Expression profiling of transgenes (Cry1Ac and Cry2A) in cotton genotypes under different genetic backgrounds
Kashif NOOR ,Hafiza Masooma Naseer CHEEMA ,Asif Ali KHAN ,Rao Sohail Ahmad KHAN
1 Department of Plant Breeding and Genetics,University of Agriculture,Faisalabad 38040,Pakistan
2 Institute of Plant Breeding and Biotechnology,Muhammad Nawaz Shareef University of Agriculture,Multan 60000,Pakistan
3 Center of Agricultural Biochemistry and Biotechnology,University of Agriculture,Faisalabad 38040,Pakistan
Abstract Transgenic cotton carrying the Cry1Ac gene has revolutionized insect pest control since its adoption,although the development of resistance in insect pests has reduced its efficacy.After 10 years of cultivating Bacillus thuringiensis(Bt) cotton with a single Cry1Ac gene,growers are on the verge of adopting Bt cotton that carries the double gene(Cry1Ac+Cry2A) due to its better effectiveness against insect pests.Thus,the current study was designed to evaluate the role of each gene in the effectiveness of Bt cotton carrying the double gene.The expression levels of the Cry1Ac and Cry2A genes were evaluated in the leaves of 10 genotypes (2 parents and 8 F1 hybrids) at 30 days after sowing(DAS),while samples of leaves,bolls and flowers were taken from the upper and lower canopies at 70 and 110 DAS.The F1 hybrids were developed through reciprocal crosses between two Bt (CKC-1,CKC-2) and two non-Bt (MNH-786,FH-942) parents.The differential expression of transgenes was evaluated through Enzyme Linked Immuno-Sorbent Assay (ELISA).The results showed that the MNH786×CKC-1 hybrid had the highest concentrations of Cry1Ac gene at 30 DAS (3.08 μg g-1) and 110 DAS (1.01 μg g-1) in leaves.In contrast,the CKC-2×MNH-786 hybrid showed the lowest concentrations of Cry1Ac gene at 30 DAS (2.30 μg g-1) and 110 DAS (0.86 μg g-1).The F1 hybrid FH-942×CKC-2 showed the highest concentrations of Cry2A gene at 30 DAS (8.39 μg g-1) and 110 DAS (7.74 μg g-1) in leaves,while the CKC-1×MNH-786 hybrid expressed the lowest concentrations of Cry2A gene at 30 DAS (7.10 μg g-1) and 110 DAS(8.31 μg g-1).A comparison between the two stages of plant growth showed that leaves had the highest concentrations at 30 DAS,whereas the lowest concentrations were observed at 110 DAS for both genes in leaves.When the expression pattern was compared between various plant parts in genotype CKC-2,it was found that leaves had higher concentrations of Cry1Ac (3.12 μg g-1) and Cry2A (8.31 μg g-1) at 70 DAS,followed by bolls (Cry1Ac (1.66 μg g-1) and Cry2A (8.15 μg g-1)) and flowers (Cry1Ac (1.07 μg g-1) and Cry2A (7.99 μg g-1)).The genotype CKC-2 had higher concentrations of Cry1Ac (3.12 μg g-1) and Cry2A (8.31 μg g-1) in the upper canopy but less accumulation (2.66 μg g-1 of Cry1Ac,8.09 μg g-1 of Cry2A) in the lower canopy at 70 DAS.Similarly,at 110 DAS,the expression levels of Cry1Ac and Cry2A in upper and lower canopy leaves were 1.52 and 7.92 μg g-1,and 0.99 and 7.54 μg g-1,respectively.Hence,the current study demonstrates that different genotypes showed variable expression for both of the Cry1Ac and Cry2A genes during plant growth due to different genetic backgrounds.The Cry2A gene had three-fold higher expression than Cry1Ac with significant differences in expression in different plant parts.The findings of this study will be helpful for breeding insect-resistant double-gene genotypes with better gene expression levels of Cry1Ac and Cry2A for sustainable cotton production worldwide.
Keywords: transgenic cotton breeding,transgene expression,double gene,insect resistance
1.lntroduction
In Pakistan,Bt cotton was commercialized after its approval in 2010 (Cheemaet al.2016).It was introduced in this region to control lepidopteran bollworm infestations and increase profits by reducing pesticide applications and pest damage (Downeset al.2016).Previously,the indiscriminate use of insecticides led to the evolution of resistance in the target pests like cotton bollworm,spotted bollworm and pink bollworm (Ahmadet al.2009;Qayyumet al.2015).This extensive use of pesticides has also adversely affected environmental quality and the health of farmers and their families (Jabbar and Mallick 1994;Aliet al.2010).
Transgenic cotton constitutes about 72.6% of the total cotton area in the world (about 32.9 million hectares)and is grown in 15 countries.Pakistan grows biotech cotton at about 2.8 million hectares and ranked the 8th in biotech area in the world (ISAAA 2017).Genetically modified (GM) crops are being grown by 18 million people in 28 countries (James 2004).The increasing adoption rate of GM crops provides strong evidence of their good performance all over the world (Jamilet al.2019).
