkC,SpkD) or several (SpkF, SpkI, and SpkL) putative transmembrane helices at their C-terminal regions, and the rest 7 protein kinases do not have a predicted transmembrane domain[20]. It was suggested that those protein kinases with only one transmembrane segment could be likely localized on the membrane[28], while others with more than one transmembrane segment could act as membrane receptors during signal transduction[29].
1.3 Research progress of Ser/Thr kinases in Synechocystis sp. PCC 6803
The physiological significance of 12 Ser/Thr kinases inSynechocystissp. PCC 6803 was investigated using inactivation mutants. Studies have shown that the products of at least 5 protein kinase genes (spkA,spkB,spkC,spkD, andspkF) belonging to the PKN2 subfamily showed kinase activity,although their knockout mutants didn’t show any obvious phenotypes inSynechocystissp. PCC 6803[30]. In the glucose-tolerant strain commonly used by most laboratories, the coding region ofspkAgene has a frameshift leading to a defect in cell movement[31].SpkA functions in phosphorylation of 2 membrane proteins with molecular weight 90 and 30 kD respectively, which are involved in cell motility[31-32].Inactivation of the rest 11 protein kinase genes (spkBthroughspkL) ofSynechocystissp. PCC 6803 did not obtain any obvious phenotypes between all single mutants and the WT in terms of the growth rate in liquid cultures and of color, size, and shape of colonies during cultivation on solid medium under standard growth conditions[33]. Still, some of these Ser/Thr kinases (STKs) have been reported playing different roles inSynechocystissp. PCC 6803 (Table 2). SpkB participates in the oxidative stress response by phosphorylatingβ-subunit of glycyl-tRNA-synthetase(GlyS)[34]and regulates cell motilityviaprotein phosphorylation[30]. ThespkBmutant is more sensitive to iron deficiency than WT[34]. SpkC might take part in the regulation of nitrogen metabolism[35]. SpkD might be involved in adjusting the pool of tricarboxylic acid (TCA) cycle metabolites[36], and the microarray data suggested that SpkD could be under the control of a transcription factor NdhR in response to the limitation of inorganic carbon inSynechocystissp. PCC 6803[37]. The transcription profile of cold-inducible genes of thespkEmutant resembled theHik33mutant, suggesting that SpkE may act as an additional component in the regulatory pathway ofSynechocystissp. PCC 6803 cells in response to cold stress[38]. SpkE also might be involved in the regulation of nitrogen metabolism[39]. SpkG played an essential role in high-salt resistance[40], and was involved in the phosphorylation of Fd5 (ferredoxin 5)protein too[41]. SpkC, SpkF, and SpkK were involved in the phosphorylation of the GroES chaperone protein[38]. SpkH was reported to take part in a signaling pathway of the hyperosmotic stress response,and its expression was under the regulation of a twocomponent system composed of a histidine kinase Hik34 and a response regulator Rre1[42]. It was suggested that Ser/Thr kinases might be involved in cyanobacterial state transition[14]. However, by creating 12 single kinase mutants, it showed that Ser/Thr kinases were not related to the cyanobacterial state transition, or no specific protein kinase was required for this process[43]. 6 Ser/Thr kinases (SpkASpkD, SpkF and SpkG) were identified to have one or more of post-translational modifications. Among them, SpkC and SpkF were found with significantly fluctuating phosphorylation dynamics in response to nitrogen starvation[44], and these 2 Ser/Thr kinases were shown to autophosphorylate in vitro by radio labeling experiments[45]. Therefore, it was conceivable that the phosphorylation dynamics of SpkC and SpkF were interconnected and could involve a protein phosphatase[46]. There are 9 genes encoding phosphatases (slr0328(PTP family),sll1771,slr1860,sll1033,sll0602,slr0114,slr1983(PPM family),sll1387(PPP family), andslr0946) inSynechocystissp. PCC 6803[43]. Thus, it is worth to investigate the activities of kinases or phosphatases and to assign the counterpart of each kinase or phosphatase,which may mediate a eukaryotic-type signal transduction pathway in cyanobacteria[30]. It has been remarked that some Ser/Thr/Tyr kinases or phosphatases could cooperate together with two-component kinase systems in the signal transduction pathways[17,46].

