ldentifying potential flavonoid biosynthesis regulator in Zanthoxylum bungeanum Maxim.by genome-wide characterization of the MYB transcription factor gene family
WANG Xiang-yuan ,TlAN Lu ,FENG Shi-jing ,WEl An-zhi
1 College of Forestry,Northwest A&F University,Yangling 712100,P.R.China
2 Research Centre for Engineering and Technology of Zanthoxylum State Forestry Administration,Yangling 712100,P.R.China
3 College of Forestry,Guizhou University,Guiyang 550025,P.R.China
Abstract Plant MYB transcription factors (TFs) play crucial roles in regulating the biosynthesis of flavonoids but current analysis on their role in Zanthoxylum bungeanum Maxim.(ZBM) is far from comprehensive. In this study,we identified 270 MYB genes in ZBM and divided them into four subfamilies. The R2R3-MYB (ZbMYB) category contained 251 genes and was classified into 33 subfamilies according to their phylogenetic results and sequence similarity. These subfamilies included 24 subgroups containing both MYBs of ZBM plants and AtMYBs,and nine subgroups containing only ZBM MYBs or AtMYBs. ZbMYBs with similar functions clustered into the same subgroup,indicating functional conservation. The subcellular localization analysis predicted that most ZbMYB genes were found in the nucleus. The transposed duplications appeared to play a major role in the expansion of the MYB gene family in ZBM. Through phylogenetic analysis and transcriptome profiling,it was found that 28 ZbMYB genes may regulate the biosynthesis of flavonoids in ZBM,and these genes expression presented distinct temporal and spatial expression patterns. In different fruit development stages of ZBM,the expression patterns of EVM0042160 and EVM0033809 genes obtained by qRT-PCR analysis are very similar to the flavonoid and anthocyanin content curves in ZBM. Further correlation analysis showed that the content of flavonoids in different fruit development stages and the transcript abundance levels of 28 ZbMYB genes have different degrees of correlation relationship. These results indicated that the ZbMYB genes might be involved in the flavonoid metabolic pathway. This comprehensive and systematic analysis of MYB family genes provided a solid foundation for further functional analysis of MYB TFs in ZBM.
Keywords:Zanthoxylum bungeanum Maxim.,MYB transcription factor,expression pattern,regulation of flavonoid biosynthesis
1.lntroduction
Secondary metabolite is one of the most valuable and important biological activities for plant growth and development,and flavonoids are one of the largest groups of secondary metabolites (Pollastri and Tattini 2011;Xuet al.2015). As an important class of natural organic compounds,flavonoids can scavenge free radicals in the body to produce an antioxidant effect,which contributes to plant health,colors of plant tissues and food quality (Bueret al.2010;Cheynieret al.2013;Guoet al.2021).
Regulating flavonoid biosynthesis involves a complex network. Researchers found that flavonoid biosynthesis is usually regulated together by two types of genes,early biosynthetic genes (EBGs) and late biosynthetic genes (LBGs) (Moritaet al.2006;Nakatsukaet al.2012). The former is involved in the synthesis of flavonoids,α-essential oils and phenol;the latter participates in the biosynthesis of proanthocyanidins and anthocyanins (Gonzalezet al.2008). Most genes from the MYB family participate in the regulation of these two types of genes,and play an important role in regulating plant flavonoid (Owenset al.2008).
The MYB family,one of the largest transcription factor (TF) families,is well researched in plants. MYB proteins typically have two discrete regions:the N-terminus and the C-terminus. The N-terminal region has a conserved MYB domain,whereas the C-terminal region is variable,which leads to MYB proteins’ diverse functions (Strackeet al.2001;Zimmermannet al.2004). Generally,the MYB domain contains 1-4 serial and no redundant incomplete repeats (R) (Lipsick 1996). According to the number of repetitions,the MYB family is divided into four main categories:1R (related to MYB),2R (R2R3-MYB),3R (R1R2R3-MYB) and 4R-MYB (Jiang and Rao 2020). Each repeat contains about 50-53 amino acids and has three α-helices,forming a helix-turn-helix (HTH) structure between the second and third α-helices,which helps to identify specific short DNA sequence in binding (Duboset al.2010). MYB protein is extremely important for plants since it uses various methods to participate in primary and secondary metabolism,such as regulating the morphogenesis of cells,epidermal hair and petals in response to biotic and abiotic stresses (Molet al.1998;Wanget al.2009).
R2R3-MYB is plants’ most common and largest MYB TF (Jiang and Rao 2020). These genes are involved in almost every stage of plant growth and development. Importantly,R2R3-MYB plays a central role in regulating the transcriptional metabolism of flavonoid synthesis in plants (Medina-Pucheet al.2014). Many members of the R2R3-MYB have been recognized and studied in various plants,such asArabidopsis,corn,soybean,citrus,wheat,pineapple and strawberry (Jianget al.2004;Matuset al.2008;Ayaet al.2009;Daiet al.2012;Duet al.2012b;Zhanget al.2012;Liuet al.2017). For example,inArabidopsis,TT2specifically regulates the accumulation of proanthocyanidins in seed coats (Routaboulet al.2006). In addition,some genes such asAtMYB11,AtMYB12andAtMYB111are flavonol-specific transcriptional regulators,and deletion or mutation of these genes normally cannot synthesize flavonols. These genes also regulate the accumulation pattern of flavonoids inArabidopsis(Strackeet al.2007). Moreover,overexpression ofAtMYB114can up-regulate the synthesis of plant pigments (Routaboulet al.2006). The artificial interference with the genesAtMYB75,AtMYB90,AtMYB113andAtMYB114can lead to down-regulation of genes expression,and anthocyanins deficiency inArabidopsis(Gonzalezet al.2008). Similar results have also been found in other species. In peach trees,PpMYB10positively regulates the upstream promoters of the structural genesUFGTandDFRin the flavonoid biosynthetic pathway (Zhaoet al.2017;Zhanget al.2018). AppleMdMYB10can up-regulate theDFRgenes expression,thereby improving the anthocyanin synthesis in apple peel,pulp and leaves (Chagnéet al.2013). All these results highlight the importance of transcriptional regulations of MYB in the control of flavonoid biosynthesis. However,how MYB genes regulate flavonoid synthesis inZanthoxylumbungeanumMaxim.(ZBM) has never been reported.
ZBM has a bright red pericarp and is the most popular commercial product in theZanthoxylumgenus. It can be regarded as an emergent and important source of biomass energy,which highly valuable in various fields (Yanget al.2008). ZBM is rich in secondary metabolites that are not only basic condiment ingredients but also used in medicines as active ingredients for its warmth,analgesic,dampness,antidiarrheal and antipruritic pharmacological properties (Yanget al.2013;Zhanget al.2017). Flavonoids are important natural organic compounds with a high concentration in ZBM secondary metabolites (Ross and Kasum 2002;Leiet al.2021),but current research on flavonoids in ZBM mainly focuses on the extraction,separation and detection (Fuet al.2018;Maet al.2019). It was reported that MYB takes part in flavonoid biosynthesis by regulating genes expression related to flavonoid synthesis,which indirectly affects the content and characteristics of flavonoids in plants (Falconeet al.2012). However,the number of the MYB family members in different species greatly varies. Homologous genes in different species also show different structures and functions (Duet al.2012a;Caoet al.2013).