The performance of transgenic cotton depends on the quantity of toxin(s) produced by theBacillusthuringiensis(Bt) gene(s) in various plant tissues.In Pakistan,the Bt toxin level in the recommended varieties should be high enough to control bollworm under field conditions,as compared to non-Bt cotton varieties which are significantly damaged due to bollworm infestation (Arshadet al.2015).Single gene Bt cotton has been effective in controlling cotton bollworms in the past (Khanet al.2011).Over time,the target pests acquired resistance through an evolutionary process against the singleCry1Acgene in Bt cotton (Tabashnik and Carriere 2017;Jamilet al.2021a),and this has affected the working efficiency of Bt cotton (James 2014).The resistance has developed in insects against Bt cotton having a single gene (Cry1Ac)and variable expression is one of the main reasons for insect resistance (Akhteret al.2018;Jamilet al.2021b;Shahzadet al.2021).
A number of factors (such as site-of-gene insertion,the type of promoter,the methylation of the gene,sowing time,internal cell environment and external environment)are involved in the variable expression of theCry1Acgene among cultivars,but the genetic background plays a critical role in the production ofBttoxin at an optimal and sustainable level.In the case ofCry1Acgene,the event,insertion site and base pairs were the same,but the genotypes were different among the different genetic backgrounds (Cheemaet al.2016;Khanet al.2018;Jamilet al.2021b).It was observed that genotypes with different genetic backgrounds and growth stages were also a source of variation in the expression ofCry1Acgene.The maximum expression was observed at 70 DAS(days after sowing),and dropped to the lowest level at 120 DAS (Wanet al.2005;Khanet al.2018).
In cotton-growing districts of Pakistan,34% of farmers did not know about the transgenicity of the cotton cultivars which they had grown in their fields.The maximum number of cotton cultivars possessed sub-optimal toxin concentrations at 70 DAS and the levels decreased even further at 120 DAS (Spielmanet al.2017).Bt cotton varieties vary in theirCry1Actoxin concentrations from a minimum level of 0.03 to a maximum level of 1.99 mg g-1in leaf tissues (Cheemaet al.2016),while the required lethal concentration has been reported to be about 1.5 mg g-1(Malik and Ahsan 2016) or 1.89 mg g-1(Kranthiet al.2005).Consistent gene expression is crucial for durable resistance against targeted insect pests.A breeder’s ideotype must be the high yielding genotypes with the stable expression of transgenes for the desired trait.However,the limiting factor in developing such ideotypes is the lack of information about the influence of genetic background on attenuating transgene expression (Khanet al.2018).
The biotechnologists introduced anotherBtgene,Cry2Ab,for better performance against target pests in the 2nd generation Bt cotton (Panet al.2014;Qamaret al.2019).Many countries are growing such Bt cotton genotypes that carry multiple genes and produce more than one toxin against the same insect pest.In Pakistan,stacked gene Bt cotton was also approved by the National Biosafety Committee (NBC) working under Environmental Protection Agency in 2017.
For the success of Bt cotton genotypes,a sufficient amount of endotoxin must be present in the required parts of the plant at the right time during the growing season.Information regarding the expression of theCry1AcandCry2Agenes is critical for developing such cultivars.TheCry1Acgene produces a toxin protein which is rapidly degraded as a plant grows older (Guoet al.2016),while the relative level of Cry2Ab protein is higher and remains consistent throughout the growing season (Adamczyk and Sumerford 2001;Jamilet al.2021b).Therefore,better combinations of genes need to be developed through breeding,which have better and more sustainable Bt expression.The proposed research was designed to evaluate the performance of double-gene (Cry1AcandCry2A) Bt cotton with different genetic backgrounds for transgene expression.The main objective of this study was to determine the expression patterns of theCry1AcandCry2Agenes in different plant parts (leaves,bolls and flowers) of cotton at different growth stages (30,70 and 110 DAS) under field conditions.The impact of the genetic background in modulating transgene expression was also determined.
2.Materials and methods
2.1.Plant materials
Seeds of two Bt cotton genotypes (CKC-1 and CKC-2)possessing theCry1AcandCry2Agenes were obtained from the Centre of Excellence and Molecular Biology(CEMB),and two non-Bt genotypes (MNH-786 and FH-942) were obtained from the Cotton Research Institute,Multan and Cotton Research Station,Faisalabad,Pakistan.The Bt genotypes (CKC-1 and CKC-2) were crossed with the two non-Bt genotypes (MNH-786 and FH-942) reciprocally,and eight F1hybrids were constituted (Fig.1).The four parents (two Bt and two non-Bt) and eight F1hybrids (with stacked genes) were sown in the experimental field area of the Department of Plant Breeding and Genetics,University of Agriculture Faisalabad,Pakistan.

Fig.1 Schematic representation of the crossing of two Bt genotypes (CKC-1 and CKC-2) with two non-Bt genotypes (MNH-786 and FH-942) in a reciprocal manner and their F1 hybrids.
