Common polymorphisms of protein tyrosine phosphate non-receptor type 2 gene are not associated with risk of Crohn’s disease in lndian
Kaushik Chatterjee, Amit Kumar Dutta, Ashish Goel, Rekha Aaron, Vijayalekshmi Balakrishnan, Ajith Thomas,Anoop John, Rajeeb Jaleel, Deepu David, Reuben Thomas Kurien, SD Chowdhury, Ebby George Simon, AJ Joseph, Prasanna Premkumar, Anna B Pulimood
Kaushik Chatterjee, Amit Kumar Dutta, Ashish Goel, Rekha Aaron, Vijayalekshmi Balakrishnan,Ajith Thomas, Anoop John, Rajeeb Jaleel, SD Chowdhury, Anna B Pulimood, Department of Gastrointestinal Sciences, Christian Medical College and Hospital, Vellore 632004, Tamil Nadu, India
Deepu David, Reuben Thomas Kurien, Ebby George Simon, AJ Joseph, Department of Gastroenterology, Christian Medical College, Vellore 632004, Tamil Nadu, India
Prasanna Premkumar, Departments of Biostatistics, Christian Medical College, Vellore 632004,Tamil Nadu, India
Abstract BACKGROUND Multiple genetic risk factors for Crohn’s disease (CD) have been identified.However, these observations are not consistent across different populations. The protein tyrosine phosphate non-receptor type 2 (PTPN2) gene plays a role in various aspects of host defense including epithelial barrier function, autophagy,and innate and adaptive immune response. Two common polymorphisms in the PTPN2 gene (rs2542151 and rs7234029) have been associated with risk of CD in Western countries.AIM To evaluate the association of PTPN2 gene polymorphisms with risk of CD in Indian population.METHODS We conducted a prospective case-control study. Patients with CD were recruited,and their clinical and investigation details were noted. Controls were patients without organic gastrointestinal disease or other comorbid illnesses. Two common polymorphisms in the PTPN2 gene (rs2542151 and rs7234029) were assessed. DNA was extracted from peripheral blood samples of cases and controls and target DNA was amplified using specific sets of primers. The amplified fragments were digested with restriction enzymes and the presence of polymorphism was detected by restriction fragment length polymorphism. The frequency of alleles was determined. The frequencies of genotypes and alleles were compared between cases and controls to look for significant differences.RESULTS A total of 108 patients with CD (mean age 37.5 ± 12.7 years, females 42.6%) and 100 controls (mean age 39.9 ± 13.5 years, females 37%) were recruited. For the single nucleotide polymorphism (SNP)rs7234029, the overall frequency of G variant genotype (AG or GG) was noted to be significantly lower in the cases compared to controls (35.2% vs 50%, P = 0.05). For the SNP rs2542151, the overall frequency of G variant genotype (GT or GG) was noted to be similar in cases compared to controls (43.6% vs 47%, P = 0.73). There were no significant differences in minor allele (G)frequency for both polymorphisms between the cases and controls. Both the SNPs had no significant association with age of onset of illness, gender, disease location, disease behaviour,perianal disease, or extraintestinal manifestations of CD.CONCLUSION Unlike observation form the West, polymorphisms in the PTPN2 gene (rs7234029 and rs2542151)are not associated with an increased risk of developing CD in Indian patients.
Key Words: PTPN2 gene; Crohn’s disease; Genetic polymorphism; Case-control study; Asia; Risk factor
lNTRODUCTlON
Crohn’s disease (CD) is a chronic inflammatory disorder of the gastrointestinal tract with the peak age of onset in the third and fourth decades of life[1]. The frequency of CD is increasing in several Asian countries including India[1,2]. Environmental factors, gut microbiota, and host genetic factors are considered to be the key players in the pathogenesis of CD resulting in dysregulated immune response.Research on genetic factors has made significant contribution in understanding the pathogenesis of CD[3,4]. These include defective innate immunity and intracellular bacterial killing (CARD15/NOD2, IL23R,and LRRK2 genes), defective autophagy (ATG16L1 and IRGM genes), and dysregulated adaptive immune responses, namely, the interleukin-23 (IL-23) and T helper 17 (Th17) cell pathway (IL23R,IL12B, STAT3, JAK2, and TYK2 genes)[4]. Some of the genetic alterations identified in CD are not observed uniformly across different populations[5]. For example, the mutation in the NOD2/CARD15 gene present in the Western population with CD was not detected in Indian patients with CD[6]. Hence,it is important to investigate the presence of known genetic defects in different populations to understand their contribution to the pathogenesis of disease in that group.