Table 2 Partly published function descriptions among 12 Ser/Thr kinases in Synechocystis sp. PCC 6803
2 Phylogenetic analysis of Ser/Thr kinases in Synechocystis sp. PCC 6803
Previous functional studies onSynechocystissp.PCC 6803 Ser/Thr kinases mainly focused on 7 members of the PKN2 subfamily[30-31,33,36,38,41,47],while less was known about the other 5 members of the ABC1 subfamily. Since ABC1 type Ser/Thr kinases were highly conserved among different species, it was expected to get some ideas of the characterization of the 5 ABC1 type Ser/Thr kinases inSynechocystissp. PCC 6803 from functional studies in plants. For this purpose, a phylogenetic tree was constructed including 12 Ser/Thr kinases inSynechocystissp. PCC 6803, 17 ABC1Ks inArabidopsis,and 15 ABC1Ks in rice (Fig. 1). ABC1 genes in rice could be induced by a wide range of abiotic factors such as H2O2, abscisic acid, low temperature, drought, darkness and high salinity[48].

Fig. 1 Phylogenetic analysis of Ser/Thr kinases in Synechocystis sp. PCC 6803, rice and Arabidopsis
As shown in Fig. 1, 2 main branches were formed in the rooted tree. One branch consisted of the seven Ser/Thr kinases inSynechocystissp. PCC 6803, SpkASpkG, belonging to the PKN2 subfamily (circled in red dashed line); while the other 5 Ser/Thr kinases(marked in red rectangle), SpkH-SpkL, were scattered in the other big group along withArabidopsisand rice kinases, all belonging to the ABC1 subfamily. It indicated that the PKN2 subfamily might take a different evolutionary path from the ABC1 subfamily with some kinase features being preserved, and that’s probably the reason why SpkA-SpkG was divided separately and kept a distance from ABC1 subfamily kinases. As for SpkHSpkL, though they are eukaryotic protein kinases from a prokaryote, they do maintain a close evolutionary relationship with ABC1Ks in higher plants. The ABC1 group was further divided into 5 subgroups (A~E), and the members ofSynechocystissp. PCC 6803 (SpkH-SpkL) were spread into subgroup A, C and D (Fig. 1). A total of 10 pairs of orthologous genes were found in all subgroups, which emphasized that the divergence of ABC1 subfamily had occurred and the main characteristics had been established before the monocot-dicot split[48].
As shown in Fig. 1 and Table 3, SpkH and SpkK were clustered in subgroup A, and the homologous genes in rice andArabidopsiswere OsABC1-4, At3g24190 and OsABC1-10, At5g24970,respectively. OsABC1-4 was predicted to localize in the chloroplast, and reacted to cold stress; while OsABC1-10 was localized in mitochondria, and was responsive to salt stress[48]. Searching for TheArabidopsisInformation Resource (TAIR-Home Page (arabidopsis.org)), it shows that At3g24190 and At5g24970 both were involved in protein phosphorylation biological process, and At3g24190 was located in chloroplast while At5g24970 was localized in mitochondria, which was consistent with OsABC1-4 and OsABC1-10 in rice.In line with this,spkHandspkKshowed different responses to salt and cold treatments on transcription levels (https://cyanoexpress.sysbiolab.eu).

Table 3 Homology of 5 Ser/Thr kinases (SpkH-SpkL)in rice and Arabidopsis, respectively
SpkI and SpkL were clustered in subgroup C.SpkI was most similar to OsABC1-12 in rice and At3g07700 (AtSIA1) inArabidopsis. OsABC1-12 was induced by cold and salt in rice and was predicted to be located in the chloroplast[48]. Gene Ontology (GO) enrichment analysis showed that OsABC1-12 was involved in the biosynthesis of isoprenoids, including chlorophylls[49]. At3g07700,also named by AtSIA1 or ABC1K7, was localized in chloroplast and might be involved in salt stress tolerance inArabidopsis[50]. SpkL was close to OsABC1-2 and At5g64940, respectively. OsABC1-2 was induced by salt and inhibited by H2O2and darkness, and was localized in chloroplast theoretically[49]. GO enrichment analysis showed that OsABC1-2 was related to energy metabolism in plant cells, like photosynthesis[49]. At5g64940 was also named by AtOSA1 or ABC1K8. Among theArabidopsisABC1K proteins, ABC1K7 (AtSIA1) was most closely related to ABC1K8 (AtOSA1) (46% identity) and showed a 50% identity with cyanobacterial ABC1K proteins[25]. AtOSA1 was responsive to cadmium and oxidative stresses[51];ABC1K7 and ABC1K8 were probably involved in oxidative stress responses, isoprenyl lipid synthesis,and distribution of iron within chloroplast as well[25].