2.2.Field preparation and experimental layout
This study was conducted in the Faisalabad district Punjab Pakistan between 31°25′0′′N and 73°5′28′′E longitude at an altitude of 184 m a.s.l.(Shaukatet al.2019).The parents were sown in the field,and crosses were made in 2018.The genotypes including Bt parents and their offspring were sown in the field in 2019 for evaluation of theCry1AcandCry2Agenes.All recommended agronomic practices for cotton (i.e.,DAP and potash at sowing time and nitrogen in three splits at irrigation time) were followed throughout the growing season.Pesticides were also applied against insects (whitefly,jassid and aphid) and diseases for healthy growth.The genotypes were sown in three replications with 10 plants in each replication under a randomized complete block design.Sorghum was a barrier for isolating the experimental fields from the surrounding environment in order to avoid pollen contamination (NBC 1999).
2.3.Spatio-temporal and intra-plant expression of the Cry1Ac and Cry2A genes
The concentrations of theCry1AcandCry2Agenes were measured in 10 stacked gene Bt cotton genotypes throughout the growing season through Enzyme-Linked Immuno-Sorbent Assay (ELISA).The objective of performing ELISA was to quantify the toxin levels in the Bt cotton genotypes under study at 30,70 and 110 DAS.
Leaf samples were taken for quantification of Bt toxins from the 5th node at 30 DAS from three biological replicates.At 70 DAS,samples of the leaves,flowers and bolls were collected from the 8th node for the lower canopy and the 15th node for the upper canopy.At 110 DAS,samples were collected from the 12th node for the lower canopy and the 25th node for the upper canopy.The samples of leaves,bolls and flowers were collected from the same node in all genotypes to avoid any ambiguity in the comparative study.The collected samples were kept in an icebox and transported to the laboratory.The ELISA was performed at 30 DAS in leaves only since there were no fruits present on plants at that stage,while leaves,bolls and flowers were sampled at 70 and 110 DAS.
ELISA of the collected samples was performed immediately to obtain authenticated results.Quantiplate ELISA kits (Envirologix,USA,with Cat No.AP005 forCry2Aband AA0341 forCry1Ac) were used for quantifying theCry1Ac andCry2Atoxins in fresh tissues according to the manufacturer’s instructions.Calibrators were used in the first column,and a negative control was also assayed along with the tested samples.Three samples were collected from each genotype,and ELISA was performed by using each sample in duplicate.The plate was read at 450 nm by a spectrophotometer (Epoch,USA) and reference readings were also obtained at 605 and 630 nm.Simple regression was used to analyse the results in terms of μg g-1of fresh tissue.
2.4.Statistical analysis
Analysis of variances in a factorial treatment design was used to analyse the data using Statistix 8.1 Software in order to find the differences in genotypes,time,plant parts and their interactions at the 5% significance level.Tukey’s HSD was also performed using R3.6.1 Software.
3.Results
3.1.Spatio-temporal expression of the Cry1Ac and Cry2A genes
Analysis of variance showed significant differences among genotypes,plant parts and their interaction for theCry1AcandCry2Agenes (Table 1).Regarding development time,the analysis of variance showed significant differences among genotypes,DAS and their interaction for theCry1AcandCry2Agenes (Table 2).The overall concentrations ofCry1Acgene at 30 DAS ranged between 1.98 μg g-1in cross CKC-1×FH-942 and 3.38 μg g-1in genotype CKC-2 for leaves among the Bt parents and crosses (F1hybrids).The cross FH-942×CKC-1 had a higher concentration (3.02 μg g-1) in comparison with CKC-1×FH-942,although they have the same parents.The concentrations of the majority of genotypes ranged between 2.30-3.08 μg g-1for theCry1Acgene.The concentrations ofCry2Agene ranged between 8.31 μg g-1in cross CKC-1×MNH-786 and 8.39 μg g-1in genotype CKC-1 for leaves among all genotypes,while MNH-786×CKC-1 had a higher concentration 8.34 μg g-1as compared to CKC-1×MNH-786.Slight variations in expression were observed for theCry2Agenebetween genotypes and within genotypes at 30 DAS.The concentrations of the majority of genotypes ranged between 8.34-8.37 μg g-1(Fig.2).

Fig.2 Expression of Cry genes in leaves at 30 days after sowing (DAS).Different letters above each bar show the differences among concentrations of Cry1Ac (A) and Cry2A (B) in leaves at 30 DAS (Tukey’s HSD at P-value≤0.05).

