OCT6 inhibits differentiation of porcine-induced pluripotent stem cells through MAPK and PI3K signaling regulation
Xin-Chun Yang, Xiao-Long Wu, Wen-Hao Li, Xiao-Jie Wu, Qiao-Yan Shen, Yun-Xiang Li, Sha Peng,Jin-Lian Hua,*
1 College of Veterinary Medicine, Shaanxi Centre of Stem Cells Engineering & Technology, Northwest A & F University, Yangling, Shaanxi 712100, China
ABSTRACT As a transcription factor of the Pit-Oct-Unc (POU)domain family, octamer-binding transcription factor 6(OCT6) participates in various aspects of stem cell development and differentiation. At present,however, its role in porcine-induced pluripotent stem cells (piPSCs) remains unclear. Here, we explored the function of OCT6 in piPSCs. We found that piPSCs overexpressing OCT6 maintained colony morphology and pluripotency under differentiation conditions, with a similar gene expression pattern to that of non-differentiated piPSCs. Functional analysis revealed that OCT6 attenuated the adverse effects of extracellular signal-regulated kinase (ERK)signaling pathway inhibition on piPSC pluripotency by activating phosphatidylinositol 3-kinase-protein kinase B (PI3K-AKT) signaling activity. Our research sheds new light on the mechanism by which OCT6 promotes PSC maintenance.
Keywords: OCT6; piPSCs; MAPK/ERK;PI3K/AKT; Pluripotency
INTRODUCTION
Octamer-binding factor 6 (Oct-6, Pou3f1, SCIP, Tst-1) is a
This is an open-access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/), which permits unrestricted non-commercial use, distribution, and reproduction in any medium,provided the original work is properly cited.
Copyright ©2022 Editorial Office of Zoological Research, Kunming Institute of Zoology, Chinese Academy of Sciences transcription factor of the Pit-Oct-Unc (POU) family (Wu et al.,2010). The DNA-binding (POU) domain of these proteins is highly conserved, consisting of a POU-specific domain and a POU homeodomain (Sock et al., 1996). POU transcription factors recognize the common DNA octamer motif(ATGCAAAT) (Jauch et al., 2011). They play distinct roles in stem cell development and function and exhibit spatiotemporal expression patterns (Kim et al., 2020).OCT6is not widely expressed in all cells but has been detected in embryonic stem cells (ESCs) (Meijer et al., 1990), Schwann cells(Friedrich et al., 2005), glial cells (Kuhlbrodt et al., 1998), and neonatal testicular cells (Wu et al., 2010). To date, most studies have explored the function ofOCT6in Schwann cells,while little is known regarding their functions in other cell types. For induced pluripotent stem cells (iPSCs),OCT6partially replacesOCT4to initiate reprogramming (Jerabek et al., 2017).OCT6is also implicated in the maintenance of iPSC pluripotency and plays a decisive role in neural differentiation(Malik et al., 2019). However, few studies have investigated the potential functions and mechanisms ofOCT6in porcine iPSCs (piPSCs).
The mitogen-activated protein kinase (MAPK) signaling pathway regulates many biological processes, including cell cycle, apoptosis, differentiation, protein biosynthesis, and tumorigenesis (Wen et al., 2022). Activation of the mitogenactivated protein/extracellular signal-regulated kinase (MEK)signaling pathway can promote differentiation of mouse ESCs(mESCs), while inhibition of MEK/ERK signal transduction can promote self-renewal and pluripotency of mESCs (Choi et al.,2017). Inhibition of MEK facilitates transformation of mouse ectodermal stem cells into ESC-like cells, however inhibition of the MEK signaling pathway reduces pluripotency in porcines(Xu et al., 2019). Previous reports have indicated that piPSCs differ from mESCs. Inhibition of MAPK impairs piPSC pluripotency and decreases cell proliferation ability (Zhu et al.,2021). Phosphatidylinositol 3-kinase (PI3K), a dimer consisting of a regulatory subunit p85 and catalytic subunit p110 (Aoki & Fujishita, 2017), activates protein kinase B(AKT), thereby regulating cell proliferation, differentiation,apoptosis, and migration (Haddadi et al., 2018). PI3K/AKT plays a vital role in maintaining ESC self-renewal (Yao et al.,2010). Interestingly, impairment of pluripotency by inhibiting MEK in mouse ESCs can be counteracted by PI3K activation(Singh et al., 2012).
