Recent progress on fluorescent probes for viruses
Siyng Shen ,Weilin Xu ,Jinxing Lu ,Shuhui Wng ,Yurou Hung ,Xioyn Zeng ,Weimin Xio,Jun Yin,∗
a Key Laboratory of Pesticide and Chemical Biology,Ministry of Education,Hubei International Scientific and Technological Cooperation Base of Pesticide and Green Synthesis,International Joint Research Center for Intelligent Biosensing Technology and Health,College of Chemistry,Central China Normal University,Wuhan 430079,China
b Shenzhen Academy of Metrology and Quality Inspection,Shenzhen 518109,China
Keywords:Fluorescent probe Detection Bioimaging Virus Health
ABSTRACT Viruses are ubiquitous in human life.Some viruses can be used as vectors of genetic engineering and specific pesticides.Other viruses trigger a variety of diseases in humans,animals and plants,resulting in high infection rates and mortality.Therefore,convenient,accurate and rapid detection of viruses is of great significance for the diagnosis and treatment of subsequent diseases.In contrast to traditional methods of detection,which rely on time-consuming and complex techniques such as polymerase chain reaction (PCR),fluorescent probes and imaging methods generate real-time results,with high specificity,and have been widely used in viral detection.In this review,the application of viral fluorescent probes in analyzing the molecular structure,detection and biological imaging is discussed.In particular,we categorized the probes based on their specificity for human and plant viruses,reviewing the latest findings and analyzing their limitations.The potential of fluorescent molecular probes in the treatment of viral disease and environmental analysis,and their possible combinations with protein and immune technology are discussed.
1.Introduction
Viruses can cause great destruction to cells,tissues and organs in humans,animals and plants [1–4].Although a few of these viruses are beneficial to humans,such as phages that can be used to treat certain bacterial infections [5],most of them are harmful.Many viruses can cause epidemics.Smallpox viruses have historically killed millions of people worldwide [6].Effective strategies to prevent the spread of the epidemic include good personal hygiene,self-isolation and developing vaccines.However,some viruses mutate,which reduces the effectiveness of vaccines [7].Therefore,it is especially important to utilize real-time,rapid,effective,convenient and economical methods of detection.
Traditional methods of virus detection,such as virus culture,are time-consuming and labor-intensive [8].Although methods such as molecular assays and PCR are sensitive and fast,they are expensive and complex,and the results may be affected by trace contaminants.With the development of nanotechnology and chemistry,new techniques have emerged,such as enzyme-linked immunosorbent assay (ELISA),agglutination assay,electrochemical sensing,and protein sequence fingerprinting [9].Despite the rapid and sensitive detection,the operational complexity is still a limitation [10].
Fluorescent probes have been increasingly used for virus detection because of their simplicity,high sensitivity,selectivity,real-time results and nanoscale features [11–16].The fluorophores can bind specifically to the target and the diversity of fluorophores facilitates viral detection accurately,with a higher signalto-noise ratio [17].Fluorescent probes are of different types,such as inorganic,nanoparticle and graphene probes,as well as quantum dots and organic fluorescent probes [18–20].Organic,smallmolecule fluorescent probes are superior to other types of fluorescent probes,based on structural diversity,significant fluorescence[21–24],easy modification,high sensitivity [25–28],high selectivity [29–33],and simplicity of operation.Therefore,organic small molecular fluorescent probes are preferred [34–39].
In the past decades,viral detection methods have been developed rapidly and studies have focused on organic small molecules as fluorescent probes to detect viruses,with significant application in real life.Herein,we reviewed the latest developments in organic small molecular fluorescent probes based on their optical structures and properties as well as applications and classified the different species of virus.
2.Fluorescence probes for human viruses
2.1. Fluorescence probes for respiratory system viruses
2.1.1.Hemagglutinin1neuraminidase1(H1N1)probes
Influenza A (H1N1) virus can spread among people through droplets and contact.The sources of infection include other infected individuals or carriers of H1N1.The main symptoms of influenza A (H1N1) include fever,fatigue and body aches.Due to its high infectivity and pathogenicity,it can lead to the global pandemic and great losses [40].Therefore,it is especially important to develop rapid,convenient and efficient methods to identify and detect H1N1 virus in advance.
(1) Hemagglutinin-targeted probes:
Human and avian sialic acid terminal glycosides bind specifically to hemagglutinin (HA) on the surface of influenza virus[41,42].Therefore,HA can be used as a target to detect influenza A virus.
Schmidt and lyer’s group reported two fluorescent probes (1–1and1–2) using biotinylated biantennaryS-sialosides to detect influenza viruses rapidly [43].The probes contain three components,including biotinylated divalent scaffold,recognition elements (Ssialosides) and oligoethylene glycol spacer (Fig.1a).1–1and1–2differ based on the sialic acid configuration.Fluorescent antibodies were used as the reporter and the biotinylated carbohydrate together with the fluorescent streptavidin was used as the detector.Using the carbohydrate analogue of a sandwich immunoassay on the surface of a waveguide-based optical biosensor (Fig.1b),the background fluorescence was eliminated when the molecules were more than 200 nm away from the surface due to the decreased effect of the evanescent field.Using1–2,H3N2 emitted a strong signal at the wavelength of 675 nm,while H1N1 had almost no signal,which indicated that the sialoside distinguished H1N1 and H3N2 clearly.However,such a phenomenon could not be observed with1–1,which suggested that1–2was a specific probe while1–1was not.The detection limit was 106 viral particles/mL.The achievement of this research is that they find the first influenza capture and reporter pair that was completely based on glycans,and the1–2probe was stable and active at ambient temperature.