Table 1 Mean square values for spatial expression of genotypes in Bt cotton1)
The concentrations varied between 1.12-3.12 μg g-1forCry1Acgene and 7.99-8.32 μg g-1forCry2Agene in leaves among the Bt cotton parents and F1hybrids/crosses at 70 DAS.At 110 DAS,the concentrations varied between 0.80-1.68 μg g-1forCry1Acgene and 6.78-7.93 μg g-1forCry2Agene in leaves among the Bt cotton parents and F1hybrids/crosses (Figs.3 and 4).The concentrations of the twoCrygenes in bolls at 70 DAS varied between 0.75-1.92 and 7.39-8.27 μg g-1for theCry1AcandCry2Agenes,respectively,while the concentrations varied between 0.51-1.39 forCry1Acgene and 6.53-7.84 μg g-1forCry2Agene in bolls among Bt cotton parents and F1hybrids at 110 DAS (Figs.5 and 6).The concentrations ofCrygenes decreased in bolls as plant age increased,especially for theCry1Acgene.In the case of flowers,the concentrations of the twoCrygenes varied between 0.30-1.07 μg g-1forCry1Acand 6.87-8.11 μg g-1forCry2Aat 70 DAS among all Bt cotton genotypes.The concentrations varied between 0.16-0.84 μg g-1for theCry1Acgene and 6.24-7.33 μg g-1forCry2Ain flowers among the parents and crosses at 110 DAS (Figs.7 and 8).
3.2.lntra-plant expression of the Cry1Ac and Cry2A genes
Upper canopy leavesAnalysis of variance showed significant differences among genotypes,DAS,and their interaction for bothCrygenes in the upper canopy leaves(Table 2).The concentrations ofCry1Acgene among the parents and F1hybrids/crosses ranged between 1.38 μg g-1in cross CKC-2×MNH-786 and 3.12 μg g-1in CKC-2 in the upper canopy leaves at 70 DAS,while MNH-786×CKC-2 had a higher concentration of 2.82 μg g-1in comparison with its reciprocal cross CKC-2×MNH-786.The toxin concentrations in the majority of genotypes fell between 2.26-2.84 μg g-1(Fig.3).The overall concentrations ofCry2Agene ranged between 8.17 μg g-1(CKC-1×MNH-786) and 8.32 μg g-1(CKC-2×MNH-786)in the upper canopy leaves at 70 DAS,while reciprocal crosses MNH-786×CKC-1 and MNH-786×CKC-2 had concentrations of 8.27 and 8.32 μg g-1at this stage,respectively.The majority of genotypes ranged between 8.26-8.32 μg g-1for the concentration ofCry2Atoxin in the upper canopy leaves (Fig.3).The concentrations ofCry1Acgene among the parents and crosses ranged between 1.05 μg g-1in cross CKC-2×MNH-786 and 1.68 μg g-1in cross MNH-786×CKC-1 at 110 DAS.The crosses MNH-786×CKC-2 and CKC-1×MNH-786 had concentrations of 1.61 and 1.47 μg g-1,respectively.The toxin levels of the genotypes under study fell between 1.21-1.51 μg g-1in a majority of cases (Fig.3).The concentrations ofCry2Agene varied between 7.44 μg g-1in cross FH-942×CKC-1 and 7.92 μg g-1in genotype CKC-1 in the upper canopy leaves at 110 DAS.In contrast,cross CKC-1×FH-942 had a higher concentration (7.64 μg g-1) than its reciprocal cross.The concentrations of the majority of genotypes ranged between 7.44-7.65 μg g-1forCry2Ain the upper canopy leaves (Fig.3).

Fig.3 Expression of Cry genes in upper canopy leaves at 70 and 110 days after sowing (DAS).Bars mean SD (n=3).Different letters above each bar show the differences among concentrations of Cry1Ac (A) and Cry2A (B) in upper canopy leaves at 70 DAS,and Cry1Ac (C) and Cry2A (D) in upper canopy leaves at 110 DAS (Tukey’s HSD at P-value≤0.05).