The present study aims to identify and explore the genes involved in regulating flavonoid biosynthesis in ZBM,and to understand the specific structural and functional characteristics of these genes for further research. We identified MYB genes throughout the ZBM genome and studied their gene structural characteristics and their evolutionary relationships. We used the transcript abundance data of tissue samples from different time and different locations. We further conducted qRTPCR for specific genes to examine the specific expression pattern of tissue and expression time of these genes.According to the comparison of flavonoid content in ZBM and taking into account the result of transcript abundance correlation analysis,MYB genes could be considered as an important index for regulating flavonoid biosynthesis.Therefore,this study provides a starting point for further analysis of the function of MYB genes in ZBM,and lays a solid foundation for selecting and utilizing candidate genes for various genetic engineering.
2.Materials and methods
2.1.Plant materials
The samples ofZ.bungeanumMaxim.‘Fengxiandahongpao’ were collected from an experiment station for Chinese pepper located in Shaanxi Province,China (106.66°E,33.98°W). For RNA-seq,fresh pericarps were collected from seven fruit development stages of 12 plants,including 10-,30-,40-,50-,60-,70-,and 80-days post anthesis (P10,P30,P40,P50,P60,P70,and P80). The leaves and stems were collected at 80 days post anthesis. There were three biological replicates for each sample during tissue collection. All samples were immediately frozen in liquid nitrogen and stored at −80°C until further analysis.
2.2.ldentification and classification of MYB proteins in ZBM
The Hidden Markov Model (HMM) profile of MYB DNAbinding domain (PF00249) was downloaded from Pfam protein families database (http://pfam.sanger.ac.uk/) (Batemanet al.2004) and was used to query all MYB sequences from our ownZ.bungeanum(ZBM) genome (Fenget al.2021) local database using HMMER 3.0 (http://hmmer.janelia.org/) (Finnet al.2011). The default parameters were employed,and the cutoff value was 0.01.Further,theArabidopsisprotein sequences of 168 MYB genes served as query sequences (Strackeet al.2001),which were downloaded from The Arabidopsis Information Resource (TAIR) (http://www.arabidopsis.org/) (Yanhuiet al.2006). The BLASTn program was applied to identify all MYB genes in the ZBM genome,using anE-value cutoff of 1.0×10−20(Altschulet al.1997). To ensure accuracy of the results,the PROSITE (http://prosite.expasy.org/scanprosite/) (de Castroet al.2006) and the SMART programs (http://smart.embl-heidelberg.de/) (Letunicet al.2004) were used to confirm the presence of MYB domain for further analysis. The protein sequences that did not contain the entire conserved domains were deleted. Since there are no reports of members of the MYB gene family in ZBM,the MYB genes were temporarily named according to their annotation of the ZBM genome. The data of the sequences analysis used in this study were submitted to NCBI and the accession numbers are BankIt2425051:MW558612-MW558881.
2.3.Sequence analysis
To analyze the sequence features of the 251 typical ZBM R2R3-MYB proteins,multiple sequence alignments were performed using ClustalX (Thompsonet al.1997) with default parameters. WEBLOGO (http://weblogo.berkeley.edu/logo.cgi) (Crookset al.2004) was used to indicate the features of the DNA-binding domains of MYB proteins. GSDS (http://gsds.cbi.pku.edu.cn/index.php) was used to sketch exon/intron organization of MYB genes. To investigate the conserved motifs of ZBM R2R3-MYB proteins,the complete amino acid sequences were subjected to MEME analysis (http://meme.nbcr.net/meme/intro.html) (Baileyet al.2009),with its optimal parameters set as the following:the maximum number of motifs was set to identify 10 motifs,and the optimal width of each motif was set from 10 to 130 residues. Wolf PSORT Software with default parameters was used to predict the subcellular localization of these MYB proteins. Furthermore,the basic characteristics of the potential MYB members in ZBM were analyzed,including the predicted proteins and the physicochemical parameters.The predicted molecular weights (MWs) and isoelectric points (pIs) of the MYB proteins were analyzed using the ExPASy proteomics website (http://web.expasy.org).
2.4.Analysis of chromosomal locations and synteny analysis for ZbMYB genes
The detailed chromosomal location information of eachZbMYBgene was obtained from ZBM genome documents. Then the data were mapped to the chromosomes of ZBM by Circos Software (Krzywinskiet al.2009). To identify WGD/segmental,tandem,proximal and dispersed duplication events in theZbMYBfamily,MCscanX Software with the default parameters was applied following the operation manual (Wanget al.2012a). The TBtools Software was adopted to exhibit the synteny relationship of the orthologousMYBgenes in ZBM.
2.5.Phylogenetic analysis and function prediction
To explore the evolutionary relationships among MYBs in ZBM and predict the functions of these MYBs,the MYB sequences ofArabidopsiswere used as the reference. A neighbor-joining (NJ) phylogenetic tree was constructed with ClustalW to align the full-length of MYB amino acid sequences (251ZbMYBsand 168AtMYBs) using MEGAX 10.0 (Larkinet al.2007). Poisson correction,pairwise deletion,and bootstrap analysis with 1 000 replicates were employed to estimate the parameters.Classification of the ZBM MYBs was performed according to their phylogenetic properties of the correspondingArabidopsisAtMYB proteins. Additionally,the biological functions of some MYB proteins can be predicted based on the aforementioned phylogenetic tree,which identified candidate MYB transcription factors that are likely to regulate flavonoid biosynthesis.
2.6.Expression analysis of ZbMYB genes
In order to study the expression pattern ofZbMYBgenes in three tissues (pericarp,leaves and stems) and seven fruit developmental stages in ZBM,the total RNA of the corresponding samples was collected for further RNAseq library construction. The transcript abundance ofZbMYBgenes was calculated as fragments per kilobase of exon model per million mapped reads (FPKM) (Trapnellet al.2010). Log2(FPKM+1) from RNA-seq data were hierarchically clustered using Cluster 3.0 and the results were graphed by TBtools.
2.7.Determination of total flavonoids and anthocyanin in different periods and tissues of ZBM
Total flavonoid content was determined according to the improved aluminum nitrate-sodium nitrite colorimetric method (Yanget al.2012;Chenet al.2017,2020).Powdered pericarp (0.25 g) was accurately weighed with three replications. A total of 10 mL of 70% ethanol solution was added in each replicate before it was ultrasonically extracted for 40 min,centrifuged at 3 000 r min-1,and the supernatant was collected. Rutin concentration C (x) and absorbance value A (y) were used as independent variables and dependent variables,respectively,to make a standard curve. The obtained equation was used to determine the absorbance value of extracts of different ZBM samples to calculate the flavonoid content. The anthocyanin content was determined using an improved method described by Neff and Chory (1998) and Solomonet al.(2006). The samples (0.25 g) were accurately weighed with three replications,and immersed in chilled 0.1% (v/v) methanolic HCl. The slurry was ultrasonically extracted for 30 min and centrifuged at 3 000 r min-1,and the supernatant was collected. The measurements of A510 and A700 were conducted with a spectrophotometer. By subtracting the A700 measurement from the A510 measurement,the amount of anthocyanin per sample was calculated (Sutharut and Sudarat 2012).