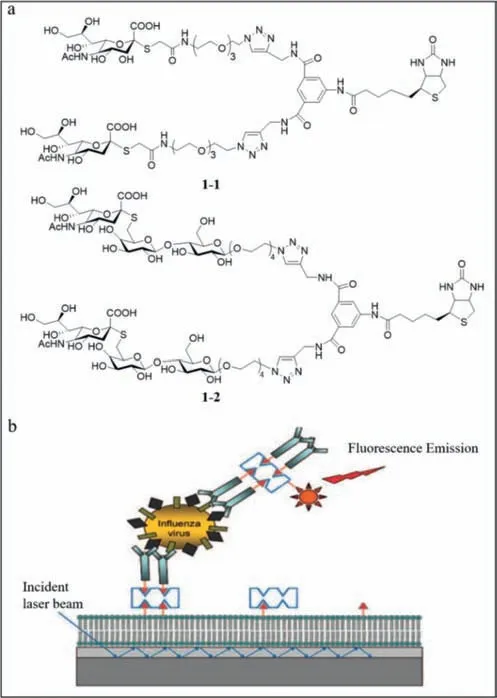
Fig.1. (a) The structures of 1–1 and 1–2.(b) Diagrammatic sketch of the sandwich“carboassay” on a planar optical waveguide biosensor.Reproduced with permission[43].Copyright 2008,ACS.
Based on the results of Schmidt and lyer’s group,Tian’s group developed a probe (probe2) with better specificity [44].They reported a series of fluorogenic glycofoldamers,including TO23,TN23,and P23,which were avian receptors,while TO26,TN26,and P26 were human receptors.Under normal conditions,the probe2was folded;however,upon binding to the virus,the probe unfolded and released the pyrene tails and emitted a strong fluorescence with a blue-shifted emission peak (Fig.2a).The fluorescence reached its maximum within 5 min and remained stable.TO23,TN23,and P23 fluoresced only upon binding to H10N8 and H7N9,whereas TO26,TN26,and P26 fluoresced only upon binding to H3N2,H1N1,and H7N9,reflecting the strong specificity of probe2to the virus.Further,H7N9 infects human beings.The green fluorescence was observed visually under ultraviolet (UV) light,enabling rapid colorimetric identification of influenza A viruses by the naked eye with a detection limit of 0.08 HA units (HAU)50 μL−1.Fluorescence spectrometry revealed an emission peak at 531 nm.The higher the virus concentration,the higher the peak and the stronger the fluorescence.Also,viral mutations affected the fluorescence intensity,demonstrating that probe2facilitated the detection of mutant viruses.Probe2also facilitated the detection of viruses in different media and at different times.Using dynamic light scattering,an increase in particle size was observed upon binding of TO23 to H10N8 and a green fluorescence emission was detected under confocal laser scanning microscopy,while TO26 did not exhibit this property (Fig.2b).
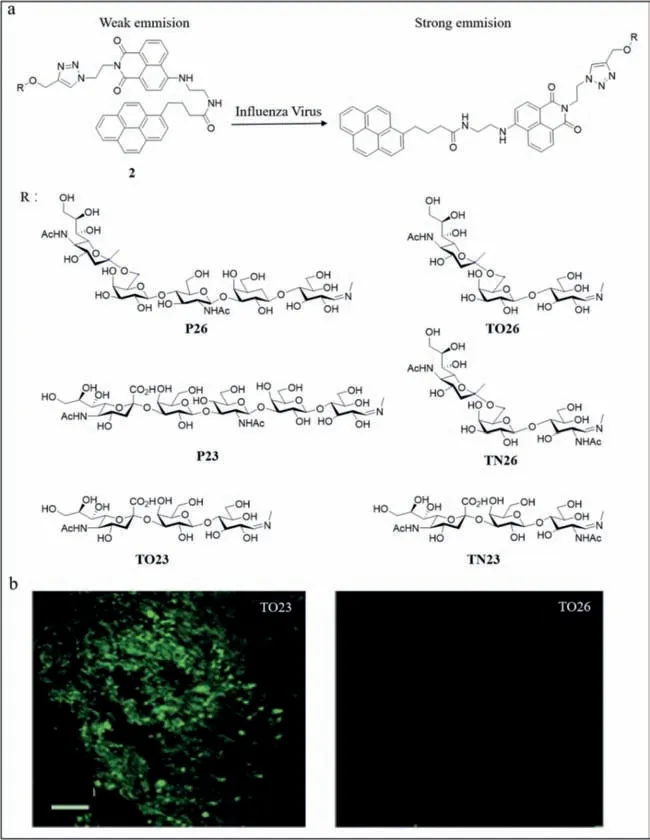
Fig.2. (a) The structure of probe 2 and the introduction of the fluorescence response to influenza A virus.(b) The confocal laser scanning microscopy of TO23 and TO26.Reproduced with permission [44].Copyright 2016,Wiley.
Miyahara’s group performed electrochemical detection to increase the sensitivity [45].Conducting polymers are sensitive biosensors due to their good carrier ability,flexibility,low toxicity and the ability to stabilize doped bioactive molecules.A conductive polymer probe (probe3) carrying a sialic acid-terminated trisaccharide was developed for the specific detection of human influenza virus.Probe3consisted of a conductive polymer made of 3,4-ethylenedioxythiophene (EDOT) carrying an oxylamine group and a sialyllactose on the side chain (Fig.3a).The human influenza virus probe was grafted with 2,6-sialyllactose and the avian influenza virus probe was grafted with 2,3-sialyllactose.The group finally selected the 25 mol% EDOT-OA (EDOT bearing oxylamine group) proportion based on cyclic voltammetry,scanning electron microscopy,X-ray photoelectron spectroscopy,and Nyquist plot.At this proportion,the conductive polymer was electrodeposited at the electrode and the low impedance at the solid-liquid interface ensured the sensitivity of the probe.Label-free detection of influenza virus was achieved by specific binding of sialyllactose to influenza virus (Fig.3b),which was monitoring using a quartz crystal microbalance (QCM) and potentiometry.QCM revealed significant changes in resonance frequency when human influenza virus H1N1 was bound to 2,6-sialyllactose,while only a weak change was observed when the virus was bound to 2,3-sialyllactose and PEDOT (poly3,4-ethylenedioxythiophene),demonstrating the specific binding of the virus to probe3with a detection limit of 0.12 HAU 50 μL−1(Fig.3c).When using potentiometry to study the combination of the probe and H1N1,they observed that the larger the HAU,the greater was the change in potential.2,6-Sialyllactosemodified surfaces emitted a significantly higher signal than 2,3-sialyllactose-modified surfaces and poly EDOT (PEDOT),which also demonstrated binding specificity,with a detection limit of 0.013 HAU 50 μL−1.Although QCM emitted a stronger signal,potentiometry was more sensitive and more convenient,and therefore expected to be detected significantly.