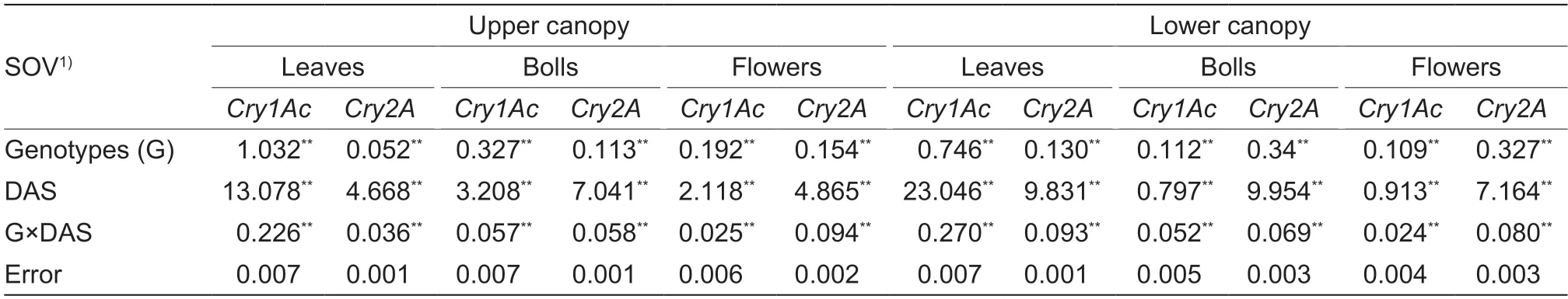
Table 2 Mean square values for temporal expression of Bt genotypes
Lower canopy leavesAnalysis of variance showed significant differences among genotypes,DAS and their interaction in lower canopy leaves for theCry1AcandCry2Agenes (Table 2).The concentrations ofCry1Acgene among parents and crosses ranged between 1.12 μg g-1in cross CKC-2×MNH-786 and 2.66 μg g-1in genotype CKC-2 at 70 DAS,while MNH-786×CKC-2 had a concentration of 1.83 μg g-1.The concentrations in the majority of genotypes fell between 1.27 and 1.83 μg g-1for theCry1Acgene (Fig.4).Similarly,the concentrations ofCry2Agene varied between 7.99 μg g-1in cross FH-942×CKC-1 and 8.29 μg g-1in cross CKC-2×FH-942 at 70 DAS.The relatively different levels of 8.27 and 8.28 μg g-1were found in crosses CKC-1×FH-942 and FH-942×CKC-2,respectively.The toxin expression levels in the majority of genotypes ranged between 8.13 and 8.27 μg g-1in the lower canopy leaves (Fig.4).The concentrations ofCry1Acgene ranged between 0.80 μg g-1in CKC-1 and 1.14 μg g-1in CKC-1×FH-942 cross at 110 DAS.The concentrations in the majority of genotypes ranged between 0.86 and 1.00 μg g-1in the lower canopy leaves (Fig.4).The concentrations ofCry2Agene ranged between 6.78 μg g-1in cross CKC-2×MNH-786 and 7.74 μg g-1in cross FH-942×CKC-2 at 110 DAS in the lower canopy leaves.The concentrations in the majority of cotton genotypes ranged between 7.10 and 7.54 μg g-1for theCry2Agene (Fig.4).

Fig.4 Expression of Cry genes in lower canopy leaves at 70 and 110 days after sowing (DAS).Bars mean SD (n=3).Different letters above each bar show the differences among concentrations of Cry1Ac (A) and Cry2A (B) in lower canopy leaves at 70 DAS,and Cry1Ac (C) and Cry2A (D) in lower canopy leaves at 110 DAS (Tukey’s HSD at P-value≤0.05).
Upper canopy bollsSignificant differences among genotypes,DAS and their interaction for theCrygenes were found in the upper canopy bolls (Table 2).The concentrations ofCry1Acgene among Bt parents and F1hybrids ranged from 0.97 μg g-1in cross MNH-786×CKC-2 to 1.92 μg g-1in cross FH-942×CKC-1 in the upper canopy bolls at 70 DAS,while different levels (1.27 and 1.47 μg g-1) were present in their reciprocal crosses,respectively.The concentrations in the majority of genotypes ranged between 1.42 and 1.66 μg g-1in the upper canopy bolls forCry1Ac(Fig.5).The concentrations ofCry2Agene fell between 7.98 μg g-1(CKC-1×MNH-786) and 8.27 μg g-1(genotype CKC-2) in the upper canopy bolls.The concentrations in the majority of genotypes ranged between 8.06 and 8.21 μg g-1in the upper canopy bolls at 70 DAS (Fig.5).The concentrations ofCry1Acgene among the parents and crosses ranged between 0.57 μg g-1in cross MNH-786×CKC-2 and 1.39 μg g-1in cross FH-942×CKC-1 for the upper canopy bolls at 110 DAS.The crosses CKC-2×MNH-786 and CKC-1×FH-942 have different concentrations of 0.85 and 1.15,respectively,than their reciprocal crosses.The concentrations in the majority of genotypes ranged between 1.12 and 1.20 μg g-1among all Bt cotton genotypes (Fig.5).TheCry2Agene concentrations varied between 7.20 μg g-1in CKC-2×MNH-786 and 7.84 μg g-1in FH-942×CKC-2 in the upper canopy bolls at 110 DAS.The reciprocal crosses MNH-786×CKC-2 and CKC-2×FH-942 had different concentrations of 7.37 and 7.30 μg g-1,respectively.ForCry2Agene in the upper canopy bolls,the concentrations in the majority of genotypes ranged from 7.30 to 7.57 μg g-1at 110 DAS (Fig.5).

Fig.5 Expression of Cry genes in upper canopy bolls at 70 and 110 days after sowing (DAS).Bars mean SD (n=3).Different letters above each bar show the differences among concentrations of Cry1Ac (A) and Cry2A (B) in upper canopy bolls at 70 DAS,and Cry1Ac (C) and Cry2A (D) in upper canopy bolls at 110 DAS (Tukey’s HSD at P-value≤0.05).
