Expression Characteristics and Potential Function of Neuropeptide MIP in Larval Settlement of the Echiuran Worm Urechis unicinctus
LU Li, ZHANG Zhifeng, 2), ZHENG Qiaojun, CHEN Zongtao,BAI Shumiao, and ZHANG Zhengrui, *
Expression Characteristics and Potential Function of Neuropeptide MIP in Larval Settlement of the Echiuran Worm
LU Li1), ZHANG Zhifeng1), 2), ZHENG Qiaojun1), CHEN Zongtao3),BAI Shumiao1), and ZHANG Zhengrui1), *
1),,,266003,2),,,572000,3).,.,266003,
Larval settlement and metamorphosis are important developmental events in marine invertebrates. Some neuropeptides may play a key role in these processes. In this study, we revealed the expression characteristics of myoinhibitory peptide (MIP) in the embryos and larvae of the echiuran wormusing RT-qPCR,hybridization and immunofluorescence tech- niques, and found its location in the top cells of the upper hemisphere was conservative in larvae. Furthermore, we used the active MIP peptide to verify its role in inducing the larval settlement of, and confirmed that MIP can trigger an obvious larval settlement when 10μmolL−1or 15μmolL−1MIP was added in the seawater where early-segmentation larvae were cultured. Our finding indicated that neuropeptide MIP participates in the larval settlement in.
; neuropeptide MIP; larval settlement
1 Introduction
Most marine invertebrates have larval settlement and metamorphosis behaviors during their life cycles (Rodri- guez., 1993; Hadfield., 2001; Marshall., 2012). Larval settlement and metamorphosis involve many important morphological, physiological and biochemical changes (Marshall., 2012).They are critical stages determining the successful survival of species because more than 90% of larvae die during these processes (Chia, 1989; Gosselin., 1997). ‘Settlement’ is the process bywhich a planktonic larva moves toward the substratum, ex- plores and attaches to the substratum, and then begins its benthic life (Qian, 1999). For commercial aquatic animals,it is of great significance to their successfulbreeding by increasing the rate of larval settlement.
In pearl oysterand Pacific oys- ter, the receptors of acetylcholine, epi- nephrine, dopamine, histamine, 5-HT and neuropeptide Y were determined to play critical roles in larval settlement based on transcriptome analyses (Zheng., 2019). Tran- scriptomic analysis was also performed to determine the differentially expressed genes (DEGs) involved in settle- ment and metamorphosis between the pelagospheric larva and creeping larva ofThese DEGs were mainly related to the Notch signaling pathway, TNF sig- naling pathway, protein digestion and absorption, thyroid hormone signaling pathway, and ECM-receptor interaction, and so on (Cao., 2020). The proteomic study inrevealed that differentially expressed proteins reflected multiple processes may participate in larval me- tamorphosis, including cytoskeleton and cell adhesion, in- gestion and digestion, stress response and immunity (Song., 2016). A differentially expressed miRNA-mRNA correlation analysis inindicated that miRNAs participating in cell proliferation, migration, apoptosis, me- tabolic regulation, and energy absorption were also con- sidered to associate with larval metamorphosis (Song.,2017). Larval settlement and metamorphosis can be af- fected by multiple factors. For example,larval metamorphosis can be induced by different chemi- cal compounds such as epinephrine, dopamine, serotonin, KCl and MeOH (Yang., 2013). Neuropeptides have received widespread attention in recent years.
Neuropeptides are produced by the cleavage of inactive precursor protein peptides called prepropeptides (Hook., 2008), and are involved in many biological pro- cesses, such as reproduction, metabolism, feeding and cir- cadian rhythms (Willows., 1997;Katsukura., 2004;Yapici., 2008; Williams., 2015; Nagataand Nagasawa, 2017).In recent years, several studies have implicated the regulation of neuropeptides in larval settle- ment and metamorphosis of marine invertebrates (Grasso., 2011; Takahashiand Hatta, 2011; Joyce and Vogeler,2018). In the coral, neuropeptides LWamide and RFamide have been suggested to play key roles in the larval settlement and metamorphosis based on a persisting expression in the planula during settlement,then disappear shortly before attachmentand are undetect- able in the newly settled polyp (Attenborough., 2019).In the hydroidand several coral spe- cies, GLWamidehas also been demonstrated to involve in the larval settlement (Erwin., 2010;Lechable., 2019). In the gastropod, FMRFamide is located in the prototrochal ciliated cells, and significant- ly depressed the larval cilia beat frequency which is criti- cal for settlement behavior (Penniman., 2013). In the polychaete,neuropeptidesRYamide, FVMamide, DLamide, FMRFamide, FVamide, LYamide, YFamide, FLamide, GWamide and MIP (Myoinhibitory peptide) are related to the frequency of cilia swinging, and MIP has been verified to regulate the settlement of the lar- vae (Conzelmann., 2011). In the starfish,eight neuropeptide precursor genesare expressed in the attachment complex which comprises brachia and ad- hesive disk in brachiolaria larvae, and mediates the larval attachment (Mayorova., 2016).
Myoinhibitory peptide (MIP), also known as allatostatin- B or prothoracicostatic peptide, is characterized by a W- X6–8-Wamide motif (Predel., 2001; Poels., 2010;Szabo., 2011; Paluzzi., 2015; Tsukamoto and Nagata, 2016). MIP was first found to inhibit spontaneous contraction ofhindgut and oviduct, so it was named myoinhibitory peptide (Schoofs., 1991).MIP has been studied in some protostomian animals, but has never been identified in deuterostomia including hu- man so far (Peymen., 2019). In, MIPsuppress ecdysteroidogenesis in the prothoracic glands (Ya-manaka., 2010).MIP also regulates aspects of thebehavioral sequence underlying ecdysis inand, as part of a peptidergic signaling cascade initiated by ecdysis triggering hormone (Hua., 1999; Davis., 2003;Santos., 2007). Moreover, researchers revealed that MIP controls and mo- difies the salt chemotaxis behavior in(Peymen., 2019). In, MIP plays an important role in the larval settlement (Conzelmann., 2013), and the similar role is also suggested in the barna- cle(Yan., 2017).