Fig.3. (a) The structure of probe 3.(b) H1N1 can be detected by probe 3.(c) QCM signals’change after injecting virus solutions of various HAU.Reproduced with permission[45].Copyright 2017,ACS.
Later,Zhou and He’s group developed a more sensitive fluorescent probe (probe4) (Fig.4a) [46].They reported a series of sialyl-glycan-functionalized vibration-induced emissive probes(VIEgens) for specific detection and identification of influenza A viruses.VIEgens V1,V3,V5,V7,V9 areα2,6-linked human receptors and VIEgens V2,V4,V6,V8,V10 areα2,3-linked avian receptors.Binding of probe4to the hemagglutinin of influenza A viruses alters the conformation of the probe,resulting in fluorescence and a blue-shifted emission peak (Fig.4b).Fluorescence spectroscopy revealed the specific binding of probe4to the receptor with a significantly altered fluorescence emission profile with a detection limit of 2.84× 10−3–9.20× 10−3HAU 50 μL−1,while the optimal detection range was 0–5.12 HAU 50 μL−1.The fluorescence emission profile also strongly correlated with the change in probe conformation,resulting in variation in response between probes.Accordingly,the ten probes were combined into a high-throughput array successfully and accurately distinguished between influenza A viruses with different specificities and from other virusesviahigh concentration principal component analysis.The potential of the sensor array to distinguish homogeneous influenza viral mutants was also determined.Finally,the adenovirus vaccine with the sensor array revealed that only the influenza adenovirus vector vaccine significantly changed the fluorescence emission profile,reflecting the probe specificity.Different vaccines were distinguishedviaprincipal component analysis based on quadrants and axes.The results contributed significantly to the development of new vaccines.This simple and effective probe is expected to optimize the medical system in underdeveloped regions.

Fig.4. (a) The structures of probe 4.(b) The sensing mechanism.Reproduced with permission [46].Copyright 2022,Elsevier.
(2) Superoxide anions-targeted probe:
The non-structural protein of influenza A virus induces the synthesis of superoxide anions,which can also be used to detect the viruses.Lee’s group developed a sulfinate-based fluorescent superoxide probe (probe5) to detect and distinguish influenza A virus subtypes rapidly and simply [47].Based on the differences in infection characteristics of influenza A viruses in each cell,cell arrays were developed to distinguish the three different influenza A viruses,H1N1,H5N9,and H9N2.The virulence of H9N2 was less than that of the other two subtypes.The virulence of influenza A viruses is not only cellular but also related to its subtypes.The non-structural protein in influenza A virus induced superoxide anions.Therefore,a fluorescent sensor was designed to detect superoxide anion radical.The nucleophilic attack by the superoxide anion on the central sulfur atom of the sulfinate group of the sulfinate-caged fluorescein (SoDA-1) sensor was followed by the departure of thetert–butyl sulfinate radical and synthesis of 2-(9,9a-dihydro-6–hydroxy-3-oxo-3H-xanthen-9-yl)benzoic acid.The interaction betweentert–butyl sulfinate radical and water resulted in the formation oftert–butyl sulfinic acid (Fig.5a).Compared with other reactive oxygen species,SoDA-1 showed good selectivity for superoxide anions and emitted significant and stable fluorescence with a detection limit of 30.4 μmol/L (Figs.5b and c).Unlike cells treated with phorbol-12-myristate-13-acetate (PMA)and lipopolysaccharides (LPS),influenza A viruses do not infect all cells uniformly (Fig.5d).Therefore,the fluorescence intensity varied and cells with strong fluorescence carried additional viruses.Using principal component analysis,H9N2,H5N9,and H1N1 were distinguished based on differences in variance.This study provided a colorimetric and simple method for identifying influenza A virus subtypes,which was expected to be used for virus detection on a large scale in the future.
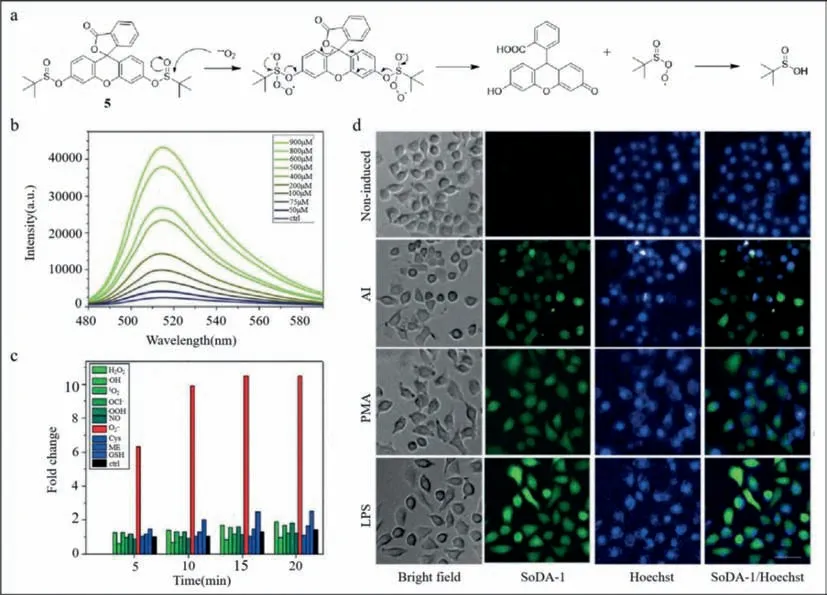
Fig.5. (a) Mechanism of action for detecting superoxide anion by nucleophilic reaction.(b) Fluorescence spectra of SoDA-1 after adding reactive oxygen species (ROS) species and bio thiols.(c) Fluorescence spectra of SoDA-1 after adding superoxide.(d) Fluorescence imaging of SoDA-1 and Hoechst in MDCK cell.Reproduced with permission [47].Copyright 2018,Wiley.