2.8.RNA isolation and quantitative RT-PCR analysis
In our study,samples (P10-P80) were collected at seven fruit development stages for qRT-PCR analysis.According to the manufacturer’s instructions,total RNA was extracted from the samples using RNAprep Pure Plant Kit (Polysaccharides &Polyphenolics-Rich,TIANGEN,Beijing,China). The HiScript II Q RT SuperMix for qPCR (+gDNA wiper) (Vazyme,Nanjing,China) was used for cDNA synthesis. The primer pairs for qRT-PCR were designed to amplify specific genes using Primer Premier 5.0 Software (Premier Biosoft International,Palo Alto,CA). qRT-PCR analysis was performed on the ABl Q6 Flex instrument using PerfectStartTMGreen qPCR SuperMix (TransGen,Beijing,China) according to the manufacturer’s protocol.
The PCR reaction conditions were as follows:2 min at 95°C,and then 40 cycles,95°C for 10 s,55°C for 10 s and 72°C for 30 s. Melting curve analysis confirmed that only one product was amplified and detected. The calculated average cycle threshold (Ct) of the sample was used to describe the difference in fold change in expression. ZBMactingene was used as the internal reference genes to normalize the expression levels of the target genes. The relative expression level was determined using the 2-ΔΔCtmethod (Livak and Schmittgen 2001). The results were graphed by OriginPro 9.1 (Seifertet al.2014).
2.9.Statistical analysis
Data collected from the three biological replicates were analyzed by the average and standard error. A correlation analysis was performed using SPSS Statistics (version 22.0;SPSS Inc.,Chicago,Illinois,USA) to assess the correlation between gene expression levels and the total flavonoid content and accumulation rate of ZBM at seven fruit development stages. The correlation analysis provides the following default results:correlate bivariate correlations,correlation coefficients,Pearson correlation,test of significance,two-tailed,and flag significant correlations.
3.Results
3.1.ldentification and sequence analysis of ZBM MYB genes
The amino acid sequence of the HMM profile of the Pfam MYB domain (PF00249) and the BLAST search were used to identify MYB genes in the ZBM genome,usingArabidopsisMYB protein as the query sequence. More than 300 deduced amino acid sequences that contain MYB or MYB-like repeats were obtained. We then used InterProScan Program to confirm the presence of the MYB domain,and used SMART (http://pfam.janelia.org/) and Pfam (http://pfam.janelia.org/) to analyze the completeness of MYB domains and exclude unqualified proteins. Finally,270 ZBM MYB proteins were identified,containing 8 1R-MYB proteins,251 R2R3-MYB proteins (2R-MYB) and 11 R1R2R3-MYB proteins (3R-MYB). But no 4R-MYB proteins were identified,which may be because of the incompleteness of the ZBM dataset. Quantitatively,the R2R3-MYB (ZbMYB) proteins constituted the largest group of MYB genes. To find out and identify the protein characteristics in ZBM MYB,the protein sequence analysis was carried out.These proteins ranged in length from 88 amino acids in EVM0007591 to 1 048 amino acids in EVM0007591.The average length of R2R3-MYB (ZbMYB) protein was about 299.1 amino acids,while that of 1R-MYB and 3R-MYB was 252.4 and 531.8,respectively. The calculated molecular weight (MW) of ZBM MYBs ranged from 10.10 to 60.28 kDa,and the theoretical pI of ZBM MYBs was from 4.3 to 9.91. The predicted subcellular localization results suggested that 268 ZBM MYB proteins located in the nuclear region,one ZBM MYB protein in the mitochondria,and one ZBM MYB protein in the cytoplasmic (Table 1).

Table 1 ZBM MYB related gene information
Multiple sequence alignments and comparisons among R2R3-MYB (251) domains ofZbMYBswere performed (Fig.1). It is found that R2 and R3 MYB repeats of the ZBM R2R3-MYB family contained characteristic amino acids (Strackeet al.2001). These amino acids are considered as a landmark of the MYB domain,whichare a series of evenly distributed and highly conserved tryptophan (Trp) residues (Duboset al.2010). The first Trp residue in the R3 repeat generally were replaced by a hydrophobic amino acid,such as Phe (F) or Ile (I),which is a common phenomenon in plants R2R3 MYB proteins (Duet al.2012b). However,substitution by the amino acids Leu (L) and Tyr (Y) was also observed at Trp-53 position of ZbMYB proteins. It was assumed that tryptophan in ZBM was replaced by a hydrophobic amino acid,but this change might have little effect on the DNAbinding activity of the MYB domain in ZBM (van Aaltenet al.1998;Diaset al.2003;Heineet al.2004). Glu-10,Asp-11,Cys-36 and Arg-39 in R2 repeat,Leu-44 in the linker region,Glu-57,Arg-81 and Thr-82 in R3 repeat were also completely conserved in the ZbMYB proteins.The insertion of Leu residues into the R2 repeats was observed in most ZbMYB proteins,which echoed the view that it was one of the important steps in the origin of the typical plant R2R3 MYB protein (Williams and Grotewold 1997). The conservative areas in the ZBM MYB DNAbinding domain were mainly located between the second and third Trp (W) of the 2R repeats (Fig.1). The third α-helix was the most conserved region in ZBM MYBs and played a role in identifying DNA binding (Duet al.2012b). In this region,residues substitution often weakened DNAbinding activity. In comparison,the remaining amino acid sequences were relatively conserved in the ZbMYB DNA binding domain.

Fig.1 Sequence logos of the R2 and R3 MYB repeats of Zanthoxylum bungeanum Maxim.(ZBM) R2R3-MYB domains. The sequence logos of the R2 (A) and R3 (B) MYB repeats are based on full-length alignments of all ZBM R2R3-MYB domains. Multiple alignment analysis of 251 R2R3-MYB domains was performed with ClustalW. The bit score indicates the information content for each position in the sequence. The position of the α-helices is marked (Helix 1 to 3). Arrows indicate the conserved tryptophan (Trp) residues in the MYB domain.
3.2.Phylogenetic analysis and classification of the ZbMYB genes
To survey the evolutionary relationships within the R2R3-MYB gene family,we performed a combined phylogenetic analysis ofZbMYBs(251 genes) and theArabidopsis(125 genes) full-length R2R3-MYB proteins,using the NJ (neighbor-joining) method in the MEGAX Program (Strackeet al.2001). Based on the sequence similarity and topological structure of the NJ phylogenetic tree (Duboset al.2010),we divided ZBM andArabidopsisMYBgenes into 33 subgroups (named S1-S33 in this study). Most of the large subfamilies in our classification were supported by the previous studies,including 25 subfamilies (S1-S25) based onArabidopsis. However,several small subgroups not represented inArabidopsiswere retrieved from the previously constructed phylogenetic tree,so we reclassified them and generated eight other subfamilies (S26-S33). Different groups contained different numbers of genes and the sizes of the subgroups were unequal. S8 constituted the largest clade with 27 genes:20 from ZBM and 7 fromArabidopsis(AtMYB20,40,42,43,80,85,and99) (Fig.2).