We previously reported that porcine PSCs can be induced in medium containing leukemia inhibitory factor (LIF), basic fibroblast growth factor (bFGF), and other cytokines, with the addition of signal transduction inhibitors (Chir099021,SB431542) and feeder cells to maintain colony morphology and pluripotency (Ma et al., 2018). Here, asOCT6is not expressed in piPSCs, we explored the function ofOCT6in piPSCs by its overexpression. Notably, piPSCs overexpressingOCT6showed smoother colony edges in the presence of feeder cells (feeders) and maintained normal colony morphology and pluripotency after feeder removal.RNA sequencing (RNA-seq) analysis revealed thatOCT6expression promoted the up-regulation of extracellular matrix(ECM)-related genes and inhibited the differentiation of piPSCs. In addition,OCT6activated PI3K and inhibited ERK in piPSCs, consistent with its ability to maintain piPSC colony morphology and pluripotency upon feeder removal.
MATERIALS AND METHODS
Overall experimental scheme
In this study, we used lentivirus packaging to transfect PCDHTeton-3×Flag-OCT6-Puro and PCDH-Teton-3×Flag-Puro into piPSCs. The piPSCs transfected with theOCT6gene vector(OE-OCT6 piPSCs) were used as the experimental group, and the piPSCs transfected with empty vector (OE-NC piPSCs)were used as the control group.OCT6expression was detected by quantitative real-time polymerase chain reaction(qRT-PCR), cellular immunofluorescence staining, and western blot analysis. We performed total RNA-seq of piPSCs and detected the expression levels of related genes using qRT-PCR, western blotting, and immunofluorescence staining.Small molecular compounds were used to activate or inhibit the ERK and PI3K-AKT signaling pathways, respectively, to compensate for the maintenance of pluripotency caused by overexpression ofOCT6.
Acquisition and production of piPSCs
The piPSCs were obtained from previous research. We established doxycycline (Dox)-induced porcine PSCs, in which the expression of transgenic genes can be switched on/off by Dox and lentiviral particles of TetO-FUW-OSKM (OCT4,SOX2,KLF4, andc-MYC) and FUW-M2rtTA can be infected into porcine embryonic fibroblasts (PEFs). Here, the somatic cells were reprogrammed into porcine iPSCs induced by Dox(DOX-iPSCs), which displayed a normal karyotype of 38 chromosomes, formed embryonic bodiesin vitro, and spontaneously differentiated into three germ layers (Ma et al.,2018).
Cell culture
HEK293T cells were cultured in 6-well plates (Thermo Fisher Scientific, USA) using Dulbecco’s Modified Eagle Medium(DMEM) (Corning, USA) and 10% fetal bovine serum (FBS)(Gibco, USA) at 37 ℃ with 5% CO2, with new medium changed each day. Mouse embryonic fibroblasts (MEFs) were cultured with DMEM supplemented with 10% FBS, then treated with mitomycin C for 3 h and cultured on a 12-well plate (1×105cells/well) at 37 ℃ with 5% CO2. The piPSCs were cultured in DMEM (Corning, USA) supplemented with 15% FBS (Vistech, New Zealand), 10 ng/mL LIF (Sino Biological, China), 10 ng/mL bFGF (10014-HNAE, Sino Biological, China), 0.1 mmol/L L-glutamine, 3 μmol/Lol/L CHIR99021 (M3148, Sigma-Aldrich, USA), 2 μmol/Lol/L SB431542 (S1067, Selleck, USA), 0.1 mmol/L βmercaptoethanol (M3148, Sigma-Aldrich, USA), 4 μg/mL doxycycline (D9891, Sigma-Aldrich, USA), 100 μg/mL streptomycin, 100 U/mL penicillin, and 0.1 mmol/L nonessential amino acids (NEAA) (Gibco, USA) at 37 ℃ with 5%CO2. After the piPSCs were grown to 80%-90% confluency,they were digested into single cells on a 12-well plate (2×104cells/well) using TrypLE Select (Gibco, USA) (Wu et al., 2021;Zhu et al., 2021).