(3) mRNA targeted probes:
The genetic material of H1N1 is RNA,which undergoes transcriptional changes when cells are infected with influenza viruses[48].Therefore,targeting mRNA is also a feasible method.The peptide nucleic acid (PNA)-based forced intercalation (FIT) probe is a fluorogenic hybridization probe that replaces fluorescent bases with asymmetric cyanine dyes and can be used for RNA imaging in living cells [49,50].Seitz’s group developed a set of PNA FIT probes in different colors for detecting transcribed RNA encoding neuraminidase (NA) and matrix protein 1 (M1) of the influenza A(H1N1) virus-infected cells [51].They inserted thiazole orange (6–1),oxazole yellow (6–2),and basic brilliant blue (6–3) into probes to modify the corresponding target DNA and RNA.The probe did not fluoresce at this time because of the low excited state as the probe was single-stranded.In the double-stranded state,the central methine bridge distortion was reduced,which increased the duration of the excited state,resulting in fluorescence (Fig.6a).6–3and6–1were thermally stable and highly selective,and used as probe sets for two-color imaging.As the temperature increased,the probe sets became more reactive and the fluorescence intensity increased significantly.Therefore,the real-time PCR measurements were conducted at 37 and 60 °C.The fluorescence enhancement was obtained at 60 °C,corroborating the effect of temperature on the probe and the wide range of probe hybridization temperatures.Also,with increased lapse of time,more specific sequences were synthesized in the samples and the fluorescence increased,reaching a maximum after 5 h,reflecting the time dependence of mRNA expression.Next,M1 mRNA imaging at 37 °C using the pegylated6–3resulted in significant fluorescence of virus-infected cells after 5 h,while the two groups of cells used as controls remained dark,demonstrating the feasibility of the probe for monitoring mRNA.The imaging of NA and M1 mRNA simultaneously with6–3and6–1turned on the corresponding fluorescence under basic brilliant blue (BO) or thiazole orange (TO) channels,respectively,demonstrating that the probe can be used to perform independent imaging.Finally,when detecting the expression of mRNA in living cells,the fluorescence of6–1bound to NA mRNA was observed after 2 h,mainly in the nucleolus,and the fluorescence also appeared in the cytosol eventually.However,the fluorescence of6–3binding to M1 mRNA was induced after 5 h in the nucleolus and cytosol (Fig.6b),demonstrating that the probe can be used to locate mRNA.

Fig.6. (a) The mechanism of probe 6.(b) The second column to the last column are the images obtained by adding the M1 specific 6–3 and the NA specific 6–1 to the cell after infection at the specified time,meanwhile the first column is the control.Reproduced with permission [51].Copyright 2012,ACS.
Carell’s group later reported a faster fluorescent probe with high signal-to-noise ratio [52].The traditional fluorescentinsituhybridization (RNA-FISH) carries only a single fluorophore and requires a large number of probes for detection,which increases background noise.Carell’s group improved the conventional probe and developed a sensitive fluorescent probe (probe7)(Fig.7a) with three fluorophores by incorporating three C8-alkynedU residues into each of the 10 oligonucleotides and used click chemistry (a method to synthesis molecules by splicing small moieties) to react with Eterneon-Red 645 azide,and then purified and finally obtained 10× 3 small FISH probes (Figs.7b and c).The fixed permeabilized cells were used in quantification,localization and analysis of the transcripts after the probe addition.Compared with the conventional 30× 1 probe,the 10× 3 probe emitted a stronger signal with a higher signal-to-noise ratio,resulting in clearer fluorescence image.Among mixed cell populations,the conventional probe could not distinguish cells of different species when the probe concentration was 0.05 ng/μL,while the 10× 3 probe separated positive green fluorescent protein (GFP) cells from negative GFP cells,and the proportions were observed under blue gating.Also,the 10× 3 probe was effective in detecting transcribed RNA of endogenous ABL.The 10× 3 probe was also effective in RNA target imaging.Despite the small number of binding sites of the RNA target,the 10× 3 probe facilitated the detection of transcribed RNA with red fluorescence after the cells were infected for 4 h.Eventually,additional transcripts were detected,and probe7emitted a green signal,reflecting the good sensitivity of probe7.Probe7greatly reduced the number of probes required for FISH and can be utilized to detect low concentrations of transcripts.
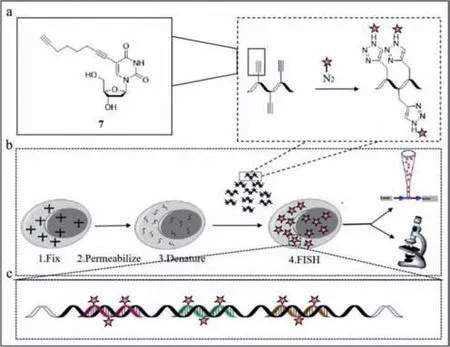
Fig.7. (a) The structure of probe 7.(b,c) The working mechanism.Reproduced with permission [52].Copyright 2020,Wiley.
2.1.2.Hemagglutinin5neuraminidase1(H5N1)probe
Unlike H1N1,the infection sources of influenza A H5N1 viruses are poultry and birds.It can be transmitted from infected birds to humans,but it is not easy to transmit from person to person.However,the high pathogenicity of H5N1 cannot be ignored.The key to preventing its transmission is fast and accurate diagnosis.The hemagglutinin proteins (H) on the surface of H5N1 virus are strong diagnostic targets [53–55].Thus,Tong and Dong’s group reported a series of fluorescent probes (probe8) based on hexaphenylbutadiene (Fig.8a) [56].The H5 of the virus is negatively charged,while the hexaphenyl-butadiene pyridine salt derivatives (HPB-Xs)are positively charged.Therefore,an electrostatic interaction occurred when they were combined,which limited the rotation of the HPB-Xs within the molecule.The aggregation-induced emission (AIE) of the probe resulted in red fluorescence,which was observed with the naked eye under UV light (Fig.8b),enabling colorimetric detection of H5N1.Four HPB-Xs were tested with 11 different proteins,such as H5,and trypsin.The results showed that probe8yielded the most significant signal when bound at the top of H5 cavity (Figs.8c and d) with HPB-I,and HPB-C being the strongest.Meanwhile,the higher the concentration of H5,the stronger was the fluorescence,and the detection limit was 179.5 ng/mL.The experiment demonstrated the strong sensitivity of probe8.The addition of other substances did not change the intensity of the fluorescence,demonstrating the selectivity of probe8.The probe emitted stable and bright fluorescence in less than 10 s when combined with H5,which showed the efficiency of probe8.This fast,efficient and sensitive probe is expected to be used in clinical testing in the future.