belongs toEchiuroidea (Xenopneu- sta,Urechidae), which inhabits in a U-shaped burrow in coastal mud flats and is a commercial species in China, Korea and Japan.larvae present an obvious larval settlement process during the development. They settle on the bottom surface and crawl for several days,eventually burrow the sediment and metamorphose into the benthic worm (Ma., 2011; Hou., 2020). Hou. (2020) have suggested that several neuropeptide precur- sors may participate in the larval settlement ofbased on the larval transcriptome data. To verify the regulation of neuropeptide MIP on the larval settle- ment of, we synthesized an active MIP pep- tide and prepared its antibody in this study, and then re- vealed the MIP expression characteristics induring the development. Furthermore, the inducing effect of MIP onlarval settlement was analyzed by adding the active MIP peptide. Our aim is to provide basic data for studying the mechanism of neuropeptide-induced larval settlement.
2 Materials and Methods
2.1 Ethics Statement
The collection and handing of thewere performed in accordance with Institutional Animal Care and Use Committee of the Ocean University of China (OUC-IACUC) and local government.
2.2 Animals and Sample Preparation
The adultswith 15.7cm±1.4cm in body length were collected from the intertidal area in Yantai, Chi-na. Mature sperms and ova were obtained from the ne- phridia (gonoducts) of adults. Artificial insemination was implemented through mixing the sperms and ova with the ratio of 10:1 in filtered sea water (FSW). Then fertilized eggs were hatched in FSW (17℃±1℃, pH 7.7±0.08, and salinity 30±1). The larvae were reared at at room tem- perature (17–20℃), and fed with single-cell algae,andThe densities of the three species of algae were all 1×106indmL−1. The feeding ratio of them was 2:1:2. The col- lected embryos included zygote (30min), 2–8 cell embryo (1hpf), blastula (8hpf), and gastrula (12hpf). The collect- ed larvae included early-trochophore larva (24hpf), mid- trochophore larva (20dpf), late-trochophore larva (25dpf), early-segmentation larva (33dpf), segmentation larva (37dpf), and worm-shaped larva (45dpf). Parts of them were fixed in 4% paraformaldehyde for 16h, dehydrated with serial methanol (25%, 50%, 75% and 100%), and stored in 100% methanol at −30℃ for whole mounthybri- dization andimmunofluorescence. The others were frozen with liquid nitrogen immediately, and then stored at −80℃for total RNA extraction.
2.3 RNA Isolation and cDNA Synthesis
Total RNA from the embryos and larvae was isolated using MicroElute®Total RNA Kit (Omega, Norcross, USA). After removal of contaminant DNA with DNase I (TaKa- Ra, Dalian, China), purified RNA was quality-checked withagarose gel electrophoresis and quantified with spectropho- tometry. Then the cDNA was reversely transcribed from mRNA using PrimeScriptTMRT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China), and diluted using distilled water with a ratio of 1:5 for the subsequent experiments. Procedures were conducted by referring to the user ma- nual.
2.4 Quantitative Real-Time PCR (RT-qPCR)
ThecDNA sequence was obtained from our pre- vious RNA-seq data (Hou., 2020). A 158bp fragmentof(GenBank number: MT162087.1) was amplified using the cDNAs as templates from the embryos and larvae at different stages with the specific primers, qMIP-F and qMIP-R (Table 1), and(123bp fragment) was used as a reference gene (Wei., 2019). The RT-qPCR was performed following the manu- facturer’s instruction with SYBR Premix Ex TaqTM (Ta- KaRa, Dalian, China) and Light Cycler 480 system (Roche, Basel, Switzerland). Data were analyzed using the Roche Light Cycler 480 system software version 1.5 (Roche, Ba- sel, Switzerland), and the 2−ΔΔCtmethod was used to cal- culate the relative levels ofmRNA. All these reactionswere performed with three sample replicates and three tech- nical replicates.
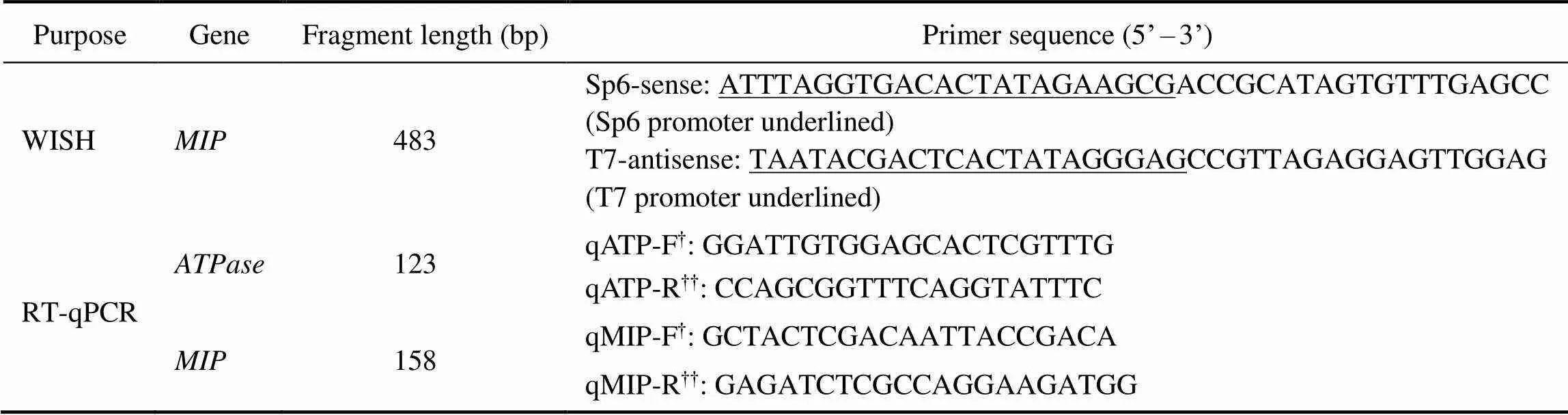
Table 1 Specific primers used in this study
Notes:†F represents the forward primer;††R represents the reverse primer.