Vector construction and cloning
To explore the function ofOCT6in piPSCs, total RNA was extracted from porcine testes using TRIzol Reagent (Takara,Japan), then reverse transcribed (Thermo Fisher Scientific,USA) into cDNA. The porcineOCT6gene was PCR-amplified from cDNA, and subcloned into an in-house PCDH-Teton-3×Flag-MCS-Puro vector using a NovoRec®plus One step PCR Cloning Kit (Novoprotein, China). All primers used in this study are provided in Supplementary Table S1, and the core coding sequence of the recombinant vector was verified by sequencing.
Lentiviral packaging
After HEK293T cells were grown to 80%-90% density in 6-well plates, lentiviral plasmids (VSV-G, PAX2) for packaging were transfected into the cells using polyethyleneimine (PEI).In total, 1 μg of VSV-G, 1 μg of PAX2, and 2 μg of lentiviral plasmids were mixed with 12 μL of PEI (1 mg/mL), rested for 15 min, followed by the addition of 200 μL of Opti-MEM to the HEK293T cells. After 12 h, the culture medium was changed to DMEM. After 48-72 h, the lentiviral particles were collected,and cell fragments were filtered with a 0.45 μmol/L filter.Viruses were used after 12 h of rest at 4 ℃.
Lentiviral particle transduction
The passaged piPSCs were inoculated into a 12-well plate covered with feeders. The viral supernatant containing lentiviral particles was mixed with the piPSC culture medium(1:1) and 4 μg/mL polystyrene. The piPSCs in mixed medium were cultured at 37 ℃ for 8-12 h, after which the medium was replaced with new piPSC medium. Puromycin was used to select purine-positive cells after 7 days of culture.
RNA extraction, reverse transcription, and qRT-PCR detection
RNAiso Plus Reagent (Takara, Japan) was selected to extract total RNA. Quality of the extracted RNA was detected using a Nanodrop microspectrophotometer (Thermo Fisher Scientific,USA) and agarose gel electrophoresis. After removing the feeders, the cells were passaged once to remove feeder interference, followed by total RNA extraction. The RNA (2 μg)was then reverse transcribed using a RevertAid First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific, USA) to obtain cDNA. Finally, qRT-PCR was performed through a three-step process using SuperReal PreMix Plus (SYBR Green)(Tiangen, China) (Wei et al., 2021). The primers for qRT-PCR are shown in Supplementary Table S1.
Immunofluorescence staining
The piPSCs were washed twice with phosphate-buffered saline (PBS) and fixed at room temperature for 15 min with 4% paraformaldehyde. The cell membrane was perforated with 0.1% Triton-100 for 10 min, then sealed at room temperature for 1 h with 10% FBS. The cells were incubated with primary antibodies (Flag; 1:1 000; Sigma-Aldrich, USA) at 4 ℃ for 12 h, then washed with PBS three times and incubated with goat anti-mouse IgG (H+L) secondary antibody AlexaFluor488 (1:500; ZSGB-BIO, China) conjugated at room temperature for 1 h. Cells were counterstained at room temperature with Hoechst33342 (1:1 000) for 5 min. Finally,images were collected using an EVOS fluorescence microscope (Thermo Fisher Scientific, USA).
Western blot analysis
The piPSCs (~80% density) were digested using TrypLETMSelect (Thermo Fisher Scientific, USA), with the same volume of DMEM and 10% FBS then added to neutralize the digestion reaction. The cell mixture was transferred into a 1.5 mL EP tube and centrifuged at 5 000 ×gfor 3 min at 24 °C to obtain the cell precipitation. RIPA lysate (Beyotime, China),10 mmol/L PMSF protease inhibitor (Sigma-Aldrich, USA), and phosphatase inhibitor were used to cleave the piPSCs on ice for 30 min. After that, 5×sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer was added for denaturation at 100 °C for 5 min. The protein sample was transferred to 8%-12% SDS-PAGE gel for separation at 100 V. The gel was then transferred to a Trans-Blot SD Cell and System (Bio-Rad, USA) and to polyvinylidene fluoride (PVDF) membranes at a voltage of 15 V. The membranes were sealed at room temperature with 8% skim milk or 5% bovine serum albumin (BSA) for 2 h, then incubated at 4 ℃ for 12 h with primary antibodies, including βactin (1:8 000; AbMole, USA), Flag (1:1 000; Sigma-Aldrich,USA), OCT6 (1:1 000; Abmart, China), phospho-p44/42 MAPK (1:1 000; Cell Signaling Technology, USA), p44/42 MAPK (1:1 000; Cell Signaling Technology, USA), PI3K-AKT(1:2 000; Abmart, China), and phospho-PI3K-AKT (1:2 000;Abmart, China). All primary antibodies were diluted with TBST buffer (20 mmol/L Tris HCl/pH 8.0, 150 mmol/L NaCl, 0.05%Tween 20).