Fig.8. (a) The structure of probe 8.(b) The pictures of HPB-Xs under 365 nm UV light.(c) Docking results and interaction between HPB-I and hemagglutinin protein.(d)HPB-I and H5’s binding site.Reproduced with permission [56].Copyright 2021,Elsevier.
2.1.3.Severeacuterespiratorysyndromecoronavirus2(SARS-CoV-2)probe
SARS-CoV-2 is highly infectious and insidious.Infected people experience fever,cough,and respiratory distress [57],but there is no specific treatment available.Therefore,the development of a simple and rapid method that can detect SARS-CoV-2 is imperative.Reverse transcription polymerase chain reaction (RT-PCR),which is a common detection method,is expensive and timeconsuming.Liu and Li’s group reported the use of a lateral flow test strip based on AIE nanoparticles to detect immunoglobulin M(IgM) and immunoglobulin G (IgG) against SARS-CoV-2 [58].They used BPBT (an AIE dye with near-infrared emission) as the fluorescence unit to eliminate background noise,and synthesized the fluorescent reporter AIE810NP,which exhibited good photostability,thermal stability and colloidal stability,and then modified the surface of AIE810NP with chicken IgY (9–1) and SARS-CoV-2 antigen (9–2).After dropping the serum containing IgM and IgG,IgM and IgG moved forward and combined with9–2respectively.Then IgM and IgG bound specifically to mouse anti-human IgM on the M line and mouse anti-human IgG on the G line,and the binding of goat anti-chicken IgY to9–1on the C line was used as the quality standard.Using a portable reader under light emitting diode(LED) illumination,the test was validated if the C line emitted fluorescence,andviceversa.The fluorescence from the M and G lines suggested the presence of the corresponding immunoglobulin and therefore yielded a positive result,otherwise,a negative result was confirmed (Fig.9).The performance of lateral flow test strip was optimal at a readout time of 10 min,while the concentration of9–2was 12.5 μg/mL and mouse anti-human IgM and IgG were 0.6 mg/mL.The threshold of the test strip for IgM was 0.200 and the limit of detection was 0.236.The threshold for IgG was 0.737 and the limit of detection was 0.125.AIE810NP test strips were comparable to SERS-based test strips and ELISA in terms of sensitivity and were greater than colloidal gold nanoparticle (AuNP)-based test strips.AIE810NP test strips can be used for early detection of SARS-CoV-2.
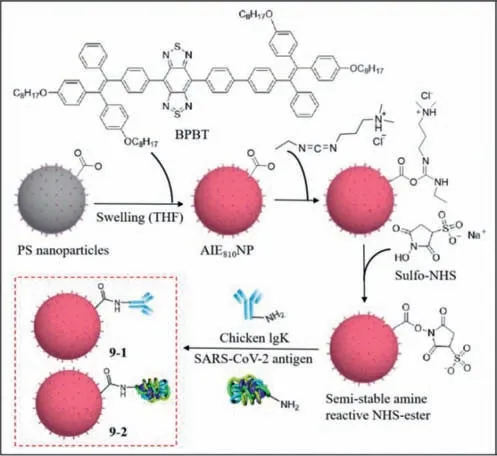
Fig.9. The structures of 9–1 and 9–2.Reproduced with permission [58].Copyright 2021,ACS.
2.2. Fluorescence probes for digestive system viruses
2.2.1.HepatitisBvirus(HBV)probe
HBV causes hepatitis and even cirrhosis and liver cancer.The viral genome can be integrated into the organism.The YMDD gene is related to the activity of HBV reverse transcriptase,including the sequences leading to tyrosine (Tyr,Y),methionine (Met,M),aspartate (Asp,D) and aspartate (D).The common drug named lamivudine cures chronic hepatitis B by inhibiting YMDD gene [59].Longterm treatment with lamivudine induces mutations from methionine (M) to isoleucine (Ile,I) or valine (Val,V).Therefore,it is important to diagnose HBV infection at the early stages.
Water-soluble fluorescent conjugated polymers,which contain a mass of absorbing units,serve as a light collector [60].When conjugated polymers interact with the target,an exciton shifts to the receptor and strong fluorescence is emitted.In addition,their optical or electrical properties are adjusted by changing their monomeric structures.Based on its unique optical characteristics,a water-soluble positively charged poly(3-alkoxy-4-methylthiophene)-based fluorescence probe (probe10) (Fig.10a),proposed by He’s group,was used to detect the YMDD gene mutation of HBV [61].The principle of detection was as follows.According to the original YMDD gene,three oligonucleotide chain targets were designed including a wild-type,a noncomplementary one emulating irrelevant viruses and a 1-bp-mismatched target for the variant YVDD gene.Probe10carried the capture probe sequence,which was completely complementary to YMDD gene.The detailed sequences of oligonucleotide chains are demonstrated in Fig.10a.The positively charged probe10combined with the negative oligonucleotide chain targets tightly,resulting in conversion of the yellow polythiophene to red.Simultaneously,the fluorescence of polythiophene was quenched.Later,the 15-fold stronger fluorescence intensity was restored with a maximum absorption at 402 nm.However,different oligonucleotide chain targets resulted in different intensities of fluorescence (Fig.10b).Polythiophene was found to recognize and distinguish YMDD and YVDD genes selectively.The detection limit of the YMDD gene was 88 pmol/L.In addition,a linear relationship was observed between fluorescence intensity and target concentration (Fig.10c).
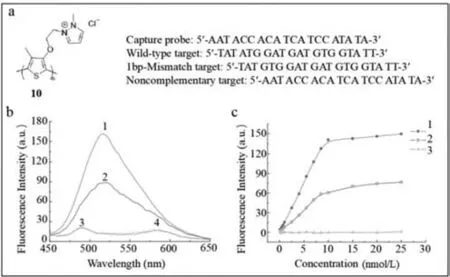
Fig.10. (a) The structure of poly(3-alkoxy-4-methylthiophene) and sequences of kinds of probe 10 and targets.(b,c) Fluorescence spectra of probe 10 with oligonucleotide chain targets parallelly 1.wild-type target;2.1 bp mismatch target;3.noncomplementary target;4.control.Reproduced with permission [61].Copyright 2009,John Wiley &Sons,Ltd.