SpkJ was divided into subgroup D and was close to At1g65950 (ABC1K14) and At2g40090 (ABC1K15)inArabidopsisand OsABC1-14, -15 in rice,respectively. The expression ofOsABC1-14was repressed by H2O2, cold, drought and darkness, the protein localization was predicted to be in the vacuole in rice[48]. Besides, At4g31390,also named as AtACDO1 and ABC1K1, located in chloroplast,played important roles in mediating chlorophyll degradation and reacting to photooxidative stress[52],and was also needed to stabilize chlorophyll-binding proteins in photosynthetic complexes, and its knockout mutant in plant showed growth defects in sugar metabolism, which suggested that ABC1K1 might integrate photosynthesis with associated metabolic pathways in chloroplasts[53]; At4g31390(ABC1K1) and At1g79600 (ABC1K3) together regulated prenylquinone metabolism, which had a significant impact on plant stress responses and chloroplast morphology[24,54]. Prediction of protein subcellular localization indicated that most of the ABC1K proteins in maize, rice, andArabidopsiswere located in either chloroplast or mitochondria[55], where photosynthesis and respiration mainly take place in organisms, considering the phylogenetic relationship betweenSynechocystissp. PCC 6803 andArabidopsis,it’s reasonable to postulate that Ser/Thr kinases inSynechocystissp. PCC 6803 probably get involved in regulating photosynthesis and respiration processes.
3 Characteristic prediction of Ser/Thr kinases in Synechocystis sp. PCC 6803
3.1 Domain and motif prediction of Ser/Thr kinases in Synechocystis sp. PCC 6803
Since SpkA-SpkG were grouped in the phylogenetic tree (Fig. 1), it’s necessary to find out the common features shared by these proteins.Multiple sequence alignment (data not shown)showed that all seven proteins contained catalytic domains Ⅰ~Ⅺ, which is consistent with previous report[26]. Moreover, most of the catalytic domains gathered at the N-terminal region.
The conserved motifs of 12 Ser/Thr kinases predicted by the MEME website (MEME-Submission form) were shown in Fig. 2. A total of 20 putative motifs of 12 Ser/Thr kinases inSynechocystissp. PCC 6803 were identified using the MEME server. Generally speaking, as with the two branches of the 12 Ser/Thr kinases in Fig. 1, two kinds of motif prediction results were also displayed. Further annotation in the InterPro database (,https://www.ebi.ac.uk/) revealed that only motifs 1, 2, 3, 4, 5, 7, 8, 19 were recorded. Motif 1 represented the protein-kinase domain, which lied in almost all 12 Ser/Thr kinases, and the homologous superfamilies for motifs 1, 3, 7, 8 all belong to the protein kinase-like domain superfamily. Besides, motifs 2, 3, 4, 5 all contain chaperone-activity of bc1 complex characteristics, which were mainly possessed by ABC1 type Ser/Thr kinases .

Fig. 2 Motif predictions of 12 Ser/Thr kinases in Synechocystis sp. PCC 6803
3.2 Transmembrane helix and protein localization prediction of Ser/Thr kinases in Synechocystis sp. PCC 6803
Transmembrane helix prediction of the 12 Ser/Thr kinases could be helpful to decide their cellular localization. It showed that 3 out of 7 PKN2 proteins(SpkC, SpkD, SpkF) had 1 or 3 transmembrane helixes, and 2 out of 5 ABC1K proteins (SpkI and SpkL) had 2 or 3 transmembrane helixes (Table 4). It suggested that those 5 Ser/Thr kinases might act as significant sensor or receptor on the membrane, or mediate signal transduction from outside to inside.While the rest without transmembrane helix was probably soluble like it was presumed before[20], or they might possess some hidden features which we didn’t find out yet.