As shown in Fig.2,mostZbMYBgenes clustered with possible homologues inArabidopsis. For instance,S10 containedAtMYB9,AtMYB107andEVM0059377,EVM0031582,EVM0024489;in S8,EVM0060641was most similar toAtMYB80. Meanwhile,the subgroup S3 includedAtMYB58,AtMYB63and sixZbMYBgenes.This indicated that theMYBgene family further expanded in ZBM,and the functional role of this family may be further differentiated (Zhouet al.2009). However,eight subfamilies (S26-S33) did not cluster in any of the identified clades or subgroups ofArabidopsis(Houet al.2014;Zhanget al.2018),showing the uniqueness of these MYBs to ZBM. The functional role of these genes needs more in-depth research and requires more genomic sequences to analyze. At the same time,the clades which only containedArabidopsisR2R3-MYB genes (S12,S15,S21,S22,S23,and S25) were observed,suggesting that these genes might have specific effects inArabidopsisthat gradually disappeared during the evolution of ZBM (Liet al.2016). In addition,similar to the case in soybean and sugar beet,a single gene inArabidopsiswas found in ZBM which has two or more putative orthologs (Strackeet al.2014). This indicated thatZbMYBsin ZBM experienced expansion events after the divergence between ZBM andArabidopsis. For example,there were fourAtMYBsand 16ZbMYBsin the subgroup S4,and threeAtMYBsand 12ZbMYBsin subgroup S2. However,regardless of these associations,the exact function of the genes involved has not been determined.

Fig.2 Classification relationships between different R2R3-MYB subgroups. The tree was inferred using the neighbor-joining method and 1 000 bootstraps with putative amino acid full length MYB sequences with ClustalW Software. The subgroups were designated through comparative analysis with Arabidopsis. The different colored arcs indicate different groups (or subgroups) of the R2R3-MYB genes.
3.3.Gene structure and motif composition of the ZbMYB genes
Structural analysis aims to interpret the structural diversity of genes and provide important evidence of related duplication events to support phylogenetic relationships in a gene family (Hajiebrahimiet al.2017). We used online analysis software to compare the exon/intron organization in the coding sequence of eachZbMYBgene. There were significant differences in gene structure betweenZbMYBmembers,including the relative position and number of introns and exons (Fig.3). Among 251ZbMYBgenes,the number of exons was determined to be 2 to 5,which indicated that exons had been lost or gained during gene evolution. It was most common that two or three exons were determined for each gene. The remaining genes had introns at different positions or altered splicing sites. TheseMYBgenes can be grouped into 20 subgroups based on intron presence and positions (named C1 to C20). For instance,MYB genes in C2 (S26) contained two introns,C3 (EVM0025547) harbored two exons and one intron,and theEVM0016868gene in C12 had four introns. However,there was a notable exception because the lengths and positions of theZbMYB’s introns and exons in C15 were different from the rest.As a result,C15 were more likely to undergo alternative cleavage,and then create new genes,probably leading to functional diversity of theZbMYBfamily. Meanwhile,the untranslated region (UTR) of most genes was short or missing directly. The coding regions were more conserved than the UTR sequence.

Fig.3 Gene structure and conserved protein motif structure in the R2R3-MYB genes (ZbMYBs) of Zanthoxylum bungeanum Maxim.(ZBM). A,a neighbor-joining tree was constructed based on the alignment of ZbMYBs full-length amino acid sequences. B,architecture of conserved protein motifs in 20 subfamilies. C,exon/intron structure of R2R3-MYB gene from ZBM.
In addition to the conserved typical MYB domain at the N-terminus of the R2R3-MYB protein,there are also many functionally important motifs located in the C-terminal region involved in protein-protein interactions and transcriptional activities (Strackeet al.2001;Li and Lu 2014). To further reveal the diversification of MYB in the ZBM genes,the conserved motifs in this family were studied using MEME. We identified ten conserved motifs with each motif ranging in size from 6 to 136.These motifs were designated as motif 1 to 10. Among the 20 subgroups,motifs 1,2,3,and 4 represented the characteristics of MYB DNA-binding domain which was the most conserved among all the 251ZbMYBgenes (Fig.3-B). Each subgroup had similar motifs. For instance,five members of C2 (S26) had five conserved motifs (motifs 1,2,3,4,and 6). 12 members of C7 (S1) had six conserved motifs,including motifs 1,2,3,4,7,or 9. C11 (S29) contained motifs 1,2,3,4,5,6,and 8. 13 members of C14 (S20) had motifs 1,2,3,4,and 10. The members of C18 had six conserved motifs (1,2,3,4,5,and 6). Most subgroups had exclusive motifs,but some motifs overlapped across subgroups. For example,motif 9 was only found in C7,but motif 5 was found in all subgroups except C2,C5,C9,C13-C17,and C20. Therefore,differences in the quantities and types of conserved motifs may cause changes in the degree and rate of evolution. Additionally,consistent with Liuet al.(2017),motif 6 directly followed the MYB R3 repeat was found to be highly conserved in certain subfamilies (C1-C6,C11,and C18;Fig.3-B). These findings further supported the classification of the subgroups. AlthoughEVM0052680(C1),EVM0043893(C3) andEVM0002171(C11) distributed in different subgroups,they contained the same motifs,suggesting that they were likely to have similar functions. The similar associations were observed inEVM0027255(C8) andEVM0036532(C12).
3.4.Genomic location and duplication events among ZBM MYB genes
Genome chromosomal location analyses showed that the ZBMMYBgenes were unevenly distributed throughout all 49 Lachesis Groups (LG) (Fig.4). In the current sequences,218MYBgenes were mapped.Some LGs had more genes than others. For example,LG7 encompassed the largest number of 14 ZBMMYBgenes,followed by 12 in LG17 and 11 in LG0,LG35,and LG65,respectively. In contrast,only oneMYBgene was found in LG21,LG36 and LG60. Substantial clustering of ZBMMYBgenes was obvious on several LGs. For example,EVM0022607,EVM0028109andEVM0006437were clustered on a little segment in LG7.EVM0003876,EVM0045634,EVM0022005andEVM0001914were clustered in LG4. Gene duplication has long been recognized to play an important role in the expansion of large gene families in plants,and it occurs throughout plant evolution (Cannonet al.2004). The collinearity of theZbMYBgenes identified by BLASTP and MCScanX methods revealed the possible relationship betweenMYBgenes and potential replication events.Finally,72 collinearZbMYBgene pairs were identified in the ZBM genome (Fig.5). TheZbMYBgenes were located within synteny blocks in almost all LGs except for LGs 4,19,23,37,and 62. A gene cluster refers to two or moreZbMYBgenes residing within 20 kb of each other. TheseZbMYBswere physically located near to each other in a syntenic region of related chromosomes,forming 29ZbMYBtandem duplication pairs (i.e.,EVM0006449/EVM0059508,EVM0042160/EVM0020636,EVM0029593/EVM0044648,and so on).The members ofZbMYBgenes were assigned to five different duplication events:WGD,transposition,proximal,tandem or dispersed.Results showed that 74.19% (184) of theZbMYBgenes were duplicated and retained from transposed duplication events,followed by 16.13% (40) from WGD,compared to only 2.02% (5) from proximal,2.42% (6) from dispersed,and 5.24% (13) from tandem (Fig.6). These results showed that someZbMYBgenes may have been produced by gene replication events,and transposition and WGD duplication events played critical roles in the expansion of theZbMYBgene family in ZBM.