The PVDF membranes were washed in TBST buffer for 30 min, and new TBST buffer was replaced every 10 min. The membranes were incubated with goat anti-mouse/rabbit IgG(H+L) (Cell Signaling Technology, USA) at 37 ℃ for 1 h, then washed with TBS-T buffer at room temperature for 15 min.Finally, the Tanon-5200 automatic chemiluminescence image analysis system (Tanon, China) was used to detect horseradish peroxidase (HRP) signals. ImageJ (v1.53c) was used to analyze the relative grayscale of western blots.
Alkaline phosphatase (AP) staining
Suitable piPSCs were fixed with 4% paraformaldehyde (pH 7.4) for 15 min, then washed 2-3 times with PBS. The cells were stained with AST Fast Red TR and α-naphthol AS MX phosphate (Sigma-Aldrich, USA) according to the manufacturer’s instructions. In brief, 1.0 mg/mL AST Fast Red TR, 0.4 mg/mL α-naphthol AS MX phosphate, and 0.1 mmol/L Tris-HCL 8.8 buffers were used to prepare the AP staining solution. The cells were incubated for 20 min with AP stain,which was then replaced with PBS. The AP-positive piPSC clones showed red and images were captured using a phase contrast microscope (Nikon, Japan).
RNA-seq analysis
To explore the molecular mechanism ofOCT6in piPSCs, we performed RNA-seq of OE-OCT6 and OE-NC piPSCs after feeder removal (Zhu et al., 2021). We extracted total RNA using RNAisoPlus Reagent (Takara, Japan) and guanidine isothiocyanate phenol-chloroform extraction. Total RNA quality was detected using a Nanodrop spectrophotometer, and total RNA integrity was detected by agarose gel electrophoresis.Total RNA was digested using DNase Ⅰ, mRNA was enriched using Oligo (dT) magnetic beads, and cDNA was synthesized by adding random primers after mRNA was interrupted(second-strand cDNA was synthesized with dUTP instead of dTTP). Next, 3’adenylated and adaptor ligation was performed at the ends of the synthesized double-stranded DNA. Joint connection and PCR amplification of the connection products were carried out. A DNA library was constructed and qualitychecked, with the resulting product then cyclized (Yu et al.,2021). The DNBSEQ platform was used for computer sequencing, and quality control (QC) of the obtained raw reads was carried out. The filtered clean reads were compared to the porcine genome Ssc11.1, and expression level was normalized to reads per kilobase per million mapped reads (RPKM) using gene annotation files. EdgeR was used to analyze differences in samples. Heat-maps were drawn using pheatmap (v1.0.12) to show specific gene expression levels. ClusterProfiler (v4.2.2) was used for Gene Ontology(GO) and Kyoto Encyclopedia of Genes and Genomes(KEGG) analysis. AdjustedP<0.05 orQ<0.05 was used to define the working threshold of statistical significance.
Statistical analysis
Studentt-test was used to determine significant differences between two groups, and univariate or multivariate analysis of variance (ANOVA) was used to determine significant differences between multiple groups. All data were expressed as mean±standard error (SE) and differences were considered significant atP<0.05.
RESULTS
Cloning and overexpression of porcine OCT6 gene in piPSCs
To determine whether overexpression ofOCT6can improve the anti-differentiation ability of piPSCs, we introduced theOCT6gene cloned from porcine testicular tissue into the lentiviral vector. TheOCT6protein sequence was highly conserved, with only three amino acid differences amongHomo sapiens,Mus musculus, andSus scrofa(Figure 1A).