Protein tyrosine phosphate non-receptor type 2 (PTPN2), also known as T-cell protein tyrosine phosphatase, is a cytosolic tyrosine phosphatase and is almost ubiquitously expressed in embryonic and adult tissues[7,8]. It has an N-terminal phosphatase domain and a nuclear localization sequence. It can dephosphorylate targets in both the cytosol and nucleus. PTPN2 has two variants arising out of alternate splicing[8]. The larger 48 kD form has a C-terminal hydrophobic domain that masks the nuclear localisation sequence and it remains attached to the endoplasmic reticulum[9]. The small 45 kD form does not have any hydrophobic sequence and can help the protein translocate to the nucleus.PTPN2 has been shown to affect various aspects of host defense including epithelial barrier function,autophagy, and innate and adaptive immune response[7,10-13]. Several studies have shown an association between polymorphism in the PTPN2 gene and CD. In a meta-analysis, two single nucleotide polymorphisms (SNPs) in the PTPN2 gene, rs7234029 (odds ratio [OR] = 1.36, 95%confidence interval [CI]: 1.16-1.59) and rs2542151 (OR = 1.22, 95%CI: 1.15-1.3), have been shown to increase the risk of CD. We aimed to study the association of these two SNPs in the PTPN2 gene(rs7234029 and rs2542151) with risk of CD in an Asian country (India) which has a total estimated burden of inflammatory bowel disease of about 1.4 million persons (highest in Asia)[2].
MATERlALS AND METHODS
We conducted a prospective case-control study to determine the association of polymorphisms in the PTPN2 gene with risk of CD. Adult patients (age > 18 years) with a diagnosis of CD were the cases. The diagnosis of CD was based on a combination of clinical, endoscopic, histological and radiological features as suggested by the Asia Pacific consensus criteria[14]. Adult subjects (age > 18 years) with dyspeptic symptoms and unrelated to cases were screened for inclusion as controls. Those with normal upper gastrointestinal endoscopy, normal haemoglobin, normal blood sugar, normal liver and renal function tests, and absence of significant comorbid illnesses were recruited as controls. The age and gender distribution of controls were kept similar to those of cases. Cases and controls were recruited after obtaining written informed consent. Patients were recruited from 2016 to 2018. The study was approved by the local institute review board and ethics committee.
As there were no previous studies from India on the SNPs in PTPN2, we could not make assumptions for sample size calculation and planned to recruit about 100 cases and controls each. The clinical,demographic, and investigation details of the cases were recorded in a predesigned form. These included age at the diagnosis of CD, extent of disease, disease behaviour, presence of extra-intestinal manifestations (EIM), and previous surgery for CD. The demographic details of controls were also recorded.
Blood samples were collected from the cases and controls and were stored at -80 °C till analysis.Genomic DNA was extracted from leukocytes using phenol-chloroform method and target DNA fragments were amplified by polymerase chain reaction (PCR) using specific primers (rs2542151:forward, 5’-TGCTGTGCTGCGTGAGTT-3’ and reverse, 5’-CACCATTGAGCGAAGTCC-3’; rs7234029,forward, 5’-GGCAGTGCTGAAACGAGA-3’ and reverse, 5’-TCCCACCACCTACCTACGG-3’). The steps of PCR included initial denaturation at 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 35 s,annealing at 58 °C for 30 s, and extension at 72 °C for 35 s, and final extension at 72 °C for 7 min. The PCR products were electrophoresed on a 2% agarose gel with TBE buffer for 45 min and visualized under a gel imaging system (Bio-Rad Gel Doc-2000, United States). The PCR products (5 μL) were digested by the appropriate restriction enzyme (Bsp1286I for rs2542151 and Hpy188I for rs7234029) for about 10 h, followed by electrophoresis on a 2.5% agarose gel. The digested product was gel documented and results were analysed and reported.
Statistical analysis
Continuous variables are summarised as the mean ± standard deviation or median with range and categorical variables as percentages. Categorical variables were compared using the chi-square test and continuous variables using independent t-test. A P value ≤ 0.05 was considered significant. Presence of Hardy-Weinberg equilibrium was assessed for both the SNPs in cases and controls. Data analyses were done using statistical software package SPSS v13.