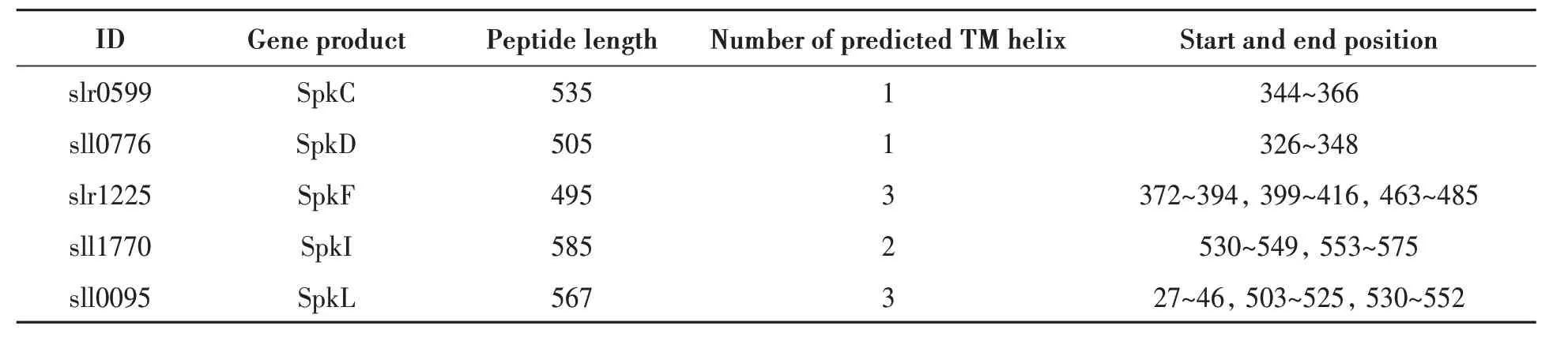
Table 4 Transmembrane helix prediction of Ser/Thr kinases in Synechocystis sp. PCC 6803
Comprehensive proteomic analyses have revealed some Ser/Thr kinases inSynechocystissp. PCC 6803.Among 12 Ser/Thr kinases inSynechocystissp. PCC 6803, 9 of them have been identified using CyanoAtlas.As shown in Table 5, most of them were exclusively localized on membrane fractions, except for SpkK and SpkL, which have a very small intensity in the soluble fraction. In any case, all these 9 proteins were mainly located on the membranes including the outer membrane, plasma membrane, and thylakoid membrane with varied intensities whether they were predicted to have transmembrane helix or not. SpkA,SpkD and SpkG were not included in the database yet,which could have several possibilities: SpkA was not completely translated inSynechocystissp. PCC 6803 because of self-mutagenesis; SpkD and SpkG were probably due to low protein abundance for proteomic analysis. SpkD was predicted to have one transmembrane helix (Table 4) suggesting most likely its membrane localization. Considerable amounts of the proteins of SpkH, SpkI, SpkK and SpkL were found in the thylakoid membrane, suggesting that they might be involved in photosynthetic functions. The majority of SpkB and SpkJ were found in outer membrane fractions, which indicated that they may have a function related to environmental sensing. Since SpkH and SpkK were clustered in subgroup A in the phylogenetic tree (Fig. 1), it makes sense that they shared a certain similarity in a way, and their subcellular localizations also showed a similar distribution trend mainly on plasma membrane and thylakoid membrane (Table 5), and same for SpkI and SpkL. SpkJ was mainly located on outer membrane and plasma membrane, which suggested that it might function on signal sensing and transducing from outside to inside. Except SpkC, SpkF, SpkI and SpkL,which had one or more transmembrane helix, the rest five Ser/Thr kinases (SpkB, SpkE, SpkH, SpkJ and SpkK), which didn’t have transmembrane domains,may locate on membranes by interacting with other membrane proteins.