To further investigate the role of porcineOCT6in piPSCs,we generated piPSC lines stably expressing porcineOCT6using the TetOn lentiviral vector (Figure 1B). Quantitative analysis showed that theOCT6gene was up-regulated(1.25×104times) compared with the negative control (NC,TetOn-3×Flag empty vector). After three consecutive passages, AP staining was performed. Results showed that overexpression ofOCT6did not affect piPSC pluripotency,and the edge of the piPSC clones was smoother than that of the NC (Figure 1D). Both protein and immunofluorescence analyses showed that the OCT6 protein expression was significantly up-regulated inOCT6piPSCs but could not be detected by Flag or OCT6 antibodies in the NC group (Figures 1E, F). These results suggest thatOCT6helps maintain colony morphology of piPSCs.
OCT6 inhibits piPSC differentiation under differentiation conditions
We then applied differentiation conditions to verify the role ofOCT6in maintaining piPSC morphology and pluripotency (Kim et al., 2020, 2021; Wu & Yao, 2005). Under differentiation conditions, the piPSCs overexpressingOCT6maintained basic colony morphology for three consecutive passages without feeders (Figure 2A). AP staining was used to determine the effects ofOCT6on piPSC pluripotency under differentiation conditions. Results showed that piPSCs overexpressingOCT6maintained pluripotency, even in the culture system without feeders (Figure 2B). Detection of core pluripotent genes showed a slight but significant increase in exogenous OSKM and endogenousOCT4expression (1.31 and 1.86 times, respectively) and an obvious up-regulation in endogenousc-Mycexpression (2.92 times) in the OE-OCT6 piPSCs (Figure 2C). Previous studies have shown thatc-Mycis the downstream target of the Wingless/Integrated (WNT)signaling pathway, and an increase inc-Mycgene expression may be related to activation of WNT (Yu et al., 2019; Zhang et al., 2021). We therefore removed the GSK3 inhibitor Chir99021 from the culture medium. However, results showed that OE-OCT6 still maintained colony morphology and pluripotency after Chir99021 removal (Figure 2D).
To further explore the results caused by overexpression ofOCT6, we performed RNA-seq of the two piPSC lines after 5 days of culture. Results showed that transcriptional changes occurred in both directions after overexpression ofOCT6(339 up-regulated genes and 639 down-regulated genes)(Figure 2E). We detected pluripotency-related genes by qRTPCR and found that compared with the OE-NC group, the OEOCT6 group showed elevatedc-MycandOCT4expression but inhibitedLIN28Aexpression, which may be related to the inhibition of differentiation (Figure 2C). The most significant differentially expressed genes (DEGs) from RNA-seq were verified by qRT-PCR. Results showed that these hypervariable genes were involved in the inhibition of differentiation and maintenance of stem cell pluripotency.Among them,PAX5,CLU, andCOL5A1affect differentiation(Baghdadi et al., 2018; Mansouri et al., 1996; Oh et al., 2020)andSOX3,SIX6, andL1CAMare involved in the regulation of stem cell pluripotency (Corsinotti et al., 2017; Son et al., 2011;Wolfrum et al., 2010) as well as decreased expression of imprinted geneNNAT(Teichroeb et al., 2011), with epigenetic erasure required for resetting the cell identity to a ground state, which may explain whyOCT6overexpression is beneficial for the cloning and maintenance of pluripotency(Figure 2F).
We further carried out KEGG and GO analyses of the RNAseq data. KEGG analysis showed that the DEGs were mainly enriched in the MAPK and PI3K-AKT signaling pathways(Figure 2G). GO analysis indicated that the DEGs were primarily related to the ECM, extracellular space, and whole composition of plasma membrane (Figure 2H). These findings suggest thatOCT6not only affects the canonical pathways but may also inhibit piPSC differentiation by regulating the components of the ECM and cell membrane.