In brief,probe10facilitates the detection of DNA virus HBV and its variant without label.Further,the probe10can be improved to detect HBV in living cells [62,63].
2.2.2.Humanenterovirus71(EV71)probe
EV71 is associated with severe hand,foot,and mouth disease(HFMD) and is especially infectious to babies and children.The current methods to detect viruses such as PCR requires sophisticated equipment and professional operators.Other methods such as ELISA and localized surface plasmon resonance (LSPR) lack sensitivity [64].Compared with the aforementioned methods,probes with fluorescent signals exhibit prominent advantages [65].
Zhang’s group developed a dual probe based on aggregationinduced emission luminogen (AIEgen) (probe11) [66,67].The structure of TPE-APP,a water-soluble multifunctional AIEgen which contains enzymatic cleavage sites,is shown in Fig.11a.With the catalysis of alkaline phosphatase (ALP),the phosphoryl group of TPE-APP was removed.TPE-DMA,the product,is hardly watersoluble with extremely strong fluorescence.However,TPE-APP decreases resulting in the formation of a silver nanolayer around gold nanoparticles (AuNP),which reveals viruses based on color [68].As a whole,a dual-mode viral analysis based on fluorescent signal and macroscopic color was built (Fig.11a).In order to validate the selectivity of probe11to EV71,several experiments using CVA2,CVA4,CVA6,CVA16,ECHO-18,and EV71 as well as the blank control were conducted.As shown in Fig.11b,high-intensity fluorescence was detected in EV71,while other samples and the blank control hardly emitted light.The yellow color was observed only in EV71 (Fig.11b),suggesting that probe11was strongly selective for EV71.In addition,the detection limit was 1.4 copies/μL.When the probe was utilized in practical applications,8 throat and cloacal samples from HFMD patients was tested positive out of 24 samples analyzed by plasmonic colorimetry.Also,two additional samples were determined to be infected by fluorescence.Probe11showed 100% precision compared with TaqMan real-time PCR [69].Probe11,free of fussy sample preprocessing and expensive testing equipment,is superior due to its convenience and reliability.It is also expected to facilitate the detection of similar viruses such as H7N9 and Zika virus.
Fig.11. (a) Chemical structure and detection principle of probe 11.(b) Fluorescence spectra and absorbance of probe 11 interacting with various kinds of viruses.Reproduced with permission [66].Copyright 2018,ACS.
2.2.3.HepatitisCvirus(HCV)probe
HCV,a type of single-stranded RNA virus,is globally prevalent and leads to chronic inflammatory necrosis and fibrosis,even cirrhosis and hepatocellular carcinoma [70].G4 (Fig.12a) is a nontypical secondary structure of HCV,which comprises G-quartets with an abundance of folded guanines.However,it was difficult to monitor RNA in living cells without artificial modification.

Fig.12. (a) Structures of G4 and probe 12 and their combination.(b) Fluorescence images of probe 12 compared to MB-C (left) and BD4 (right) in different cells.(c)Schematic diagram for dynamic monitoring of serum infection with HLCZ01 cells in patients with hepatitis C.Reproduced with permission [71].Copyright 2019,ACS.
Zhu’s group developed a benzothiazole-based G-quadruplex(G4)-targeted fluorescent probe (called ThT-NE and described as probe12),which can track HCV RNA in real time [71,72].Probe12consisted of electron-withdrawing methylbenzothiazole and electron-donatingN,N-diethylaniline (Fig.12a).The energy-stable angle between methylbenzothiazole andN,N-diethylaniline was 90°,resulting in twisted intramolecular charge transfer (TICT),thereby eliminating the fluorescence of the probe [73].G4 resisted the twist in ThT-NE and changed the angle to 14° and thus emitted the fluorescence.However,the fluorescence intensity of the mixture of probe12and variant HCV or other cell compounds remained at very low level.As a result,probe12was used to selectively detect the RNA G4 sequenceinvitro.In practice,blood serum sample from hepatitis patient was extracted and stained on probe12.The single infected cell was observed based on fluorescent signals (Fig.12b).Compared with the current probe,a molecular beacon with the Cy5/BHQ3 (fluorophore/quencher) pair (MB-C) [74],probe12was specific to the G4 structure,as shown in Fig.12c.To further verify the accuracy of probe12,an antibody specifically combined with G4 structure (BG4) was used.As shown in Fig.12c,the fluorescence emitted by probe12was almost consistent with that of BG4,which demonstrated accurate recognition.Given that probe12could detect HCV without mark and decoration,it could be used to monitor the multiplication of HCV in live cells.As the G4 structure is discovered in other pathogenic RNA viruses,probe12can be expected to play an important role in viral analysis in single cells.
2.3. Fluorescence probes for immune system viruses
2.3.1.Sudanvirus(SUDV)probe
SUDV,a subtype of ebolaviruses causing fever and bleeding,is characterized by high mortality of 53.76%.Efficient diagnosis and control of SUDV is necessary.Therefore,Chen’s group has developed a fluorescent probe based on the metal frame (probe13) [75].Probe13comprised a metal-organic framework (MOF) and two carboxyfluorescein (FAM)-labeled single-stranded DNA sequences(probe ss-DNA,described as P-DNA).The MOF was synthesized using the metal ion Cu(II) [76] along with organic ligands [77] including asymmetric H3CmdcpBr and symmetric dps,contributing to a unique macroporous structure in three dimensions.The structures of ligands are shown in Fig.13a.The water-stable compound was prepared from the mixture of CuSO4and H3CmdcpBr,followed by the addition of dps.Compared to CuSO4and H3CmdcpBr and dps,the water-stable compound shows the best fluorescence quenching result.Therefore,the water-stable compound was chosen to be the important part of probe (Fig.13b).As shown in Fig.13c,the water-stable compound combines with P-DNAviaelectrostatic interaction,π-stacking effect or/and hydrogen bonds,resulting in fluorescence quenching.In the presence of HIV ds-DNA,rigid triplex structures of HIV ds-DNA and P-DNA were formed by reverse Hoogsteen base pairing.SUDV RNA was also complementary to P-DNA.The rigid triplex structures led to the dissociation of P-DNA from the water-stable compound,resulting in fluorescence.In addition,the limits of detection were 196 pmol/L for HIV-1 and 73 pmol/L for SUDV.