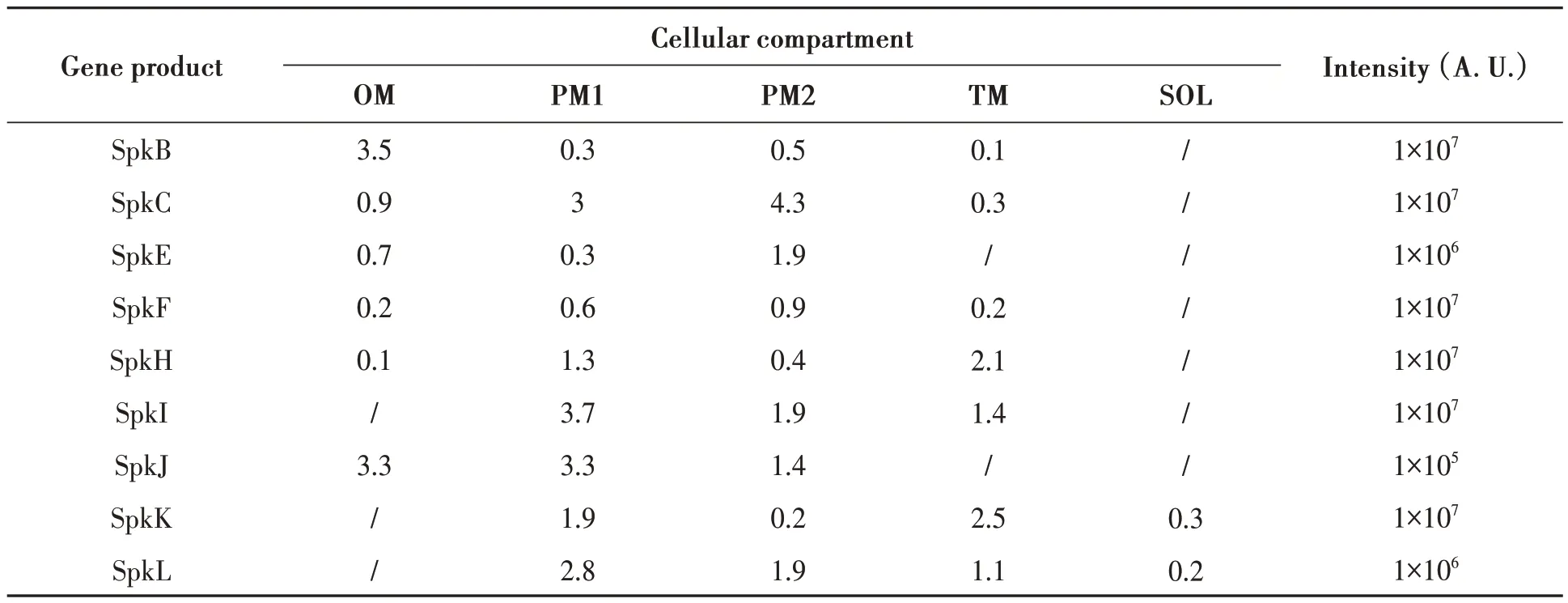
Table 5 Subcellular localization prediction of Ser/Thr kinases in Synechocystis sp. PCC 6803
4 Conclusion
Although kinase activity has been proved for the 11 out of 12 Ser/Thr kinases (except SpkE) inSynechocystissp. PCC 6803, only 2 substrates (GlyS and GroES) were identified and were assigned to certain Ser/Thr kinases so far[20]. Hence, to assign the substrates to each of the kinases inSynechocystissp. PCC 6803, probably via a comprehensive phosphoproteomic analysis by collecting samples under various treatments would be taken into consideration. Since all single mutant could grow under the photoautotrophic mode, and a majority of these mutants showed undetectable physiological changes[20], it suggested that the 12 Ser/Thr kinases either had functional redundancy, or were dispensable under standard growth conditions. We couldn’t exclude the possibility that certain Ser/Thr kinases function only under specific conditions during certain time scale. It might also be worth to try to create different combinations of double mutants,triple mutants or even mutants with more than 3 genes for a better understanding of their unrevealed functions.