Fig.4 Chromosomal locations of ZbMYB genes. The chromosome number is indicated at the left of each chromosome.

Fig.5 Schematic representations of interchromosomal relationships of the ZbMYB genes. Gray lines indicate all synteny blocks in the Zanthoxylum bungeanum Maxim.(ZBM) genome,and the red lines indicate duplicated MYB gene pairs. The chromosome number is indicated at the top of each chromosome.

Fig.6 Numbers of different duplication events in ZbMYBs.WGD,whole-genome duplication;TD,tandem duplication;PD,proximal duplication;TRD,transposed duplication;DSD,dispersed duplication.
3.5.Excavation of potential regulatory genes in flavonoid synthesis in ZbMYB gene
Currently,the most extensive annotative assessment of plant MYB comes from the model plantArabidopsis.R2R3-MYB inArabidopsiscan be divided into 25 clades,and the function of each clade has been annotated.Therefore,based on the combined results of phylogenetic analysis and subfamily classification,it is assumed that the R2R3-MYB genes in ZBM are likely to have similar functions with those ofArabidopsis,AtMYBgenes.
Previous research has found that the S4,S5,S6,and S7 subfamilies of the R2R3-MYB family inArabidopsiswere mainly involved in the regulation of structural genes in the flavonoid pathway (Grotewoldet al.1991;Strackeet al.2007). We obtained 20 related genes in the S4 subgroup,including fourAtMYBgenes and 16ZbMYBgenes. Among these regulatory genes,16 are likely to function as the fourth S4 subfamily and mainly used as a transcriptional repressor to negatively regulate the expression of related enzymes such asCHSandC4Hin the anthocyanin synthesis pathway (Jinet al.2000;Aharoniet al.2001;Ma and Constabel 2019). Similarly,through evolutionary relationships,we obtained two related genes,EVM0017457andEVM0027718,in S5 subfamily. These genes may regulate downstream related genes of proanthocyanidin biosynthesis by forming a ternary complex withbHLHandWD40(Petroni and Tonelli 2011;Liuet al.2015;Xuet al.2015). Moreover,subgroup S7 included 11ZbMYBsand threeAtMYBs(AtMYB11,AtMYB12,andAtMYB111). The threeAtMYBgenes are well known for their specifical action on the flavonoid synthesis genesCHS,CH1,F3H,andFLSand crucial roles in the early regulation of flavonol biosynthesis inArabidopsis(Strackeet al.2014). Thus,these genes in this clade may play similar roles in regulating flavonol biosynthesis and can also provide clues for solving the difference in flavonols content in leaves and fruits of ZBM. Subgroup S6 comprised of fourAtMYB(PAP1,AtMYB90,AtMYB113,andAtMYB114) genes and fourZbMYBgenes (EVM0033809,EVM0052497,EVM0047123,andEVM0026041),which are known to take part in anthocyanin biosynthesis.
3.6.The total flavonoids and anthocyanin content in pericarps at different development stages of ZBM
ZBM gradually turns red towards maturity (Fig.7). Fig.8 shows the total flavonoids and anthocyanin of ZBM at seven development stages. Total flavonoids generally increase as the ZBM fruits mature. During the fruit development stages of ZBM,the accumulation of total flavonoids experiences an initial rapid increase followed by a slow increase,whereas the anthocyanin content shows a gentle decrease initially,followed by a sharp increase,and finally decrease. In the P30 period,the accumulation rate of flavonoid content reached a maximum value of 41.29%,and decreased steadily to 12.57% in the P40 period. The rate gradually decreased to about 5% after the P50 period (Fig.8-B). P10 to P40 were considered as a rapid increase stage with the flavonoid accumulation increasing from 12.570 to 19.992 mg g-1,an increase of 59.04%.

Fig.7 Seven developmental stages of Zanthoxylum bungeanum Maxim.(ZBM). The first row is P10,P30-P50 from left to right,the second rows are P60-P80,and the leaf and stem at the P80. P10 and P30-P80,10 and 30-80 days post anthesis.
For anthocyanin,it enters a rapid growth period from the P50 stage,but begins to decline after reaching a peak at the P70 stage (Fig.8-D). In contrary to flavonoid accumulation,anthocyanin content gradually decreased,from 0.165 to 0.09 mg g-1,from P10 to P40. From P50 to P80,the accumulation of flavonoids in ZBM increased slowly by 11.75%,from 21.130 to 23.613 mg g-1,and decreased slightly to 22.092 mg g-1,at the maturity stage (P80) (Fig.8-A). On the contrary,anthocyanin content entered a rapid accumulation mode since P50,rising from the lowest 0.09 to the highest 0.235 mg g-1in P70,and dropped to 0.143 mg g-1at the mature period (Fig.8-C).There was no significant difference in the flavonoid content of ZBM between P70 and P80 when it matured,indicating that the flavonoid contents in ZBM after P70 was still high.
3.7.Expression patterns for the target ZbMYB genes of ZBM
To obtain greater insights into the potential functions ofZbMYBgenes in the regulation of flavonoid pathway,the temporal and spatial expression patterns ofZbMYBgenes were examined by constructing a heat map using transcriptome data. We analyzed their transcript abundance at seven developmental stages and three tissue parts (pericarp,stem and leaf 80 days after flowering) in ZBM. A hierarchical cluster analysis was performed with the logarithm of average values for the 33ZbMYBfamily genes of all four subgroups (S4,S5,S6 and S7) involved in the flavonoid regulation. The genes with zero expression in all plant tissues and stages were deleted (Fig.8).

Fig.8 The growth rhythm of flavonoids in various developmental stages of Zanthoxylum bungeanum Maxim.(ZBM). A,the flavonoid content curve of ZBM. B,the increase rate of flavonoid content of ZBM. C,the anthocyanin content of ZBM. D,the increase rate of anthocyanin content. P10 and P30-P80,10 and 30-80 days post anthesis.
The results showed that 28ZbMYBgenes expressed in all tested samples,about 50% were more pronounced,and 28.5% (8) expressed at a very low level. The gene expression profiles for the 28 MYB genes were grouped into five clusters (Fig.9;Z1-Z5),and genes in the same cluster possessed highly similar transcription profiles.

Fig.9 Hierarchical clustering of expression level of 28 ZbMYB genes in Zanthoxylum bungeanum Maxim.(ZBM) different stages and tissues. Transcriptome data were used to measure the expression levels of each gene. Heatmap showing expression patterns of ZbMYB genes in seven developmental stages and three different tissue parts. P10 and P30-P80,10 and 30-80 days post anthesis. Color scale at the top represents log2transformed RPKM values. Blue indicates low expression and red indicates high expression. The colored lines indicate the group with similar expression levels (Z1 to Z5). The expression data were hierarchical clustered based on Pearson correlation.