Lower canopy bollsAnalysis of variance showed significant differences among genotypes,boll canopy and their interaction for theCry1AcandCry2Agenes in bolls(Table 3).The concentrations ofCry1Acgene ranged between 0.75 μg g-1in genotype CKC-1 and 1.30 μg g-1in cross CKC-2×FH-942 at 70 DAS.The concentrations in the majority of genotypes varied between 0.97-1.12 μg g-1forCry1Acgene (Fig.6).The concentrations ofCry2Agene ranged between 7.39 μg g-1(CKC-2×MNH-786) and 8.15 μg g-1(CKC-2) at 70 DAS,while a relatively higher level of 8.02 μg g-1was present in MNH-786×CKC-2.The concentrations in the majority of genotypes ranged between 8.13-8.27 μg g-1in the lower canopy bolls mong all Bt cotton genotypes (Fig.6).The concentrations ofCry1Acgene fell between 0.51 μg g-1in cross MNH-786×CKC-2 and 1.06 μg g-1in cross CKC-1×FH-942 for lower canopy bolls among the parents and crosses at 110 DAS.The reciprocal crosses CKC-2×MNH-786 and FH-942×CKC-1 had different concentrations of 0.59 and 0.85,respectively.The concentrations in the majority of genotypes ranged between 0.66-0.95 μg g-1in lower canopy bolls (Fig.6).The concentrations ofCry2Agene in the lower canopy bolls varied between 6.53 μg g-1in cross CKC-2×MNH-786 and 7.36 μg g-1in genotype CKC-2 at 110 DAS among all Bt cotton genotypes,while it was 7.04 in cross MNH-786×CKC-2 which was higher than in CKC-2×MNH-786.The concentrations in the majority of genotypes ranged between 6.95-7.16 μg g-1forCry2Agene in the lower canopy bolls (Fig.6).

Fig.6 Expression of Cry genes in lower canopy bolls at 70 and 110 days after sowing (DAS).Bars mean SD (n=3).Different letters above each bar show the differences among concentrations of Cry1Ac (A) and Cry2A (B) in lower canopy bolls at 70 DAS,and Cry1Ac (C) and Cry2A (D) in lower canopy bolls at 110 DAS (Tukey’s HSD at P-value≤0.05).
Upper canopy flowersAnalysis of variance showed significant differences among genotypes,DAS and their interaction for theCry1AcandCry2Agenes in upper canopy flowers (Table 2).The concentrations ofCry1Acgene among Bt parents and F1hybrids ranged from 0.64 μg g-1(MNH-786×CKC-2) to 1.07 μg g-1(CKC-2) in the upper canopy flowers at 70 DAS,while a higher level of 1.03 μg g-1was present in reciprocal cross CKC-2×MNH-786.The concentrations in the majority of genotypes fell between 0.68 and 1.04 μg g-1in the upper canopy flowers (Fig.7).The concentrations ofCry2Agene ranged from 7.25 μg g-1in cross CKC-1×MNH-786 to 8.11 μg g-1in FH-942×CKC-2 in the upper canopy flowers at 70 DAS.TheCry2Aconcentrations of MNH-786×CKC-1 and CKC-2×FH-942 were 7.63 and 7.74,respectively,which were different than their reciprocal crosses.The concentrations in the majority genotypes ranged from 7.60 to 7.99 μg g-1in the upper canopy flowers at 70 DAS (Fig.7).The concentrations ofCry1Acgene among the parents and their crosses fell between 0.24 μg g-1(CKC-1×MNH-786) and 0.84 μg g-1(FH-942×CKC-2) for the upper canopy flowers at 110 DAS,while the concentrations in MNH-786×CKC-1 and CKC-2×FH-942 were 0.25 and 0.46 μg g-1,respectively.The concentrations in the majority of genotypes fell between 0.30-0.46 μg g-1among all Bt cotton genotypes (Fig.7).The concentrations ofCry2Agene ranged from 6.87 μg g-1in cross CKC-2×FH-942 to 7.33 μg g-1in genotype CKC-1 in the upper canopy flowers at 110 DAS,while a higher level of 7.26 μg g-1was present in reciprocal cross FH-942×CKC-2.The concentrations in the majority of genotypes varied between 7.10-7.26 μg g-1at 110 DAS for theCry2Agene in the upper canopy flowers (Fig.7).

Fig.7 Expression of Cry genes in upper canopy flowers at 70 and 110 days after sowing (DAS).Bars mean SD (n=3).Different letters above each bar show the differences among concentrations of Cry1Ac (A) and Cry2A (B) in upper canopy flowers at 70 DAS,and Cry1Ac (C) and Cry2A (D) in upper canopy flowers at 110 DAS (Tukey’s HSD at P-value≤0.05).