2.5 Whole Mount in situ Hybridization (WISH)
The 1428bp ORF (open reading frame) sequence ofwas amplified with PCR using the late-tro-chophore cDNA and the specific primers. Specific primers, T7-antisense and Sp6-sense with T7 or Sp6 promoter se- quence were designed (Table 1) and the Digoxigenin-la- beled RNA probes were prepared using the DIG RNA Labeling Kit SP6/T7 (Roche, Basel, Switzerland). WISH was carried out as following: the larvae were rehydrated in a gradient methanol series (75%, 50%, and 25%) andphosphate buffer solution tween (PBT, PBS+0.1% Tween- 20) for 5min respectively, and treated with 100ngμL−1pro- teinase K for 20min (trochophores) or 400ngμL−1(seg- mentation larvae) for 30min to optimize hybridization, re-spectively. The pre-hybridization (50% formamide, 5×SSC,0.1% Tween, 9.2mmolL−1citric acid for adjustment to pH6.0, 50µgmL−1heparin, 500µgmL−1yeast RNA) was car- ried out for 3h at 60℃, then probes (1ngμL−1) were add- ed to the reaction system and incubated overnight at 60℃. After removing the excess probes by several rinses in hy- bridization buffer (2×SSC and 0.1molL−1maleic acid), the non-specific binding sites in larval cells were blocked using the blocking buffer (1.5g Blocking reagent, 19mL DEPC, 1mL EDTA (0.5molL−1)) and the samples were in-cubated with the Anti-DIG-AP antibody (Roche, Basel, Switzerland) for 16h at 4℃. Finally, the samples were stained in an NBT/BCIP staining solution (Roche, Basel, Switzerland) in dark for 1h, and then dehydrated with a gradient ethyl alcohol series (25%, 50%, 75%, and 100%) and cleared (ethanol:xylene=1:1, 5min; xylene twice, 5mineach time). The results were observed and photographedusing a Nikon E80i microscope (Nikon, Tokyo, Japan). Drawings and final panels were designed using Adobe Pho- toshop CC (San Jose, CA, USA).
2.6 Western Blotting
According to the characteristics ofMIP precursor sequence (Hou., 2020), the MIP1 sequence (KWGSNSMRVW) which shows the highest homology with the other mature MIP peptide sequences was select- ed to synthesize the MIP1 peptide by Sangon Biotech. Thenan active MIP1 peptide was obtained by coupling globulinto the naked MIP1 peptide, and then the rabbit-derived poly- clonal antibody of the active MIP1 was prepared by San- gon Biotech.
Total proteins from late-trochophore larvae, early-seg- mentation larvae, segmentation larvae were isolated using a protein extraction kit (Solarbio, Beijing, China). Each sample containing 20mg of total protein was loaded on 12% SDS-PAGE and transferred to a PVDF membrane (Millipore, MA, USA). The samples were blocked with QuickBlock™ Blocking Buffer for Western Blot (Beyo- time, Shanghai, China) overnight at 4℃ and then incubatedwith MIP1 antibody as primary antibody. The solutions were diluted at a ratio of 1:1000 in QuickBlock™ Primary An- tibody Dilution Buffer for Western Blot (Beyotime, Shang- hai, China) for 2h at room temperature. Afterwards, the membranes were washed three times in tris buffer solu- tion tween (TBST, TBS+0.1% Tween-20) and incubated with HRP-labelled goat antirabbit IgG (H+L) (Beyotime, Shanghai, China) at room temperature for 1h. Antigen- antibody complexes were detectedthe enhanced chemi- luminescence method (P0018S, Beyotime, Shanghai, Chi- na). Negative controls were performed by replacing the pri- mary antibody with a preimmune serum to check the anti- body specificity.
2.7 Immunofluorescence (IF)
The rabbit-derived polyclonal antibody of the active MIP1 was prepared by Sangon Biotech as described in Sec- tion 2.6.
The samples were rehydrated in the gradient methanol and incubated 1h in PBT containing 3% bovine serum al- bumin (BSA). Subsequently, they were transferred intoMIP1 antibody solutions which were diluted 1:2000 in BSA and incubated overnight at 4℃. After washed eight times in PBT, they were incubated with secondary fluoro-chrome-conjugated antibodies (donkey anti-rabbit Alexa Fluor 488, Invitrogen, Waltham, USA) in PBT for 2h. At last, the sam- ples were incubated in PBT containing 2.5% DAPI (4-6- diamidino-2-phenylindole) (Solarbio, Beijing, China) at room temperature for 15min in dark to label cell nucleiafter washed six times in PBT. Negative controls were per- formed by replacing the MIP1 antibody with the preim- mune serum to check the antibody specificity. The samples were analyzed with the confocal laser-scanning microscope Nikon A1RSi (Nikon, Tokyo, Japan).
2.8 The Effect of MIP on U. unicinctus Larval Settlement
In order to determine the appropriate time to add MIP peptide,we first tested water layer distribution oflarvae. Because the trochophore larvae mainly dis- tributed on the surface of the water, we employed the ear- ly-mid segmentation larvae to initiate the water layer dis- tribution observation. Fifty early-mid segmentation larvae were randomly drawn from the same batch, and then put them respectively in 50 glass test tubes (diameter=1.5cm) with 20cm height of sea water (19℃±1℃, pH 7.7±0.08, and salinity 30±1), and reared them withand renewed seawater one time per 12h. The spatial distribu- tion and development of each larva were observed and ta- ken picture every 12h. The height (H) of the larvae in sea- water distribution was measured, and the relative depth (RD) of larval distribution was calculated according to the formula RD=(H/20)%. This experiment lasted for 5d.