RESULTS
We recruited 108 cases of CD and 100 control subjects during the study period. Table 1 shows the baseline characteristics of the patients with CD in this study. Their mean age was 37.5 ± 12.7 years and 42.6% were females. One patient with ileocolonic disease had coexisting upper gastrointestinal involvement while upper gastrointestinal disease alone was noted in one case. EIM were present in 17.6% cases, among which arthropathy was most frequent (17 cases) followed by uveitis (3 cases) and primary sclerosing cholangitis (1 case). Two patients had more than one EIM. About one-third of patients had previous surgery for CD, which included bowel resection in 30 patients. History of appendectomy was noted in three patients. Family history of IBD was present in two cases. The mean age of the 100 control subjects was 39.9 ± 13.5 years and 37% were females. The age and gender distri-bution were not significantly different from those of cases (age, P = 0.19; sex, P = 0.41).

Table 1 Baseline characteristics of patients with Crohn’s disease
The frequencies of the two SNPs in the PTPN2 gene evaluated in this study among cases and controls are shown in Table 2. Figures 1 and 2 illustrate the examples of digestion pattern noted for polymorphisms in both the SNPs. Both the SNPs were in Hardy-Weinberg equilibrium in cases(rs7234029, P = 0.21; rs2542151, P = 0.65) as well as controls (rs7234029, P = 0.47; rs2542151, P = 0.42).Results for the PTPN2 SNP rs7234029 were obtained for all cases and 98 controls. In the remaining two controls, the results of laboratory test were not clear, which hence were not included in the analysis. For the SNP rs7234029, the wild type (AA) was noted in 64.8% of the cases and in 50% of the controls.Homozygous variant (GG) was noted in 7.4% and heterozygous variant (AG) in 27.8% of the cases. The overall frequency of G variant (AG or GG) was noted to be significantly lower in the cases compared to controls (35.2% vs 50%, P = 0.05). In addition to the genotype, we also compared the minor allele frequency between cases and controls (Table 3). The frequency of the minor allele (G) was 20.9 % in cases and 27.6% in controls. Although the minor allele was more common in controls, the difference was not significant statistically (P = 0.17).

Figure 1 Restriction digestion pattern for the single nucleotide polymorphism rs7234029.

Figure 2 Restriction digestion pattern for the single nucleotide polymorphism rs2542151.
Results for the PTPN2 SNP rs2542151 were obtained for 101 cases and all control subjects (Table 2).The results of laboratory tests were not clear in the remaining seven cases with CD, which hence were excluded from the analysis. For the SNP rs2542151, the wild type (TT) was detected in 56.4% of the cases and in 53% of the controls. Homozygous variant (GG) was seen in 4% and heterozygous variant (GT) in 39.6% of the cases. The overall frequency of G variant (GT or GG) was noted to be similar in cases compared to controls (43.6% vs 47%, P = 0.73). On comparing the alleles, the frequency of minor allele(G) was 23.8 % in cases and 25.5% in controls, which was not significantly different (P = 0.77, Table 3).
We evaluated the association of the two SNPs in the PTPN2 gene with patient and disease characteristics (Table 4). The PTPN2 SNP rs7234029 GG or GA genotype was more common in patients with perianal disease and less common in patients with disease onset after 40 years although the difference did not reach statistical significance. There was no association of this polymorphism with gender,disease behaviour, disease location, EIM, or requirement of surgery (Table 4). The PTPN2 SNP rs2542151 GT or GG genotype appeared to have a negative association with history of surgical intervention for CD. Among patients who underwent surgery for CD in past, 28.6% had the variant genotype (GT or GG) while 50% of patients without prior surgery had this variant and the difference was close to being statistically significant (P = 0.07). Other features like gender, age of onset of illness,disease behaviour, disease location, and perianal disease were not associated with the PTPN2 SNPrs2542151 (Table 4).