OCT6 maintains morphology and reduces pluripotency of piPSC colonies by inhibiting MAPK-ERK pathway
KEGG analysis was performed to explore the possible pathways regulated byOCT6. Results indicated that the main enriched pathways were the MAPK, PI3K-ATK, and ECM pathways. As MAPK and PI3K-AKT play important roles in piPSCs, we compared the OE-OCT6 and OE-NC groups with piPSCs cultured with (MOCK group) or without feeders (Fgroup). The heat-map of MAPK-related genes showed that the expression patterns in the OE-NC and F-groups and the OEOCT6 and MOCK groups were similar (Figure 3A). The qRTPCR results further confirmed that the gene expression pattern in the OE-OCT6 group was basically the same as that in the MOCK group, indicating that the OE-OCT6 group maintained the characteristics of piPSCs after feeder removal,and OE-OCT6 may inhibit the ERK pathway via activation ofDUSP10and inhibition ofPTPRR,RRAS, andFGFR3(Figure 3B, C).
Under feeder removal conditions, the ERK and phosphorylated ERK (p-ERK) protein expression levels in the OE-OCT6 piPSCs relative to the OE-NC piPSCs significantly increased and decreased, respectively (Figure 3D). We verified the changes in the MAPK-ERK pathway in piPSCs with and without feeders, which indicated that the ERK pathway was inhibited by the removal of feeders (Figure 3E).To verify this, we tested PD0325901 (ERK inhibitor) and 12-Otetradecanoyl phorbol-13-acetate (PMA, ERK activator) in the piPSCs. When 1 μmol/L PD0325901 was added to the OEOCT group, the colonies became smaller; in contrast, piPSCs showed obvious differentiation after the addition of PMA(Figure 3F). However, the piPSCs maintained basic colony morphology after the addition of PD0325901 (Figure 3G).These findings confirm that inhibition of ERK may be of significance for the maintenance of piPSCs. After the concentration gradient test, 1 μmol/L PD0325901 and 1 μmol/L PMA were selected as the optimum concentrations for ERK inhibition and activation, respectively (Supplementary Figure S1A, B).
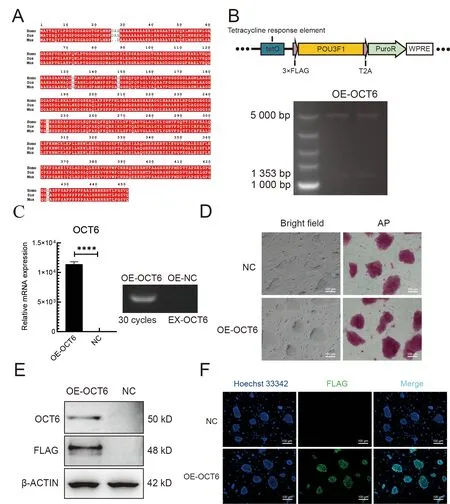
Figure 1 Cloning and overexpression of OCT6 gene in piPSCs

Figure 2 OE-OCT6-piPSCs maintained good colony morphology independent of feeders compared with OE-NC piPSCs
PI3K-AKT pathway compensates for adverse effects caused by MAPK-ERK suppression
We next asked howOCT6maintains piPSC pluripotency after feeder removal. Heat-map and qRT-PCR analysis showed that the expression patterns of PI3K-AKT pathway-related genes in the OE-OCT6 group were similar to those in the MOCK group (Figure 4A). Notably, expression of theMAGIfamily,LPAR1, andIGTB4genes increased, while PPP2R3A expression decreased. These genes are associated with cell adhesion and improvement in piPSC colony formation (Gong et al., 2019; Li et al., 2017b, 2019) (Figure 4B, C). In the absence of feeders, AKT and phosphorylated AKT were significantly decreased and increased, respectively, in the OEOCT6 group relative to the OE-NC group (Figure 4D). We also compared the protein level of AKT in the OE-NC group with or without feeders and found that AKT and phosphorylated AKT were significantly decreased and increased, respectively, in the presence of feeders (Figure 4E). This change in AKT level in the OE-OCT6 group disappeared after the addition of the PI3K inhibitor LY294002, consistent with the addition of PD0325901 in the OE-NC group, resulting in colony reduction(Figure 4F). These results suggest that elevated AKT signaling maintains the pluripotency of piPSCs under differentiation conditions. After adding the PI3K activator 740Y-P to the OE-NC group, pluripotency also recovered under differentiation conditions (Figure 4G). Therefore, the PI3K/AKT signaling pathway is beneficial for maintaining piPSC pluripotency under differentiation. After the concentration gradient test, 5 μmol/L LY294002 and 5 μmol/L 740Y-P were selected as the optimum concentrations for PI3K inhibition and activation, respectively (Supplementary Figure S1C, D).