The early-segmentation larvae (33 days post fertiliza- tion, dpf) with an average body length of 380μm , which were floating in the upper layer (19℃±1℃, pH 7.7±0.08, and salinity 30±1), were employed in the larval settlement experiment. The experiment was carried out in the tubular container (diameter=1.5cm) with a depth of 20cm of sea- water. The three experimental groups were set with the final MIP concentrations of 5, 10, 15μmolL−1, respective- ly, and the control was seawater without MIP. Each tube contained 30 larvae. The water layer distribution of larvae was observed and taken picture every 2min. The relative depth of each larva was calculated according to the formula RD=(H/20)%, and the larval percentage in each water layer were counted according to the formula. Each experi- ment was kept 30min.
2.9 Statistical Analysis
All data were presented as means±standard deviation (SD) and the one-way ANOVA was employed to analyze the significant difference using SPSS software 18.0 (SPSS Inc., Chicago, IL, USA) with Tukey’s HSD test. Different letters (a, b, c,.) were marked to illustrate the statisti- cally significant differences (<0.05).
3 Results
3.1 Temporal Expression Characteristics of the MIP in U. unicinctus Larvae
The RT-qPCR showed that the expression of() was extremely low in the embryos at different stages (Fig.1).was first obviously ex- pressed in newly hatched larvae (the early-trochophore lar- va), and then the expression level increased significantly and reached a peak in segmentation larvae which was 5- fold or 2-fold higher than that in the early-trochophore lar- vae or early-segmentation larvae (Fig.1). At last, the ex- pression level ofin worm-shaped larvae which had bur- rowed in the sediment decreased significantly (Fig.1).

Fig.1 Expression of MIP in U. unicinctus embryos and lar- vae at different stages.Data are indicated as the mean±SD from three independent samples with three technical rep- licates. The expression level of MIP in early-trochophore larvae is set as 1.00 to calibrate the relative expression of embryos and other larvae. Different letters indicate signi- ficant differences (P<0.05) among the embryos and lar- vae at different stages. Z, zygote; C, 2–8 cellembryo; B, blastulae; G, gastrulae; ET, early-trochophore larvae; MT, mid-trochophore larvae; LT, late-trochophore larvae; ES, early-segmentation larvae; S, segmentation larvae; W, worm- shaped larvae.
3.2 Spatial Expression Characteristics of MIP in U. unicinctus Embryos and Larvae
The whole mounthybridization was performed for determining the location ofmRNA inembryos and larvae at different stages. As the RT-qPCRresults showed, the positive signal ofmRNA was also invisible in the embryos detected by WISH (Figs.2A–E). At the newly hatched early-trochophore larvae, the posi- tive signal was located in the top cells of the upper hemi- sphere (Fig.2G), and the expression characteristic was main- tained from early-trochophore larva to worm-shaped larva, although the number of positive cells varied among the lar- vae at different stages (Figs.2G–L). No positive signal withthe sense probe was detected in the embryos and larvae (Figs.2c, d, e, g, k, l).
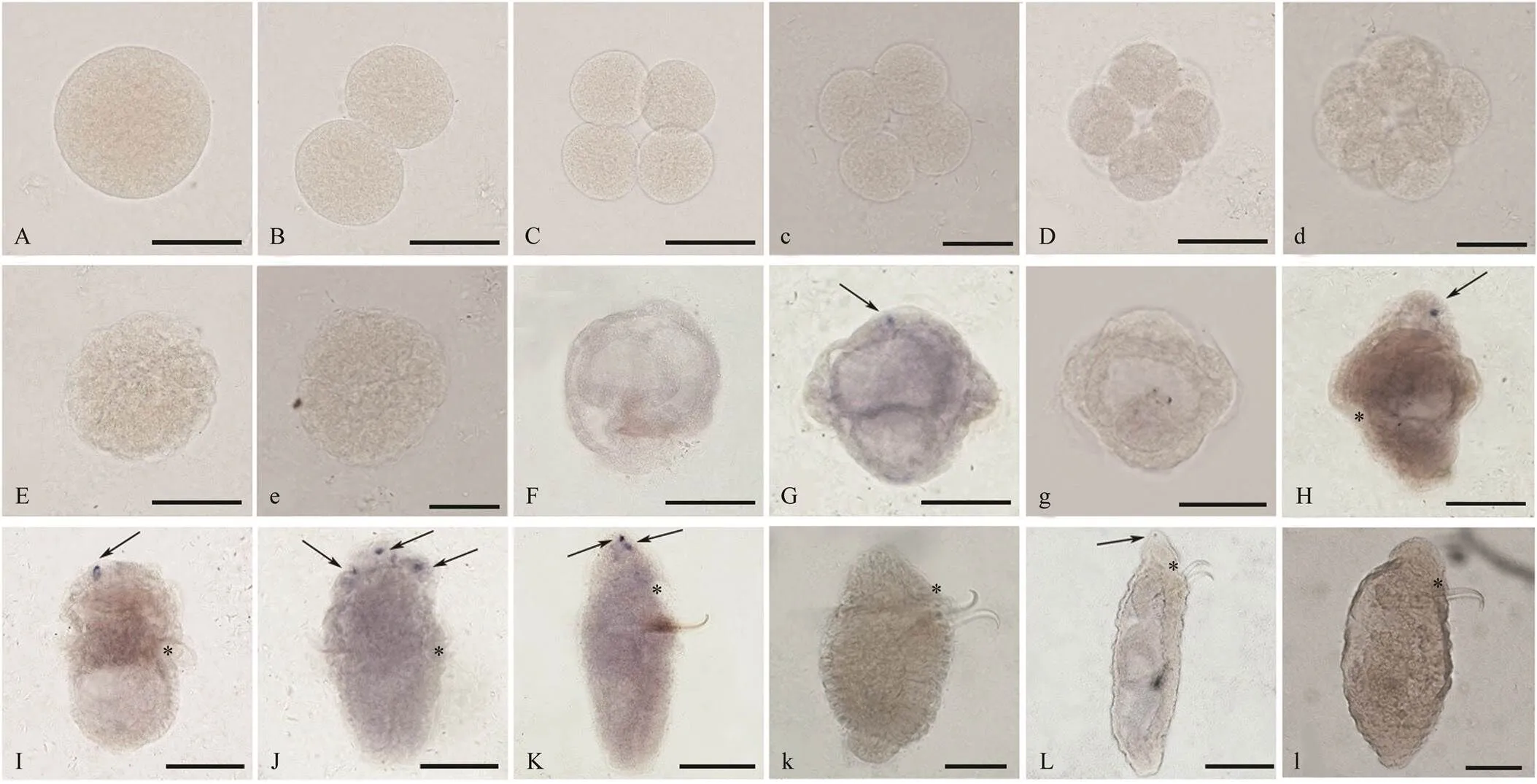
Fig.2 The location of MIP mRNA in U. unicinctus embryosandlarvae detected by whole mount in situ hybridization. A–L: the positive signals with MIP mRNA anti-sense probe are in blue; c, d, e, g, k, l: negative controls are showed with MIP mRNA sense probe. All larvae are in lateral view and oriented anterior up. A: zygote; B: 2-cell embryo; C, c: 4-cell embryo; D, d: 8-cell embryo; E, e: blastula; F: gastrula; G, g: early-trochophore larva; H: mid-trochophore larva; I: late- trochophore larva; J: early-segmentation larva; K, k: segmentation larva; L, l: worm-shaped larva. *, mouth; arrow, the positive signal. Scale bars, 100μm.