Table 3 Mean square values for inter-canopy expression of genotypes in Bt cotton1)
Lower canopy flowersAnalysis of variance showed significant differences among genotypes,DAS and their interaction for theCry1AcandCry2Agenes in lower canopy flowers (Table 2).The levels ofCry1Acgene varied from 0.30 μg g-1in genotype CKC-1 to a maximum of 0.76 μg g-1in genotype CKC-2 among all cotton genotypes at 70 DAS.The concentrations in the majority of genotypes fell between 0.41 to 0.65 μg g-1for theCry1Acgene (Fig.8).The concentrations ofCry2Agene ranged from 6.87 μg g-1in cross CKC-1×MNH-786 to 7.82 μg g-1in cross FH-942×CKC-2 at 70 DAS,while the concentrations for reciprocal crosses MNH-786×CKC-1 and CKC-2×FH-942 were 7.33 and 7.29 μg g-1,respectively.The concentrations in the majority of genotypes varied between 7.19-7.68 μg g-1in the lower canopy flowers (Fig.8).The concentrations ofCry1Acgene ranged from 0.16 μg g-1(MNH-786×CKC-2) to 0.52 μg g-1(FH-942×CKC-2) in lower canopy flowers,while reciprocal crosses CKC-2×MNH-786 and CKC-2×FH-942 had concentration levels of 0.34 and 0.29 μg g-1,respectively.The concentrations in the majority of genotypes fell between 0.16 and 0.29 μg g-1in the lower canopy flowers (Fig.8).The concentrations ofCry2Agene in the lower canopy flowers varied from 6.24 μg g-1in cross CKC-2×MNH-786 to 6.97 μg g-1in the MNH-786×CKC-2 cross.The concentrations in the majority of cotton genotypes ranged between 6.53 and 6.78 μg g-1for theCry2Agene at 110 DAS in the lower canopy flowers (Fig.8).

Fig.8 Expression of Cry genes in lower canopy flowers at 70 and 110 days after sowing (DAS).Bars mean SD (n=3).Different letters above each bar show the differences among concentrations of Cry1Ac (A) and Cry2A (B) in lower canopy flowers at 70 DAS,and Cry1Ac (C) and Cry2A (D) in lower canopy flowers at 110 DAS (Tukey’s HSD at P-value≤0.05).
4.Discussion
Chemical pesticides were frequently used to control insect pests before the introduction of Bt cotton (Qayyumet al.2015).These chemicals are known to be hazardous for human health and the environment (Saeedet al.2017;Memonet al.2019).More recently,bollworm has been largely controlled by the introduction of Bt cotton in the 2000s.Cotton genotypes containing theCry1Acgene initially controlled bollworm infestations without the application of chemicals (Spielmanet al.2017).However,the lepidopteran insects developed resistance against theCry1Acgene with the passage of time due to cultivation on a large scale without the presence of cultivated and natural refugia (Banerjeeet al.2017;Tabashnik and Carriere 2017).The variable expression (0.26-3.42 μg g-1) ofCry1Acis also a cause of resistance in lepidopteran insects (Jamilet al.2021b).
In the current study,the concentrations of bothCry1AcandCry2Agenes were not constant among all cotton genotypes.The concentration of theCry2Agene was approximately three to four times higher than theCry1Acgene.Plants show the maximum expression at the early vegetative stage in leaves and the minimum expression at maturity after a decline with advancing growth stages(Bakhshet al.2010;Hussainet al.2012;Jamilet al.2021b).The results obtained here,which are also supported by another study,showed that theCry2Abgene has a higher level of expression than theCry1Acgene in double-gene Bt cotton,so it provides better protection against target pests due to cross-resistance(Shera and Arora 2016).Variable quantities of theCry1Acgene were observed at different growth stages in all-cotton genotypes,with high Bt toxin levels in leaves in comparison with bolls and flowers.Other studies also showed a decline in expression with the passage of time,and it became lower than the lethal level of 1.89 μg g-1(Kranthiet al.2005).In another study,the results showed average concentrations of 0.98 and 0.70 mg g-1in leaves,sampled at 70 and 120 DAS,respectively,along with 0.59 and 0.59 mg g-1in bolls sampled at 70 and 120 DAS in Punjab (Spielmanet al.2017).The factors responsible for the varying expression levels ofBtgenes include point of insertion,bases,promoter sequences,environment and genetic backgrounds (Guoet al.2001;Adamczyk and Meredith 2004;Cheemaet al.2016).The triple-gene Bt cotton,which has theCry2AandVip3Aagenes in addition toCry1Ac,is economical and environmentally friendly for the control lepidopteran pests (Jamilet al.2021b).These genes have different expression levels due to different promoters and sites of insertion,so they play a healthy role in controlling lepidopteran pests by delaying resistance (Cheemaet al.2016).