Figure 3 OCT6 maintains piPSC colony morphology by inhibiting MAPK/ERK signaling pathway
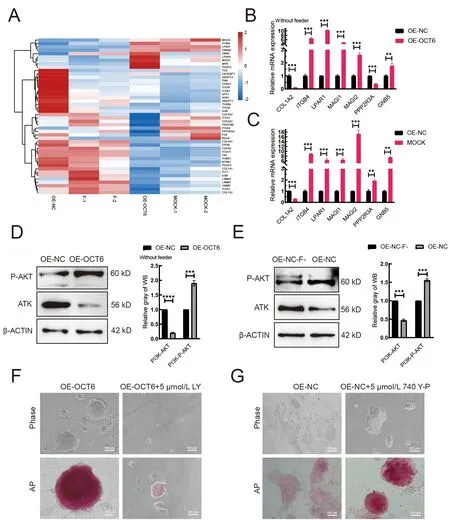
Figure 4 OCT6 counteracts the negative effects of ERK inhibition on pluripotency by activating the PI3K-AKT signaling pathway
DISCUSSION
PiPSCs can be effectively obtained by transferringOCT4,SOX,KLF4, andc-Mycinto porcine fetal fibroblasts using lentiviral vectors (Ezashi et al., 2009). However, existing culture systems cannot be maintained long-term without feeders (Montserrat et al., 2012). In the absence of feeders,existing piPSCs show obvious differentiation, indicating the existence of several defects in piPSCs compared with humans and mice (Esteban et al., 2009). Here, we attempted to modify gene expression patterns to create a piPSC line that can maintain pluripotency and colony morphology without feeders.We found that introduction of theOCT6gene enabled piPSCs to achieve maintenance with no xenogeneic cells.
OCT6andOCT4are both members of the POU family. To date, however, most studies have focused on their functions in neurons and neural progenitor cells (Kawasaki et al., 2003).Although the structures of OCT6 and OCT4 are similar, the reprogramming response is weaker in OCT6 (Jerabek et al.,2017).OCT6can cause extensive chromatin opening (Malik et al., 2019) and is reported to be expressed in early embryos(Meijer et al., 1990). As a pluripotency inducer,OCT6may play a potential role in the maintenance of pluripotency.Interestingly, in the present study, afterOCT6overexpression,piPSCs maintained their pluripotency without feeders, and their expression pattern was essentially the same as regular piPSCs cultured on feeders. Differentiation-related gene expression (e.g.,L1CAM(Bao et al., 2008)) and cell adhesion and ECM-related gene expression (e.g.,ITGB4(Zhang et al.,2020b),LPAR1(Li et al., 2017a)) were decreased and enhanced, respectively, which was beneficial for the maintenance of piPSC colony morphology. The expression ofc-Mycalso increased, which may provide additional maintenance of self-renewal and pluripotency of piPSCs.
Our study showed thatOCT6regulated the MAPK-ERK and PI3K-AKT pathways to help maintain piPSC pluripotency.Inhibition of MAPK signaling can induce PSCs to transform into the immature state (Hackett & Surani, 2014). There are many conflicting reports regarding the role of the MAPK signal transduction pathway in human embryonic stem cells(hESCs), including maintenance of pluripotency (Armstrong et al., 2006) and promotion of differentiation (Ding et al., 2010).In pigs, inhibition of MAPK can lead to the rapid loss of pluripotency (Gao et al., 2014). Furthermore, whenOCT6is overexpressed, the expression ofDUSP10(Hiratsuka et al.,2020) increases significantly, whereas the expressionPTPRR(Su et al., 2013),RRAS(Michael et al., 2016), andFGFR3(Blick et al., 2013) decreases significantly, potentially inhibiting ERK. Surprisingly, under feeder withdrawal-stimulated differentiation, we found that low-dose PD0325901 led to colony formation, although proliferation ability was significantly decreased. Furthermore, the addition of a MAPK activator abolished the effects ofOCT6, resulting in complete differentiation of piPSCs.