3.3 Specific Detection of MIP1 Antibody by Western Blotting
The specificity of the MIP1 antibody was detected by Western blotting (Fig.3A). A distinct main band was pre- sented, whose molecular mass was consistent with the theo- retical prediction of MIP precursor protein (55kDa). No band was presented with the preimmune serum (Fig.3B).

Fig.3 Specificity of MIP1 antibody detected by Western blotting. A, samples incubated with MIP1 antibody; B, samples incubated with preimmune serum. M, marker; LT, late-trochophore larva; ES, early-segmentation larva; S, segmentation larva.
3.4 Expression Characteristics of MIP Precursor in U. unicinctus Larvae
Animmunofluorescence assay was performed to further determine the location of MIP peptide. The result showed that MIP positive signals were also detected inlarvae, which were located at the top of the upper he- misphere of the larvae at different stages (Figs.4A3, B3, C3, D3, E3, F3). One cell with the strong positive signal was visible in the larvae from early-trochophore to mid-trocho- phore, and number of the strong positive cells increased to 2 in the late-trochophore, early-segmentation larva and segmentation larva, and then one positive cell was show-ed in the worm-shaped larva. The result of immunofluo- rescence was consistent with that of WISH. No positive signal was detected in the negative controls (Figs.4d3, f3). In addition, the green signals appeared in chaeta of the lar-vae in the MIP1 antibody group and negative control (Figs.4 D3–F3, d3, f3), which should be the non-specific adsorp- tion of the secondary fluorochrome-conjugated antibody.
3.5 Water Layer Distribution of U. unicinctus Segmentation Larvae
Fig.5A presented the water layer distribution of 50 lar- vae during the experiment. The results showed that 78% larvae were found in the upper layer of water with a rela- tive depth of 0.76 within one day. Most larvae settled gra- dually toward the bottom with the larval development, and can be observed in each water layer. After one and a half to four days, the average relative depth of the larvae was 0.40. After four and half days, 97% larvae settled to the bottom, and the average relative depth was 0.14 (Fig.5A).

Fig.4 MIP in U. unicinctus larvae detected by fluorescent immunofluorescence assay with the MIP1 antibody. A3, B3, C3, D3, E3, F3: the positive signals with MIP1 antibody are in green; d3, f3: negative controls are showed with preimmune serum. A1–A3, B1–B3, E1–E3, F1–F3, f1–f3: lateral view; C1–C3, D1–D3, d1–d3:ventral view. The green signal shows the MIP peptide, and blue is DAPI positive signal to show the nucleus; A3–f3 are the merges between A1–f1 and A2–f2, respectively. A1–A3: early-trochophore larva; B1–B3: mid-trochophore larva; C1–C3: late-trochophore larva; D1–D3, d1–d3: early-segmentation larva; E1–E3: segmentation larva; F1–F3, f1–f3: worm-shaped larva. *, mouth; Scale bars, 100μm.

Fig.5 Water layer distribution of U. unicinctus segmentation larvae. A, larval settlement process, n=50; B, the water layer distribution of U. unicinctus segmentation larvae varies with developmental schedule, n=30. Each dot represents 1 larva; the horizontal line indicates the average relative depth of the larvae at a specific time. ES, early-segmentation larvae; E- MS, early-mid-segmentation larvae; MS, mid-segmentation larvae; LS, late-segmentation larvae.
Based on the larval morphological characteristics and the height in the water layer, we determined the water layer distribution of segmentation larvae at the different stages (Fig.5B). The early-segmentation larvae(35dpf) were main- ly distributed on the upper surface of water layer and the average relative depth was 0.81. Morphologically, the lar- vae had been segmented only in the ventral body wall of hyposphere. The early-mid segmentation larvae (36dpf) began to settle, but were still on the upper surface of wa- ter layer while the average relative depth was 0.79. The hy- posphere of larva had been elongated, and its segmented area was extended to both sides. The mid-segmentation larvae (37dpf) distributed in a wide range of the water layer while the average relative depth was 0.45. By this time, the larval segmentation had been completely formed. The late- segmentation larvae (38dpf) were mainly at the bottom of the water layer with average relative depth of 0.31. The epi- sphere of larvae with the most obvious segmentation was significantly degraded.
3.6 Function of MIP on the Settlement of U. unicinctusLarvae
Based on the water layer distribution of segmentation larvae, the early-segmentation larvae were used to study the MIP function in larval settlement. The results showed that the larvae in three MIP groups exhibited the obvious settlement behavior within 2min (Fig.6), and their rela- tive heights in the water layer were significantly lower thanthat in the control (Fig.6B). In 10μmolL−1and 15μmolL−1MIP groups, the larvae showed a continuous settlement be- havior, and the relative height of larvae in water layer did not show significant difference in the same group. How- ever, the larvae in 5μmolL−1group did not exhibit an ob- vious settlement behavior after the experiment for 2min, and the relative height of water layer gradually returned to the same level as the control group.