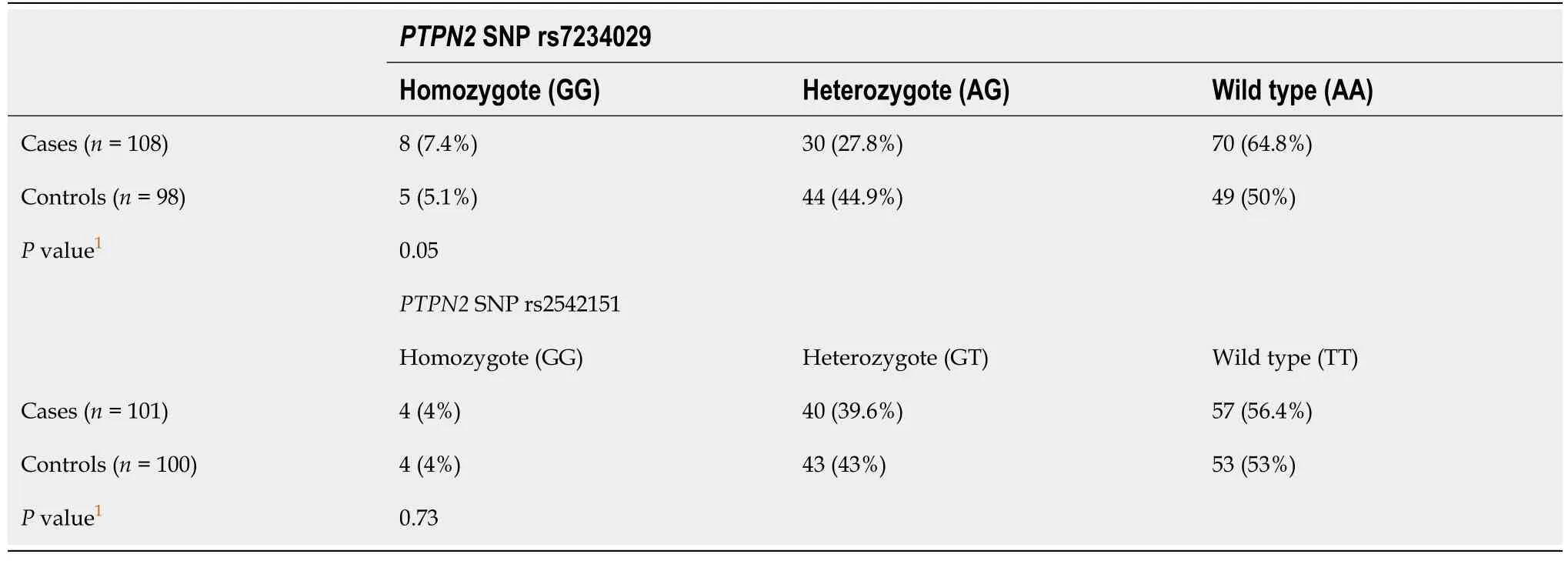
Table 2 Frequency of the single nucleotide polymorphisms in protein tyrosine phosphate non-receptor type 2 gene in cases and controls

Table 3 Minor allele frequency of the single nucleotide polymorphisms in protein tyrosine phosphate non-receptor type 2 gene in cases and controls

Table 4 Association of the single nucleotide polymorphisms in protein tyrosine phosphate non-receptor type 2 gene with patient and disease characteristics
DlSCUSSlON
The two common SNPs in the PTPN2 gene (rs2542151, rs7234029) were not associated with an increased risk of CD among Indian patients based on the observation made in the current study. In fact, the PTPN2 SNP rs7234029 was more frequent in control subjects compared to cases and hence had a negative association with CD. This highlights the heterogeneity of genetic risk factors for CD in different populations.