Fig.13. (a) Ligand structures of MOF.(b) Fluorescence quenching level of probe DNA interacting with different compounds.(c) Fluorescence quenching and restoration mechanism of probe 13 with SUDV RNA.Reproduced with permission [75].Copyright 2015,ACS.
In the future,probe13can be expected to be utilized in earlystage diagnosis of HIV and ebolavirus disease as well as other virus-associated infectious diseases in the absence of effective vaccines and treatments.
2.3.2.Denguevirus(DV)probe
Dengue hemorrhagic fever (DHF),triggered by DV serotypes 1–4,result in hemorrhage,decreased blood platelets and seepage of plasma,even dengue shock syndrome (DSS) with a death rate of 1%–2.5% [78].C-type lectin domain family 5,member A (CLEC5A)acts as a regulator of DV-induced proinflammatory cytokines and can be detectedviaimmunological testing.However,the combination of CLEC5A and DV is weak.Fluorescent detection of DV leads to quenching of usual labeling dyes.
Hence,Chang’s group developed a fluorescence probe based on double-generation gold nanoparticle chip (probe14) to detect DV[79].CLEC5A was fixed on the gold nano hemisphere on the surface of anodic aluminum oxide (AAO).Thus,the combination of DV and CLEC5A was observed by the variation of impedance [80].In particular,the nano materials must be located in the hemisphere due to the absence of changes in impedance on a flat plane.The grooves between adjacent nano hemispheres enhanced the interaction between DV and CLEC5A.The probe14involved fluorescent sensors to lysine,arginine and cysteine amino acids.The structures of specific probes of cysteine (naphthalimide derivatives,14–1) and lysine/arginine (1,8-naphthalic anhydride derivatives,14–2)were shown in Fig.14a.The compound14–3is one of the materials to synthesize14–2.To further establish the feasibility of the kit,Chang’s group compared probe14and antibody anti-mouse (anti-M) IgG-568 in Fig.14b,which revealed similar fluorescence emission.Also,the size and shape of the fluorescence spots revealed by probe14were similar to DV,as shown in Fig.14c.
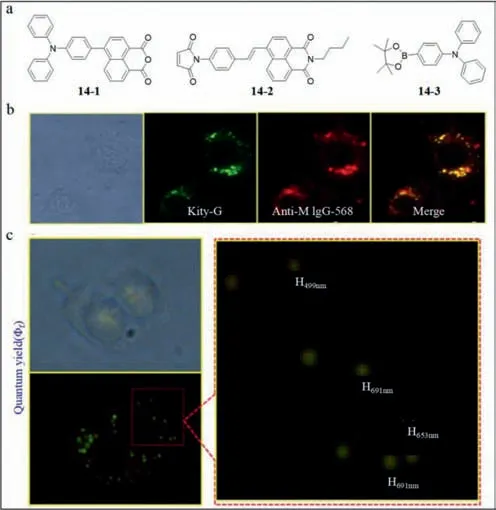
Fig.14. (a) The structures of probe 14–1, 14–2,and 14–3.(b) Fluorescent images of probe 14 and anti-M IgG-568 as well as their merge.(c) Sizes of fluorescent dots detected by probe 14.Reproduced with permission [79].Copyright 2015,Elsevier.
3.Fluorescence probes for plant viruses
3.1. Tobacco mosaic virus (TMV) probes
TMV,a single strand plus RNA virus,is the most transmissible virus specially infecting tobacco.It causes distortion of leaf shape and even necrosis of leaf tissues.When TMV invades cells,H2S is released.In addition to NO and CO,H2S is the third biological gaseous signal [81] especially released under threat.H2S inhibited the activity of RNA-dependent RNA polymerase (RdRp) and coat protein (CP).Further,the H2S donor GYY4137 exhibits significant antiviral activity.Therefore,the recognition of H2S and its donor can be used to detect TMV.In addition,fluorescence probes can be utilized to trace H2Sinvivo.
Xi’s group developed a TMV probe (described as probe15)containing NBD amine [82] as H2S receptor and 4,4-difluoroboradiazaindacene (BODIPY) as fluorophore [83].As shown in Fig.15a,when H2S excited probe15and disrupted C–N bond simultaneously,the BODIPY emitted green fluorescence light and a mercapto group was added to NBD.At 520 nm and under 5 mmol/L,the fluorescence was 40-fold stronger and in the range of 5–50 μmol/L H2S,a linear relationship was observed between fluorescence intensity and H2S concentration.Compared with other probes based on NBD,probe15exhibited 560-fold strong fluorescence and>400-fold selectivity of F/F0s.The ability of probe15for the detection of live entities was investigated by Xi’s group in live tobacco plants.As shown in Fig.15b,the tobacco leaf alone was nonblooming.The addition of probe15to the leaf without TMV resulted in weak fluorescence.However,when probe15was mixed with the TMV infected leaf,the fluorescence was of high intensity.

Fig.15. (a) The structure and response mechanism of probe 15 with H2S.(b) Fluorescence images of probe 15 on the surface of leaves without/with TMV.Reproduced with permission [83].Copyright 2018,Wiley.
As a whole,probe15facilitated the visualization and monitoring of H2S,and thereby enable the detection of TMV and elucidation of the antiviral mechanism of H2S.
Then,another TMV probe is introduced below.
TMV is a typical viral nanoparticle measuring 300 nm in length,18 nm in external diameter and 4 nm in lumen diameter [84].The single-standard RNA of TMV was surrounded by 2130 duplicate coat proteins.Due to its favorable shape,available modifiable surface and considerable transfection efficiency,TMV is used to diagnose diseases and transport drugs,highlighting the need to monitor TMV.