Fig.6 MIP triggers the larval settlement behavior in U. unicinctus. Thirty larvae were used in each group. In (A), each dot represents 1 larva; the horizontal line indicates the average relative depth of the larvae at a specific time. In (B), different letters indicate significant differences (P<0.05) among the control and three MIP groups.
4 Discussion
MIP belongs to an ancient neuropeptide family of Wa- mides, and has only been identified in protostomia so far. The expression characteristics ofprecursor gene have been revealed in several invertebrates during the early de- velopment. In,expression is weak in the embryos, but it becomes stronger in the larvae and lo- cated in the most anterior neurosecretory region of the de- veloping brain (Williamson., 2001).Inthe polychaetes,is first located specifically in the apical organ (a cluster of sensory neurons) at early larvae stage (20hpf), and then the number of thepositive cells increasesin the apical organ of the larvae at the settlement stage(Conzelmann., 2013). Thesimilar location of MIP in the larval apical organ is also observed in(Conzelmann., 2013).In the barnacle,() expression level increases significantly in the cyprid, a larva exploring appropriate substrate cues for settlement and metamorphosis using its antennae, and then significantly down-regulated in adult (Yan., 2017). In our study, thewas revealed to be first expressed in the early-trochophore larvae of, and the ex pression level increased significantly and reached a peak in the segmentation larvae (Fig.1).hybridization and immunofluorescence showedmRNA and MIP precur- sor were mainly located in the top cells of the upper hemis- phere in larvae (Fig.2, Fig.4).The location corresponded to nerve perikarya cells oflarvae (Hou., 2020). It can be observed from the above that the MIP ex- pression is generally up-regulated in larvae at the stage of settlement and metamorphosis, and the localization in the anterior or apical region of the larvae is conservative.
Conzelmann. (2013) demonstrated that MIP incan induce the settlement and exploring behavior on the substrate by arresting the ciliary beating of mid-me- tatrochophre larvae. Our data showed that neuropeptide MIP can trigger the larval settlementof(Fig.6), which is similar to that in the polychaetes. How- ever, it is different in the effective concentration of MIP and the treatment time of the larvae between the two spe- cies. In this study, MIP1 at the concentration of 10μmolL−1or 15μmolL−1can induce an obvious settlement of the ear- ly-segmentation larvae (33dpf), but the larvae in the 5μmolL−1group did not exhibit an obvious settlement be- havior (Fig.6). In, the effective MIP concentra- tion is lower, while 5μmolL−1MIP7 can arrest the ciliary beating of the mid-metatrochophore larvae (55–60hpf), and induce and trigger the exploring behavior on the substrate (Conzelmann., 2013). Schmidt(2018) reported that different MIP mature peptides inlarvae have different affinities with MIP receptor, and MIP7 has the highest affinity with the receptor. Therefore, it is pre- liminarily speculated that the difference in effective con- centration of MIP betweenandmay be caused by different affinities of MIP mature pep- tides, which needs to be verified by further experiments.
Moreover, we found that the early-segmentation larvae(33dpf) ofcould effectively sense the MIP signals for settlement (Fig.6), but the trochophore larvae ofcould not be induced to settle by the MIP signals even at the concentration of 20μmolL−1. Obviously,the larvae’s response to MIP are different betweenand, while the former is the mid-me- tatrochophore larvae 50–60hpf, and the latter is the early- segmentation larvae 33dpf. This might be due to the dif- ferent larval development processes of the two species. In, the entire floating process time of the larvae lasts for 6 days, and the larvae completes the settlement pro- cess for approximately 2 days (Fischer., 2010).But in, the planktonic life of the larvae lasts for at least half a month according to the different culture condi- tions, and completes the settlement process for approximate-ly 3 days. Additionally, thelarvae are non-feed- ing during the floating period (Fischer., 2010), thelarvae have already started to eat at the early-trochophore stage (24hpf). Therefore, the conditions of MIP inducing the larval settlement were species-specific.
In summary, our results demonstrate that neuropeptide MIP is specifically expressed at the top of upper hemisphereinlarvae, and it can initiate the settlement be- havior of early-segmentation larvae. The finding provides basic data for further exploring the molecular mechanism of neuropeptide regulating the larval settlement and meta- morphosis in Echiura, and also provides scientific data forthe establishment of regulation technology. Although we ex- tended the larvae in 10μmolL−1group rearing to 8h afterthe test and found that all larvae were alive, we did not study its potential long-term toxic effects of MIP on larval develop ment and survival. Further toxicity study should be carried out to better reveal the roles of MIP inlarvae.
Acknowledgements
This work was support by the Fundamental Research Funds for the Central Universities (Nos. 202064006, 2021 13045), the National Natural Science Foundation of Chi- na (Nos. 31572601, 42176122), the Key Research & Deve- lopment Project of Hainan Province (No. ZDYF2021XD NY180), and the Postdoctoral Applied Research Project of Qingdao.
Abbreviations
MIP: Myoinhibitory peptide;
dpf: days post fertilization;
hpf: hours post fertilization;
C: 2–8 cell embryo;
B: blastula;
G: gastrula;
ET: early-trochophore larva;
MT: mid-trochophore larva;
LT: late-trochophore larva;
ES: early-segmented larva;
S: segmented larva;
LS: late-segmented larva;
W: worm-shaped larva.
Attenborough, R. M., Hayward, D. C., Wiedemann, U., Forêt, S., Miller, D. J., and Ball, E. E., 2019. Expression of the neuro- peptides RFamide and LWamide during development of the co- ralin relation to settlement and metamor- phosis., 446(1): 56-67, DOI: 10.1016/j.ydbio.2018.11.022.
Cao, F., Zhong, R., Yang, C., Hao, R., Wang, Q., Liao, Y.,., 2020. Transcriptomic analysis of differentially expressed genes in the larval settlement and metamorphosis of peanut worm., 18: 100475, DOI: 10. 1016/j.aqrep.2020.100475.