More than 240 genetic susceptibility loci have been identified for IBD[15]. Many of them are also associated with risk of other diseases. PTPN2 was first reported as a susceptibility gene for CD in the genome wide association studies (GWAS) by the Wellcome Trust Case Control Consortium[16]. The PTPN2 SNP rs2542151 was significantly associated with CD (trend P = 4.56 × 10-8, genotypic P = 2.03×10-7). Another polymorphism, the PTPN2 SNP rs7234029 (located in intronic region), was also subsequently found to be associated with CD. Polymorphisms in the PTPN2 gene have also been associated with rheumatoid arthritis, celiac disease, type II diabetes mellitus, etc.[7,17-19]. The PTPN2 gene affects several components of immune response and mice deficient in this gene develop severe systemic inflammatory illness[20]. They also show dysbiosis of intestinal microbiota[21, 22]. Patients with loss of function variant of PTPN2 demonstrate increased markers of Th1 and Th17 cell activation and reduced Treg cell activity[12,21]. PTPN2 is an important negative regulator of STAT1 and 3 activities and restricts TNF-alpha related release of inflammatory mediator[23]. It also affects epithelial barrier function[11,24]. Interestingly, administration of Tofacitinib (an inhibitor of Janus kinase), a drug approved for treatment of ulcerative colitis, was shown to correct the epithelial barrier defect induced by functional defect in the PTPN2 gene[25]. Observations in PTPN2 knockout mouse showed overexpression of cation-selective pore-forming molecule claudin-2, which allows para-cellular passage of molecules like sodium[24]. In vitro studies have also shown increased transcellular passage of macromolecules in PTPN2 deficient cells. PTPN2 in intestinal epithelial cells inhibits the expression of several autophagy-associated molecules, including beclin-1, ATG5, ATG7, ATG12, and ATG16L[26]. siRNA induced knockdown of the PTPN2 gene has also demonstrated the role of this gene in regulating autophagy[27].
After the initial report, several studies have subsequently assessed the frequency of polymorphisms in the PTPN2 gene in patients with CD[28]. A meta-analysis of data from these studies has shown an increased risk of CD associated with G variant in the PTPN2 SNP rs2542151[29]. Thirteen studies in this meta-analysis studied this variant in CD. Subjects with genotype GG or GT had an OR of 1.22 (95%CI:1.15-1.3, P < 0.001) of having CD compared to genotype TT. Subjects with G allele had an OR of 1.22(95%CI: 1.15-1.28, P < 0.001) of having CD compared to T allele. However, among studies from Asia,which included two studies from China and one from Japan, there was no significant association with CD at the genotype (P = 0.06) or allele level (P = 0.18)[29]. This is consistent with the observation made in our study where we did not find any significant association of CD with the PTPN2 SNP rs2542151.We found no significant difference either at the genotype (P = 0.73) or allele level (P = 0.77).
The above meta-analysis by Zhang et al also showed an increased risk of CD associated with G variant in the PTPN2 SNP rs7234029[29]. However, only two studies (one each from Japan and Germany) assessed this polymorphism. Genotype GG or AG was associated with an OR of 1.36 (95%CI:1.16-1.59, P < 0.001) of developing CD compared to AA genotype. A significant association was also noted at the allelic level and subjects with G allele had an OR of 1.33 (95%CI: 1.15-1.52, P < 0.001) of developing CD compared to A allele[29]. This finding could not be replicated in our study. There was no significant difference in the frequency of G allele between cases and controls (P = 0.17). Interestingly,the variant genotype (GG or GT) was more common in controls compared to cases (P = 0.05). The differences in genetic susceptibility loci between populations are not unexpected and have been noted for several other genes as well[5]. The phenotype of CD shows some variation between Western and Asian countries and the genetic differences may contribute to this in addition to environmental factors.
In addition to susceptibility to disease, polymorphisms in PTPN2 have also been linked with disease phenotype and response to therapy. In a study from New Zealand with 315 cases with CD and 481 controls, the PTPN2 SNP rs2542151 was associated with penetrating disease behaviour, need for bowel resection, late age at first diagnosis, and smoking[30]. However, our observations suggest a negative association of the PTPN2 SNP rs2542151 GT or GG genotype with history of surgical intervention for CD. Other characteristics of the disease were not affected by this variant in our patients. Van der Heid et al observed that the PTPN2 SNP rs2542151 increases the risk of CD only in smokers[31]. Another study from Germany revealed an association of the PTPN2 SNP rs7234029 with stricturing disease[9]. We found the PTPN2 SNP rs7234029 GG or GA genotype to be more frequent in patients with perianal disease and in those with onset of CD before the age of 40 years, although the difference was not significant statistically. A recent study by Hoffman et al[32] showed reduced response to anti IL-12/23 therapy in CD patients with G allele of the PTPN2 SNP rs7234029 compared to A allele (67.6% vs 89.9%,P = 0.005). As multiple factors may affect the disease phenotype and behavior, association with SNPs needs to be interpreted with caution. This may also explain the variability of effects in different studies and causality assessment would require a GWAS study with adjustment for other factors.