Tian’s group designed a pH-sensitive single-wavelength excited rated fluorescence probe (probe16) [85,86],which revealed the site of TMV in different biological environments at different pH levels.In addition,the limit of detection was 12.9 nmol/L at 37 °C.Probe16was made up of three functional sections: TMV,a pHsensitive fluorophore fluorescein isothiocyanate (FITC) and another fluorophore rhodamine B isothiocyanate (RBITC) insensitive to pH.Upon excitation at 488 nm,FITC and RBITC showed fluorescence peaks at 515 and 575 nm,respectively.The fluorescence intensity emitted by FITC was enhanced as pH increased in the pH range of 5.7–7.4.The peak at 575 nm persisted and was used as an internal standard.Under multiple excitations,probe16was characterized by simplicity and low background noise.In addition,to retain the original morphology of TMV as far as possible,the Tian’s group assembled exogenous fluorophore on the inner surface of coat protein (Fig.16a).The two chemical reactive positions on the inner surface were glutamic acid 97 residue (97 Glu) and glutamic 106 residue (106 Glu).The procedures to synthesize probe16are described in Fig.16a.First,the amino groups were inserted between carboxyl groups of 97 and 106 Glu-and amino groups of ethylenediamine.The isothiocyanate groups of FITC and RBITC were combined with the amino groups of TMV-NH-2.To validate the distribution of probe16,50 nmol/L LysoTracker® Deep Red was used to label lysosomes.The observed yellow was merged with the red fluorescence from TMV-F&RB and the green emission from LysoTrackerR○Deep Red at the coincident sites (Fig.16b).Thus,the TMV track into lysosomes was determined to be true.To monitor the track of TMV in live cells,co-culture experiments of probe16and HeLa cells were conducted.The results presented in Fig.16c showed that FITC and RBITC emitted rapid fluorescence initially.Subsequently,the green fluorescence from FITC was strengthened,while the red fluorescence from RBITC remained at specific intensity.The total fluorescence turned from green to blue,indicating reduction in the environmental pH.The observation was consistent with endocytosis in lysosomes.
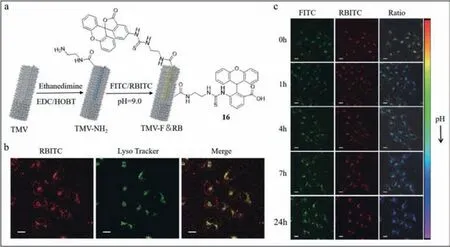
Fig.16. (a) The structures of probe 16.(b) Fluorescence images of probe 16 and LysoTracker as well as their merge in HeLa cells.(c) Fluorescence images in real time of probe 16 in HeLa cells.Reproduced with permission [85].Copyright 2019,Springer Nature.
3.2. H. armigera nuclear polyhedrosis virus (HaNPV) probe
Helicoverpaarmigera(Hubner) is a type of damaging pest infecting more than 300 plant species ranging from vegetables like tomato to grains such as wheat.H.armigeranuclear polyhedrosis virus (HaNPV) is an environmentally friendly biopesticide with high specificity to Hubner.HaNPV holds a complex structure,with coat protein and all substances inside constituting the “occlusion body” (OB).Before the wide-spread application of HaNPV,it is essential to determine whether adequate levels of HaNPV are synthesized.The currently used method to determine the amount of virus is based on a hemocytometer with enhanced Neubauer,which requires high levels of instrumental sophistication and samples.
Therefore,Bhattacharya’s group designed an amphiprotic fluorescence probe (probe17) to conduct on-field detection.Probe17,carried willowy benzimidazole moieties [87] at the 3,6-positions of the carbazole unit and hydrophilic piperazine unit at the nitrogen end [88].After interacting with HaNPV,the willowy parts were restrained,which converted the fluorescence from blue to cyan (Fig.17a).The limit of detection was 103 POBs/mL.Due to the formation of favorable nanoaggregates of probe17at pH 4.5 to 6.5 in both nonpolar/aprotic solvents and aqueous solution,probe17was pHsensitive.Additionally,the assembly of nanoaggregates was thermally reversible.At a high temperature of 85 °C,the assembled nanoaggregates were separated into single units and reassembled when the temperature was reduced to 15 °C.Finally,Bhattacharya’s group conducted experiments to simulate the practical application of probe17.They directly sprinkled HaNPV on the surface of leaves and immersed them into the water solution of probe17.Based on the results shown in Fig.17b,compared with the nonfluorescent black control group,the leaf covered with HaNPV and interacting with probe17emitted cyan fluorescence.Further,as the concentration of HaNPV increased,the intensity of fluorescence was strengthened.

Fig.17. (a) The structure of probe 17.(b) Fluorescence images of probe 17 with HaNPV on the surface of the mango leaves.Reproduced with permission [88].Copyright 2019,ACS.
In brief,Bhattacharya’s group developed a probe to detect HaNPV based on visual color change,which was easy to synthesize and obviated the need for complex pretreatment of sample or precision equipment.
4.Conclusion
In summary,the review highlighted the latest developments in viral molecular fluorescent probes,demonstrating their optical structures and properties as well as applications,with the classification depending on the different viral species.Most of the mechanisms for detection are related to the highly-selective combination of viral proteins and nucleic acids.Probes emitting fluorescent signals have the advantages of convenience and low consumption.Rapid and accurate screening and detection of human viruses associated with infectious diseases like HBV and HCV is essential for definitive diagnosis and timely treatment and control.The use of inexpensive and user-friendly methods for the identification of plant viruses causing large-scale damage such as TMV are essential for economic benefit and food security.
Although the techniques based on the use of fluorescent probes are still at different stages of development [89–96],the difficulties associated with variant detection and observationinvivohave yet to be resolved.The challenges limiting the development of variant detection include distinguishing the possible viral structures or containers,which suggests the need for highly specific probes.Also,viruses may be visualized by detectable fluorescent signals.In the future,we hope advanced viral fluorescent probes will be widely utilized in clinical diagnosis,tumor resection,studies of viral proliferation and assembly,as well as biological pest management.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors acknowledge financial support from National Natural Science Foundation of China (No.22274061),the 111 Project B17019.Supported by the Fundamental Research Funds for the Central Universities (No.CCNU22QN007).Opening fund of Hubei Key Laboratory of Bioinorganic Chemistry &Materia Medica (No.BCMM202101).
 Chinese Chemical Letters2023年12期
Chinese Chemical Letters2023年12期
- Chinese Chemical Letters的其它文章
- Spin switching in corrole radical complex
- Benzothiadiazole-based materials for organic solar cells
- Mono-functionalized pillar[n]arenes: Syntheses,host–guest properties and applications✰
- Recent advances in two-step energy transfer light-harvesting systems driven by non-covalent self-assembly✩
- From oxygenated monomers to well-defined low-carbon polymers
- Doping-induced charge transfer in conductive polymers