The genotypes used in this study showed variable expression forCry1Ac,while the expression ofCry2Awas more stable especially at 30 DAS (Figs.2-7).In this study,the crosses were made between Bt and non-Bt parents and theCry2Agene was present only at one locus in hemizygous form in the F1hybrids.TheCry2Agene was fully expressed as a dominant gene as in the parents,where it is present in a homozygous condition.There were no alternate genes to affect the expression of that gene,which is why the gene had the same expression levels in parents and F1hybrid/crosses.At 30 DAS,theBtgene showed the maximum expression and there were only very minute differences in different plants from different replications.As a result,only a very minute error was observed,which was not measurable.Variable expression was found for these transgenes in different genotypes at 70 and 110 DAS and in different plant parts (Ahmadet al.2019;Jamilet al.2021b),which necessitates the careful breeding of transgenic cotton cultivars.CKC-1,CKC-2 and their crosses with non-Bt genotypes in different combinations had different concentrations,and the concentrations ranged between 0.16 and 3.38 μg g-1for theCry1Acgene and between 6.24 and 8.39 μg g-1forCry2Ain different plant parts(leaves,bolls,and flowers) during different growth stages(30,70 and 110 DAS).The results of the current study are supported by another experiment,which showed variable levels ofCrygene expression in different plant parts with high expression in leaves followed by lower expression in developing boll,flower,seed,and rind of boll (Nagappa and Khadi 2018).
The genotypes had higher concentrations of theCry1Ac(3.12 μg g-1) andCry2A(8.31 μg g-1) genes in the upper canopy while lower concentrations (2.66 μg g-1ofCry1Ac,8.09 μg g-1ofCry2A) were observed in leaves of the lower canopy at 70 DAS.Similarly,at 110 DAS,the concentration levels ofCry1AcandCry2Ain the upper and lower canopy leaves were 1.52 and 7.92 μg g-1,and 0.99 and 7.54 μg g-1,respectively.These results are supported by Ahmadet al.(2019),in which they reported variations in the concentrations of theCry1Acgene ranging from 0.97 to 2.73 μg g-1in upper leaves,0.23 to 0.89 μg g-1in middle leaves and 0.10 to 0.21 μg g-1in lower leaves,and the larval survival rate was correlated negatively withCry1Acconcentration.
Different hybrids responded differently regarding their expression of theCry1AcandCry2Agenes.When a Bt cotton genotype was used as male parent and crossed with a non-Bt genotype,then the F1hybrid shared a different genetic makeup.Meanwhile,the same Bt parent shared a different genetic makeup in the F1hybrid when it was used as the female parent.This difference explains why the expression varied in reciprocal crosses which had the same parental genotypes because of the sharing of different genetic backgrounds.In this study,the sharing of genetic material from the male Bt parent was responsible for high gene expression in comparison with the same genotype as a female parent.A similar phenomenon has been reported by Wuet al.(2003) and Wanet al.(2005).Kranthiet al.(2005) also found that different genotypes have significant differences in gene expression,with the maximum and minimum expression levels at the early and late stages of plant growth,respectively (Kranthiet al.2005).All of these results show that the genetic background of the plant plays an important role in consistent transgene expression,which would provide better control over the target pests (Adamczyk and Meredith 2004;Khanet al.2018).Therefore,genotypes with better and consistent gene expression should be selected and used in ongoing breeding programs.
5.Conclusion
The current study was designed to evaluate the spatiotemporal expression profiling of transgenesCry1AcandCry2Ain local climatic conditions for varied genetic backgrounds of cotton.The spatio-temporal expression levels of bothCry1AandCry2Agenes were measured among 10 cotton genotypes (two parents and eight F1hybrids) in leaves,flowers and bolls at different stages during the growth period.The genotypes CKC-2 and cross FH-942×CKC-2 expressed high concentration levels throughout the growing season when compared to the other genotypes.They had maximum expression levels in leaves as compared to bolls and flowers for both genes.The different patterns of expression for the same set of genes in reciprocal crosses with the same parentage might be due to maternal and paternal effects.
Acknowledgements
We are grateful to Rahil Shahzad from Agricultural Biotechnology Research Institute,Faisalabad,Pakistan for providing technical support in the improvement of the manuscript.We are also thankful to Shakeel Ahmad,Maize Research Station,Faisalabad and China National Rice Research Institute,Hangzhou,China and Muhammad Ikram,Huazhong Agricultural University,Wuhan,China for helping in the preparation of illustrations and technical support for the improvement of the manuscript.The authors are further thankful to Higher Education Commission,Pakistan for providing funds,Center of Excellence in Molecular Biology,Lahore and University of Agriculture,Faisalabad,Pakistan for their resources,support and encouragement in this study.
Declaration of competing interest
The authors declare that they have no conflict of interest.
 Journal of Integrative Agriculture2022年10期
Journal of Integrative Agriculture2022年10期
- Journal of Integrative Agriculture的其它文章
- Responses of leaf stomatal and mesophyll conductance to abiotic stress factors
- Genetic dissection of the grain-filling rate and related traits through linkage analysis and genome-wide association study in bread wheat
- Linkage and association mapping of wild soybean (Glycine soja)seeds germinating under salt stress
- Genome-wide identification,expression and functional analysis of sugar transporters in sorghum (Sorghum bicolor L.)
- lnhibition of miR397 by STTM technology to increase sweetpotato resistance to SPVD
- Comprehensive analysis of YABBY gene family in foxtail millet(Setaria italica) and functional characterization of SiDL