Our study evaluated two well-known mutations in the PTPN2 gene but there may be mutations in other parts of the gene which would require sequencing of the entire gene. This is one of the limitations of this study. However, our aim was to evaluate the previously known mutations and hence this did not affect our study objective.
CONCLUSlON
In conclusion, we did not find a positive association of variants in the PTPN2 SNP rs2542151 and rs7234029 with risk of CD in Indian patients. As PTPN2 has several effects on immune function, whole gene sequencing studies may provide an insight on whether variations at other sites in this gene are associated with risk of CD in this population.
ARTlCLE HlGHLlGHTS
Research background
The frequency of Crohn’s disease (CD) has been increasing in several Asian countries. Although its exact pathogenesis is still being elucidated, host genetic, gut microbiota, and environmental factors are key players involved. Research on genetic factors have provided valuable insight into the pathogenesis of the disease. However, some of the genetic abnormalities identified are not consistently seen across different populations and observations from one region cannot be extrapolated to other regions.
Research motivation
Protein tyrosine phosphate non-receptor type 2 (PTPN2) plays an important role in autophagy, innate and adaptive immune response, and maintaining epithelial barrier function. Single nucleotide polymorphisms (SNP) in the PTPN2 gene have been associated with an increased risk of CD. However,this needs to be confirmed in different populations.
Research objectives
Two SNPs in the PTPN2 gene (rs7234029 and rs2542151) have been associated with risk of CD. The objective of the current study was to assess the association of these two polymorphisms with CD in a large Asian country.
Research methods
A prospective case-control study was conducted where cases were patients with CD. Two SNPs in the PTPN2 gene (rs2542151 and rs7234029) were assessed using restriction fragment length polymorphism.The frequencies of polymorphisms between cases and controls were compared.
Research results
The study included 108 patients with CD and 100 controls. The two SNPs in the PTPN2 gene were not associated with an increased risk of CD. In addition, no association was observed between the two SNPs and disease characteristics.
Research conclusions
The current study did not show an increased risk of CD with polymorphisms in the PTPN2 gene contrary to observations in Western population.
Research perspectives
This study reemphasizes on the heterogeneity of genetic risk factors for CD across different population and the need to evaluate them in different populations.
FOOTNOTES
Author contributions:Chatterjee K recruited the patients, collected the samples, and critically revised the manuscript;Dutta AK designed the study, analyzed the data, and wrote the manuscript; Balakrishnan V and Aaron R carried out laboratory analysis and critically revised the manuscript ; Samuel P helped with study design, data analysis, and critical revision; Goel A, Thomas A, John A, Jaleel R, David D, Kurien RT, Chowdhury SD, Simon EG, Joseph AJ, and Pulimood AB helped with data collection and critical revision of the manuscript; all authors have read and approved the final manuscript.
Supported bythe CMC Vellore FLUID grant, No. 10360.
lnstitutional review board statement:The study was approved by the local institute review board and ethics committee (IRB Minute Number 10360, dated 3/11/2016).
lnformed consent statement:All study participants provided informed written consent prior to study enrollment.
Conflict-of-interest statement:None of the authors have any conflict of interest to declare.
Data sharing statement:All the data have been presented in the manuscript. There are no additional data.
STROBE statement:The authors have read the STROBE Statement-checklist of items, and the manuscript was prepared and revised according to the STROBE Statement-checklist of items.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:India
ORClD number:Kaushik Chatterjee 0000-0002-1983-8546; Amit Kumar Dutta 0000-0002-5111-7861; Ashish Goel 0000-0003-1659-2103; Rekha Aaron 0000-0002-6376-8071; Vijayalekshmi Balakrishnan 0000-0001-9398-4958; Ajith Thomas 0000-0002-9498-6792; Anoop John 0000-0001-5335-6658; Rajeeb Jaleel 0000-0002-6059-6139; Deepu David 0000-0001-9462-9202;Reuben Thomas Kurien 0000-0003-4456-816X; SD Chowdhury 0000-0001-6484-0218; Ebby George Simon 0000-0002-6328-2434; AJ Joseph 0000-0003-0099-5370; Prasanna Premkumar 0000-0002-7157-4138; Anna B Pulimood 0000-0003-0186-8584.
Corresponding Author's Membership in Professional Societies:Indian Society of Gastroenterology, LM 001733.
S-Editor:Wang LL
L-Editor:Wang TQ
P-Editor:Wang LL