The expression of Z5 genes presented an increasing trend throughout the fruit development stages. In particular,the expressions ofEVM0033809andEVM0052497presented an obvious increase at the end stage (P80). While other genes,especially those in Z2 cluster,showed different expression patterns. For example,approximately 75% (6) of Z2 genes showed a downward trend.EVM0012080andEVM0027255both showed higher levels of gene expression in the early stages of development,but their expression levels decreased gradually in the middle and late stages. It is therefore can be concluded that the six genes of Z2 might be involved in important regulatory mechanisms at early stages of ZBM fruit development. We also noticed that the Z3 and Z4 clusters presented a minor change in gene expressions across development stages. However,except forEVM0055016,the otherZbMYBgenes such asEVM0052680andEVM0004503in Z3 and Z4 displayed low expression levels in all stages,indicating that these genes possibly expressed under only special conditions or are pseudogenes. During all the states,the Z1 genes expression was significantly higher than that of other clusters,suggesting that they play a more important regulatory role at multiple developmental stages.
Further,some genes showed significant tissue specificity. Most of them were detected in leaves and stems,more than half of the genes highly expressed in ZBM leaves,and the expression level in the pericarp tissue was the lowest. For example,four genes of Z1 showed superior expression in almost all tissues.Moreover,Z2 and Z4 showed dominant expression patterns in leaves and stems,but relatively low expression in pericarps. In stems,four genes (EVM000020636,EVM0042160,EVM0046410andEVM0013896) had high expression levels. In particular,EVM0009827predominantly expressed in stems,whereasEVM0027718andEVM0013896mainly expressed in leaves. In the pericarp tissue,five genes (EVM0033809,EVM0052497,EVM0020636,EVM0036148andEVM0042160) showed the highest expression. Surprisingly,the expressions ofEVM0033809andEVM0052497in pericarps were much higher than those in leaves and stems. Interestingly,their orthologRuby1,a key positive regulator of anthocyanin biosynthesis in sweet orange and a member of Rutaceae family (Butelliet al.2012;Huanget al.2020),encoded an MYB transcription factor and showed a strongly divergent expression between sweet oranges and ZBM. The expression ofEVM0052497peaked at P80,while the anthocyanin content peaked at P70. The reasons may be multifold. In addition to anthocyanin,EVM0052497might form other intermediates to control anthocyanin biosynthesis in flavonoid. Therefore,there two genes may play an important role in regulating the synthesis of anthocyanin biosynthesis in ZBM pericarps. The specificity of these genes suggested that they were ideal candidates for further functional analysis.
3.8.Confirmation of ZbMYB genes by using real-time PCR
In order to verify the RNA sequence data and further explore the role ofZbMYBgenes in regulating the biosynthesis of flavonoids in ZBM,the representative genes of four subgroups (S4,S5,S6,and S7) involved in regulating flavonoid metabolism were selected as ideal candidates for further analysis.EVM0042160,EVM0027718,EVM0033809,andEVM0021667might play a special role in regulating the flavonoids biosynthesis in ZBM. So,these genes were suitable for further functional analysis. Their expression levels quantified by qRT-PCR analysis exhibited a similar increasing trend corresponding to RNA-seq data at different developmental stages of ZBM.
The results showed significant differences in expression pattern among these genes. The overall expression level ofEVM0042160was very high,at least 20 times that of the other three genes. The expression ofEVM0042160experienced an initial sharp decline,rose later,and finally fell. Its expression peaked in P10,encountered a cliff-like decline,reaching the lowest close to 0 in P50,slightly rebounded in P60 and P70,and finally fell again in P80.
The expression pattern ofEVM0027718showed an overall downward trend,peaking in P10 but dropping to an extremely low level after P40. The expression ofEVM0033809first declined slightly,then fluctuated followed by a sharp rise,and eventually declined slightly and steadily as towards the maturity stage of ZBM. It is worth mentioning that,in addition to showing accumulated transcripts at P10 and P40,the gene expression level was the highest in P70. During the transition periods from P10 to P30,gene expression was mild and slightly decreased. However,the expression gradually increased in P50. Although the increase was moderate,it clearly showed the accumulation of transcripts (Fig.10). From P60 to P70,the expression was dramatically induced and showed a sharp and rapid up-regulation pattern.Compared with P30,the relative expression in P70 was up regulated by more than 61 times. But the expression was again decreased slightly in P80. In addition,this gene showed obvious expression characteristics of transcript accumulation during P70,indicating that it may play a developmental role at this stage.

Fig.10 Expression analysis of genes in different development stages using quantitative real-time PCR. Data were normalized to ZBM actin gene and vertical bars indicate standard deviation (n=21). P10 and P30-P80,10 and 30-80 days post anthesis.
EVM0021667showed a distinctive expression pattern from other genes. Its expression level was much higher in P10,P40,P60,and P80 than in other stages. In sum,the four genes showed vastly different expression levels:EVM0042160had the highest overall expression level,followed byEVM0027718,andEVM0033809,andEVM0021667had the lowest.
3.9.Correlation analysis
In order to explore the expression correlation between the 28ZbMYBgenes and the total flavonoid content and the increase rate of flavonoids content in the seven developmental stages of ZBM,a correlation analysis was performed (Fig.11).
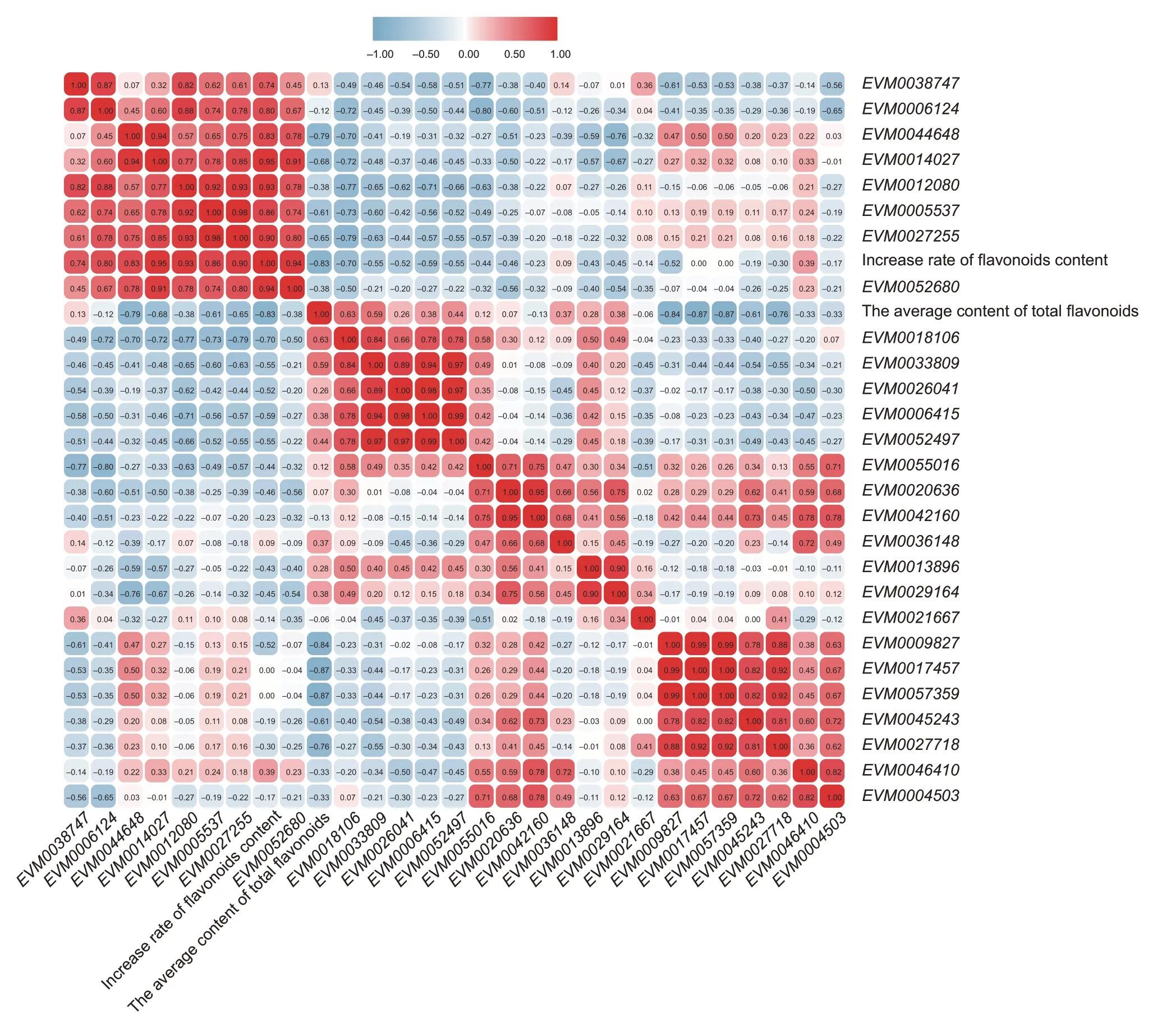
Fig.11 The correlations of ZbMYB genes expression. Red,positively correlated;blue,negatively correlated.