Conzelmann, M., Offenburger, S. L., Asadulina, A., Keller, T., Münch, T. A., and Jékely, G., 2011. Neuropeptides regulate swimming depth of., 108(46): 1174-1183, DOI:10. 1073/pnas.1109085108.
Conzelmann, M., Williams, E. A., Tunaru, S., Randel, N., Shahi- di, R., Asadulina, A.,., 2013. Conserved MIP receptor-li- gand pair regulateslarval settlement., 110(20): 8224-8229, DOI: 10.1073/pnas.1220285110.
Davis, N. T., Blackburn, M. B., Golubeva, E. G., and Hildebrand, J. G., 2003. Localization of myoinhibitory peptide immunore- activity inand, with indications that the peptide has a role in molting and ecdysis., 206(9): 1449-1460, DOI:10.1242/jeb. 00234.
Erwin, P. M., and Szmant, A. M., 2010. Settlement induction ofplanulae by a GLW-amide neuropeptide., 29(4): 929-939,DOI:10.1007/s00338-010-0634-1.
Fischer, A. H., Henrich, T., and Arendt, D., 2010. The normal de- velopment of(Nereididae, Annelida)., 7(1): 31, DOI: 10.1186/1742-9994-7-31.
Gosselin, L. A., and Qian, P. Y., 1997. Juvenile mortality in ben- thic marine invertebrates., 146: 265-282, DOI:10.3354/meps146265.
Grasso, L. C., Negri, A. P., Foret, S., Saint, R., Hayward, D. C., Miller, D. J.,., 2011. The biology of coral metamorphosis: Molecular responses of larvae to inducers of settlement and metamorphosis., 353(2): 411-419, DOI:10.1016/j.ydbio.2011.02.010.
Hadfield, M. G., Carpizo-Ituarte, E. J., Del Carmen, K., and Ned- ved, B. T., 2001. Metamorphic competence, a major adaptive convergence in marine invertebrate larvae., 41(5): 1123-1131, DOI:10.1093/icb/41.5.1123.
Hook, V., Funkelstein, L., Lu, D., Bark, S., Wegrzyn, J., and Hwang, S. R., 2008. Proteases for processing proneuropep- tides into peptide neurotransmitters and hormones., 48(1): 393-423, DOI:10. 1146/annurev.pharmtox.48.113006.094812.
Hou, X., Qin, Z., Wei, M., Fu, Z., Liu, R., Lu, L.,., 2020. Identification of the neuropeptide precursor genes potentially involved in the larval settlement in the echiuran worm.,21 (1): 1-13, DOI:10.1186/s12864-020-07312-4.
Hua, Y. J., Tanaka, Y., Nakamura, K., Sakakibara, M., Nagata, S., and Kataoka, H., 1999. Identification of a prothoracicostatic peptide in the larval brain of the silkworm,., 274(44): 31169-31173, DOI: 10. 1074/jbc.274.44.31169.
Joyce, A., and Vogeler, S., 2018. Molluscan bivalve settlement and metamorphosis: Neuroendocrine inducers and morphoge- netic responses., 487: 64-82, DOI:10.1016/j.aqua culture.2018.01.002.
Katsukura, Y., Ando, H., David, C. N., Grimmelikhuijzen, C. J., and Sugiyama, T., 2004. Control of planula migration by LWa-mide and RFamide neuropeptides in., 207(11): 1803-1810, DOI: 10.1242/jeb.00974.
Lechable, M., Jan, A., Duchene, A., Uveira, J., Weissbourd, B., Gissat, L.,., 2020. An improved whole life cycle culture protocol for the hydrozoan genetic model., 9(11): bio.051268, DOI:10.1242/bio.051 268.
Ma, Y. B., Zhang, Z. F., Shao, M. Y., Kang, K. H., Tan, Z., and Li, J. L., 2011. Sulfide: Quinone oxidoreductase from echiuran worm., 13(1): 93-107, DOI:10.1007/s10126-010-9273-3.
Marshall, D. J., Krug, P. J., Kupriyanova, E. K., Byrne, M., and Emlet, R. B., 2012. The biogeography of marine invertebrate life histories.,43: 97-114, DOI:10.1146/annurev-ecolsys-102710-145004.
Mayorova, T. D., Tian, S., Cai, W., Semmens, D. C., Odekunle, E. A., Zandawala, M.,., 2016. Localization of neuropeptide gene expression in larvae of an echinoderm, the starfish., 10: 553, DOI:10. 3389/fnins.2016.00553.
Nagata, S., and Nagasawa, H., 2017. Calcitonin-like peptide hor-mone (CT/DH) in the frontal ganglia as a feeding regulatory peptide of the silkworm,., 98: 23-28, DOI:10.1016/j.peptides.2016.05.002.
Paluzzi, J. P. V., Haddad, A. S., Sedra, L., Orchard, I., and Lange, A. B., 2015. Functional characterization and expression ana- lysis of the myoinhibiting peptide receptor in the Chagas diseasevector,.,399: 143-153, DOI:10.1016/j.mce.2014.09.004.
Penniman, J. R., Doll, M. K., and Pires, A., 2013. Neural corre- lates of settlement in veliger larvae of the gastropod,., 132(1): 14-26, DOI:10.1111/ivb.12014.
Peymen, K., Watteyne, J., Borghgraef, C., Van Sinay, E., Beets, I., and Schoofs, L., 2019. Myoinhibitory peptide signaling mo- dulates aversive gustatory learning in., 15(2): e1007945, DOI:10.1371/journal.pgen. 1007945.
Poels, J., Van Loy, T., Vandersmissen, H. P., Van Hiel, B., Van Soest, S., Nachman, R. J.,., 2010. Myoinhibiting peptides are the ancestral ligands of the promiscuoussex peptide receptor., 67(20): 3511-3522, DOI:10.1007/s00018-010-0393-8.
Predel, R., Rapus, J., and Eckert, M., 2001. Myoinhibitory neu- ropeptides in the., 22(2): 199-208, DOI:10.1016/S0196-9781(00)00383-1.