Activation of PI3K caused byOCT6overexpression can overcome the damage caused by inhibition of MAPK (Singh et al., 2012). As an important pathway for controlling ESC function, PI3K-AKT signaling plays a crucial role in cell selfrenewal and pluripotency maintenance in pigs (Welham et al.,2011). Here, we found that overexpression ofOCT6significantly activated the PI3K-AKT signaling pathway, with an increase inMAGI1/2(Zhang et al., 2020a),LPAR1(Cui et al., 2019), andITGB4(Leng et al., 2016) gene expression,and decrease inPPP2R3Agene expression (Chen et al.,2019). These genes can promote PI3K-AKT signaling pathway activation. When PI3K-AKT signaling pathway inhibitors were added to the OE-OCT6 group, pluripotency was lost, and clones became smaller. These results indicate that inhibition of the PI3K-AKT signaling pathway damages piPSC pluripotency, suggesting thatOCT6maintains piPSC pluripotency in the differentiated state via the PI3K-AKT signaling pathway.
Taken together, our results showed that overexpression ofOCT6enhanced piPSC colony formation in the differentiated state and enhanced piPSC pluripotency, primarily by inhibition of the MAPK-ERK signaling pathway and activation of the PI3K-AKT signaling pathway.
It is worth highlighting that the genetically engineered (OEOCT6) iPSCs, whose capability to differentiatein vitrowas considerably attenuated by transgenically increasing the relative abundance ofOCT6transcripts and extrinsic OCT6 proteins, may provide an excellent source of nuclear donor cells (NDCs) suitable for somatic cell nuclear transfer (SCNT)-based assisted reproductive technologies in pigs and other mammalian species (Gorczyca et al., 2021; Olivera et al.,2016; Samiec & Skrzyszowska, 2010; Secher et al., 2017;Zhang et al., 2014). Highly reprogrammable NDCs may exhibit an enhanced capacity to sustainably perpetuate stemnessand pluripotency-related molecular attributes. The latter, in turn, could enhance the epigenetic dedifferentiation capacity of OE-OCT6 iPSCs in SCNT-derived embryos, conceptuses,and progenies generated for transgenic and biomedical research, not only for pig-to-human xenotransplantation and tissue engineering, but also for regenerative and reconstructive surgical treatments in humans and other mammalian species (Berthelsen et al., 2021; Podstawski et al., 2022; Song et al., 2016; Wiater et al., 2021a, 2021b; Xu et al., 2022; Yu et al., 2021; Zhao et al., 2020).
DATA AVAILABILITY
Raw sequencing reads were deposited in the NCBI Sequence Read Archive (SRA) database (BioProjectID PRJNA854625),GSA database (PRJCA011011), and Science Data Bank database (DOI: 31253.11.sciencedb.j00139.00029).
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
X.C.Y., X.L.W., and J.L.H. designed the research. X.C.Y.,X.L.W., W.H.L., and X.J.W. performed the research. X.C.Y.and X.L.W. wrote the paper. X.C.Y., Q.Y.S., Y.X.L., and S.P.analyzed the data. X.C.Y. and X.L.W. modified the manuscript. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
The authors thank Dr. Yong Tang for help revising the manuscript, and Dr. Zhen-Shuo Zhu and Ju-Qing Zhang for helpful comments on this paper.
- Zoological Research的其它文章
- Optimization of sgRNA expression strategy to generate multiplex gene-edited pigs
- Virome in healthy pangolins reveals compatibility with multiple potentially zoonotic viruses
- Chronic lithium treatment ameliorates ketamineinduced mania-like behavior via the PI3K-AKT signaling pathway
- A glimpse into the biodiversity of insects in Yunnan: An updated and annotated checklist of butterflies(Lepidoptera, Papilionoidea)
- Unveiling the functional and evolutionary landscape of RNA editing in chicken using genomics and transcriptomics
- Animal models of Alzheimer’s disease: Applications,evaluation, and perspectives