The phylogenetic analysis identified fourZbMYBgenes with the highest homology to specific functional genes inArabidopsis,includingEVM0027718,EVM0042160,EVM0033809andEVM0021667. The correlation analysis results showed thatEVM0027718,EVM0042160andEVM0021667were negatively correlated with the total content and the accumulation rate of flavonoid in ZBM;EVM0033809was positively correlated with flavonoids content,while negatively correlated with the accumulation rate of flavonoids. In addition,EVM0044648was significantly negatively correlated with flavonoid content;EVM0018106andEVM0052497were positively correlated with flavonoid content;EVM0052680,EVM0012080andEVM0014027had a very significantly positive correlation with the accumulation rate of flavonoids. In contrast,genes such asEVM0017457,EVM0057359andEVM0009827were significantly negatively correlated with the total flavonoid content of each developmental stage of ZBM;genes such asEVM0018106,EVM0006415andEVM0052497were negatively correlated with the flavonoid accumulation rate.
At the same time,we studied the correlation between the expression patterns ofZbMYBsgenes.The correlation coefficients betweenEVM0042160andEVM0020636,EVM0017457andEVM0057359,EVM0033809andEVM0006415,andEVM0052497andEVM0026041were all greater than 0.08,showing an extremely significantly positive correlation.EVM0042160andEVM0046410,EVM0004503andEVM0046410,EVM0052497andEVM0018106showed a significantly positive correlation. Conversely,the correlations betweenEVM0038747andEVM0055016,EVM0006124andEVM0055016,andEVM0044648andEVM0029164were significantly negative.
4.Discussion
The MYB family,one of the largest and most significant TF families in plants,is involved in regulating various growth and development processes of plants (Liet al.2019;Yanget al.2021). Most importantly,MYB TFs play a vital role in regulating the flavonoids biosynthesis. In this study,270 ZBM MYB genes were identified in ZBM,which is more than those in other dicotyledonous and monocotyledonous plants,such as 198 inArabidopsisand 183 in rice (Oryza sativa) (Yanhuiet al.2006;Wilkinset al.2009). The large number of the transposed duplication events indicated that transposed duplications played a major role in the expansion of the MYB gene family in ZBM (Chenet al.2006). The transposed duplication (TRD) is presumed to generate a gene pair comprised of an ancestor and a novel locus that arises through distantly transposed duplications through DNAbased or RNA-based mechanisms (Wanget al.2012b;Qiaoet al.2019). Environmental factors may accelerate the divergence in the expression between duplicate genes (Haet al.2007). The frequent occurrence of transposition duplication may play an important role for plants in adapting to dramatic environmental changes (Xuet al.2009;Dassanayakeet al.2011;Tamateet al.2014). We believe that the expansion of this gene family greatly benefits the growth,development and adaptability of ZBM.The increase in MYB members can enrich the function of this gene family and promote the growth and development of ZBM,enabling it to adapt to multiple environments and enhancing its survival (Conant and Wolfe 2008;Flagel and Wendel 2009). From this point of view,the increase in MYB members is crucial during the evolution of ZBM as it can enhance ZBM’s adaptive capacity to varying environmental conditions.
In the present study,the phylogenetic tree characterized the intron/exon distribution and conserved domains of MYBs,revealing the conservation of gene structure and domains in the same group of members. Intron size differed between MYB subfamilies. Interestingly,most members of the same group had relatively conservative exons. This similar phenomenon has also been reported for other gene families,such as the WRKY gene family in cassava and cacao (Weiet al.2016;Waqaset al.2019). In addition,it has been shown previously that divergences in exon are less common in orthologs than in paralogs at the same pace of duplications. Previous studies also have shown that homologous MYB proteins clustered in a subgroup have similar or identical genetic structures. This finding suggests that these members are evolutionarily rigid and may have similar functions. However,there are some exceptions,that is the genetic structure of these genes is different from that of otherZbMYBgenes in the same subgroup,pointing out that the evolution may differ among the MYB family members. This finding further supports the diversity of MYB functions,an important component of gene family evolution,diversification,and new functionalization.
In this study,the phylogenetic analysis betweenArabidopsisand ZBM was conducted following the model previously described inArabidopsis. The phylogenetic analysis divided the R2R3-MYB genes in ZBM into 33 subgroups,of which the subgroups S1-S25 were the same as in theArabidopsis,and the remaining eight subgroups contained only ZBM genes. These eight subgroups were most likely to be separated fromArabidopsisand have earned unique directions and functions in the ZBM evolution process. At present,the exact functions and potential mechanisms of these genes are unclear,and they are worth further investigations.Notwithstanding,it is safe to ascertain that the MYB genes have a unique role in the specific functions and traits ofArabidopsisand ZBM,which proves that there is an evolutionary trend of gene duplication in ZBM.