Qian, P. Y., 1999. Larval settlement of polychaetes. In:... Dorresteijn, A. W. C.,and Westheide, W., eds., Springer, Dordrecht, 239-253, https:// doi.org/10.1007/978-94-017-2887-4_14.
Rodriguez, S. R., Ojeda, F. P., and Inestrosa, N. C., 1993. Settle- ment of benthic marine invertebrates., 97(2): 193-207, DOI:10.3354/meps097193.
Santos, J. G., Vömel, M., Struck, R., Homberg, U., Nässel, D. R., and Wegener, C., 2007. Neuroarchitecture of peptidergic sys- tems in the larval ventral ganglion of., 2(8): 695, DOI:10.1371/journal.pone.0001848.
Schmidt, A., Bauknecht, P., Williams, E. A., Augustinowski, K., Gründer, S., and Jékely, G., 2018. Dual signaling of Wamide myoinhibitory peptides through a peptide-gated channel and a GPCR in., 32 (10): 5338- 5349, DOI: 10.1096/fj.201800274R.
Schoofs, L., Holman, G. M., Hayes, T. K., Nachman, R. J., and De Loof, A., 1991. Isolation, identification and synthesis of locus- tamyoinhibiting peptide (LOM-MIP), a novel biologically ac- tive neuropeptide from., 36(1): 111-119, DOI:10.1016/0167-0115(91)90199-Q.
Song, H., Qi, L., Zhang, T., and Wang, H. Y., 2017. Understand- ing microRNA regulation involved in the metamorphosis of the veined rapa whelk ()., 7(12): 3999-4008, DOI:10.1534/g3.117.300210.
Song, H., Wang, H. Y., and Zhang, T., 2016. Comprehensive and quantitative proteomic analysis of metamorphosis-related pro- teins in the veined rapa whelk,., 17(6): 924, DOI:10.3390/ijms17060924.
Szabo, T. M., Chen, R., Goeritz, M. L., Maloney, R. T., Tang, L. S., Li, L.,., 2011. Distribution and physiological effects of B-type allatostatins (myoinhibitory peptides, MIPs) in the stomatogastric nervous system of the crab., 519(13): 2658-2676, DOI:10.1002/cne.22654.
Takahashi, T., and Hatta, M., 2011. The importance of GLWamide neuropeptides in cnidarian development and physiology.,2011 (5): 424501, DOI: 10.4061/2011/424501.
Tsukamoto, Y., and Nagata, S., 2016. Newly identified allatosta- tin Bs and their receptor in the two-spotted cricket,., 80: 25-31, DOI:10.1016/j.peptides.2016.03.015.
Wei, M., Lu, L., Wang, Q., Kong, D., Zhang, T., Qin, Z.,., 2019. Evaluation of suitable reference genes for normalization of RT-qPCR in echiura () during develop- mental process., 45(6): 464-469, DOI:10.1134/S1063074019300023.
Williams, E. A., Conzelmann, M., and Jékely, G., 2015. Myoin- hibitory peptide regulates feeding in the marine annelid., 12(1): 1-16,DOI:10.1186/s12983-014-0093-6.
Williamson, M., Lenz, C., Winther, M. E., Nässel, D. R., and Grim-melikhuijzen, C. J., 2001. Molecular cloning, genomic organi-zation, and expression of a B-type (cricket-type) allatostatin preprohormone from., 281(2): 544-550, DOI:10.1006/bbrc.2001.4402.
Willows, A. O., Pavlova, G. A., and Phillips, N. E., 1997. Modula-tion of ciliary beat frequency by neuropeptides from identifiedmolluscan neurons., 200(10):1433-1439.
Yamanaka, N., Hua, Y. J., Roller, L., Spalovská-Valachová, I., Mi-zoguchi, A., Kataoka, H.,., 2010.prothoracico- static peptides activate the sex peptide receptor to regulate ec- dysteroid biosynthesis., 107(5): 2060-2065, DOI:10.1073/pnas.090747- 1107.
Yan, G., Zhang, G., Huang, J., Lan, Y., Sun, J., Zeng, C.,., 2017.
Comparative transcriptomic analysis reveals candidate genes and pathways involved in larval settlement of the barnacle., 18(11): 2253, DOI:10.3390/ijms18112253.
Yang, J. L., Li, S. H., Li, Y. F., Liu, Z. W., Liang, X., Bao, W. Y.,., 2013. Effects of neuroactive compounds, ions and or- ganic solvents on larval metamorphosis of the mussel., 39(6): 106-112, DOI: 10.1016/j.aquaculture.2013.02.039.
Yapici, N., Kim, Y. J., Ribeiro, C., and Dickson, B. J., 2008. A receptor that mediates the post-mating switch inreproductive behaviour., 451(7174): 33-37,DOI:10.1038/nature06483.
Zheng, Z., Hao, R., Xiong, X., Jiao, Y., Deng, Y., and Du, X., 2019. Developmental characteristics of pearl oyster: Insight into key molecular events related to shell for- mation, settlement and metamorphosis., 20(1):1-11, DOI: 10.1186/s12864-019-5505-8.
December 21, 2020;
May 17, 2021;
June 22, 2021
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2022
. E-mail: zhangzhengrui@ouc.edu.cn
(Edited by Qiu Yantao)
 Journal of Ocean University of China2022年4期
Journal of Ocean University of China2022年4期
- Journal of Ocean University of China的其它文章
- The Role of Bottom Currents on the Morphological Development Around a Drowned Carbonate Platform, NW South China Sea
- Controls on the Gas Hydrate Occurrence in Lower Slope to Basin-Floor, Northeastern Bay of Bengal
- Effective Elastic Thickness of the Lithosphere in the Mariana Subduction Zone and Surrounding Regions and Its Implications for Their Tectonics
- Geophysical Evidence for Carbonate Platform Periphery Gravity Flows in the Xisha Islands, South China Sea
- Seismic Characteristics and Hydrocarbon Accumulation Associated with Mud Diapir Structures in a Superimposed Basin in the Southern South China Sea Margin
- Simulation and Analysis of Back Siltation in a Navigation Channel Using MIKE 21