Orthologous gene pairs,such asZbMYBandAtMYB,usually can maintain their functions after species formation,so phylogenetic trees can be used to infer the gene functions ofZbMYB(Jianget al.2004). As a model plant,mostAtMYBgenes have detailed functional characteristics. It has been found inArabidopsisthat genes in four subgroups S4-S7 are necessary for regulating the flavonoid biosynthesis,so the corresponding orthologous genes in ZBM have the same function. For instance,in group S4,AtMYBand its orthologs in ZBM can interact with bHLH transcription factors and repress regulatory elements (Sampathet al.2013). TT2 protein (ortholog of EVM0027718) in S5 is involved in regulating the synthesis of proanthocyanidins in plants (Nesiet al.2001;Xuet al.2015). It has been observed that the ortholog ofEVM0021667in S7,namelyAtMYB111,is involved in the synthesis of flavonoids inArabidopsis. However,it is worth noting that according to the homology ofAtMYBs,the functions differ among MYB members even when some members are from the same clade. For example,AtMYB4,AtMYB7,andAtMYB32in S4 are involved in regulating not only the flower development and anthocyanin biology,but also the secondary cell wall synthesis (Prestonet al.2004;Zhonget al.2008).
In addition,based on transcriptome data,we found that most orthologs ofZbMYBgenes and the content of flavonoids in ZBM expressed at different levels at different times and tissues under natural growth conditions. For example,the expression ofEVM0055016was high mainly in leaves and stems but not in pericarps. In contrast,the expression ofEVM0052497was high in pericarps while low in other tissues. Moreover,there was a positive correlation between the expression ofEVM0033809in pericarps and the accumulation of flavonoids,while a negative correlation was found forEVM0027255.It suggests that these genes played a role in the accumulation of flavonoids in ZBM pericarps. Difference expression also occurs in other species. Subgroup 7 inArabidopsiscontainsAtMYB12,AtMYB11andAtMYB111,which are known for controlling flavonol biosynthesis in different organs. In addition,GtMYBP3andGtMYBP4strongly expressed in the early developmental stages of petals in gentian flowers,and the expression profile is almost consistent with the accumulation profile of flavone derivatives. In grapevine,theVvMYBF1andVvMYBA1transcripts detected during flowering and in the skin of mature berries are associated with accumulation of flavonols (Walkeret al.2007;Nakatsukaet al.2012).
It is noteworthy that the expression of some genes was highly related to the accumulation pattern of flavonoid content in ZBM pericarps. With the increase of flavonoids in the pericarp,some genes (e.g.,EVM0029164,EVM0033809andEVM0052497,etc.) were significantly up regulated,showing that they might be the positive regulators of flavonoid biosynthesis. On the contrary,the expression trend of some genes (e.g.,EVM0044648andEVM0005537) remained relatively low after a sharp decline,suggesting that they might be involved in the negative regulation of flavonoid biosynthesis. Some other genes (e.g.,EVM0046410) that always keep a steadily high and medium expression are likely to participate in the entire regulation process.
IdentifyingAtMYBorthologs in ZBM can help verify their function in ZBM. We thus unearthed four genes (EVM0027718,EVM0042160,EVM0033809,andEVM0021667) as potential targets for studying the regulation of flavonoid biosynthesis in ZBM pericarps. In this study,qRT-PCR was used to detect the expression of MYB genes at different developmental stages of ZBM. The selected four genes are phylogenetically closer to the function-known regulation of flavonoid biosynthesis related to MYB proteins in model plants. In our research,at various stages of fruit development,the expression of the four genes varied across stages,which is very similar to the flavonoid and anthocyanin curve in ZBM. For example,the expression pattern ofEVM0042160was basically the same as the flavonoid accumulation curve.It can be assumed that this gene plays a significant role in regulating the accumulation rate of flavonoids in ZBM.EVM0033809was highly expressed during P70,and simultaneously the content of anthocyanin in ZBM peaked at P70,which shows that this gene may promote the biosynthesis of anthocyanin. These results strongly support our hypotheses on gene function. Future studies should investigate whether these genes are involved in important regulatory processes related to flavonoid biosynthesis in order to identify a network mechanism for gene regulation flavonoid biosynthesis in ZBM. The candidate MYB genes presented in this study are particularly useful for future exploration of the biological functions ofZbMYBgenes.
5.Conclusion
The analyses presented in the current study provide new evidence on the MYB gene family in ZBM and its regulation of flavonoid metabolism during ZBM development and maturation. The ZBM familywide characterization of MYBs provides insights into the evolution of this TF and has implications for the understanding of the temporal-spatial change of flavonoid. Our identification of the set of MYBs will aid the development of new genes that regulate flavonoid metabolism in ZBM. New molecules of a total of 270 ZBM MYB genes were identified,including 8 1R-like MYB,251 typical R2R3-MYB,and 11 R1R2R3-MYB.The analysis of conserved domains showed thatZbMYBgenes shared the R2R3 domain with 97 basic residues and five conserved Trp residues. The MYB genes were divided into 33 subgroups (S1-S33) according to their phylogenetic relationship,which was well supported by additional intron/exon structures and conserved motifs. Analysis of intron/exon structure indicated that the splice sites and lengths of most introns were highly conserved in the MYB domain,especially in the same subgroup. The transposed duplications appeared to play a major role in the expansion of the MYB gene family in ZBM. The expression ofEVM0027718,EVM0042160,EVM0033809,andEVM0021667was validated by qRTPCR,which revealed that their transcript abundance levels changed significantly with the increase of flavonoid and anthocyanin content in ZBM. This suggested that these four genes may play an important role in flavonoid metabolism. Moreover,transcriptome sequencing confirmed the expression levels of 28ZbMYBgenes and predicted that they might be involved in the regulation of flavonoid biosynthesis. Genes such asEVM0042160andEVM0033809have been established as potential targets for studying regulation of flavonoid biosynthesis.However,the key regulatory genes we screened were merely based on phylogenetic analysis,transcriptome data and correlation tests. Hence,for future research,the effect of these genes on the expression of anthocyanin biosynthesis-key structural genes,and how these genes interact with each other in ZBM need to be further investigated through a variety of experiments.
Our results enhance the understanding of the molecular mechanism ofZbMYBthrough which flavonoid biosynthesis is regulated in ZBM. Our results also provide a theoretical basis for further research on the MYB gene family of ZBM and other crops. The findings will help to develop and use the potential functions of flavones in ZBM,and establish a solid foundation for comprehensive functional analysis ofZbMYBin the future.
Acknowledgements
This research was financially supported by the National Key R&D Program of China (2018YFD1000605) and the Project of Science and Technology Development Center,National Forestry and Grassland Administration,China (KJZXSA202025).
Declaration of competing interest
The authors declare that they have no conflict of interest.
 Journal of Integrative Agriculture2022年7期
Journal of Integrative Agriculture2022年7期
- Journal of Integrative Agriculture的其它文章
- Roles of mushroom polysaccharides in chronic disease management
- lnfluence of high-molecular-weight glutenin subunit deletions at the Glu-A1 and Glu-D1 loci on protein body development,protein components and dough properties of wheat (Triticum aestivum L.)
- Variations in the quality parameters and gluten proteins in synthetic hexaploid wheats solely expressing the Glu-D1 locus
- ldentification of candidate genes related to soluble sugar contents in soybean seeds using multiple genetic analyses
- ldentification and characterization of long-lnDels through whole genome resequencing to facilitate fine-mapping of a QTL for plant height in soybean (Glycine max L.Merr.)
- Photosynthetic properties of the mid-vein and leaf lamina of fieldgrown,high-yield hybrid rice during senescence
