Genome-wide identification and characterization of the abioticstress-responsive lipoxygenase gene family in diploid woodland strawberry (Fragaria vesca)
Ll Zhi-qi ,XlE Qian ,YAN Jia-hui, ,CHEN Jian-qing ,CHEN Qing-xi
1 College of Horticulture,Fujian Agriculture and Forestry University,Fuzhou 350002,P.R.China
2 Horticultural Plant Biology and Metabolomics Center of Haixia Institute of Science and Technology,Fujian Agriculture and Forestry University,Fuzhou 350002,P.R.China
Abstract Lipoxygenase (LOXs) is a kind of dioxygenase without heme and iron,which plays an important role in the development and adaptation of many plants to the environment. However,the study of strawberry LOX gene family has not been reported. In this study,14 LOX genes were identified from the diploid woodland strawberry genome. The phylogenetic tree divides the FvLOX gene into two subfamilies:9-LOX and 13-LOX. Gene duplication event analysis showed that whole-genome duplication (WGD)/segmental duplication and dispersed duplication effectively promoted the expansion of strawberry LOX family. QRT-PCR analysis showed that FvLOX genes were expressed in different tissues. Expression profile analysis showed that FvLOX1 and FvLOX8 were up-regulated under low temperature stress,FvLOX3 and FvLOX7 were up-regulated under drought stress,FvLOX6 and FvLOX9 were up-regulated under salt stress,FvLOX2,FvLOX3 and FvLOX6 were up-regulated under salicylic acid (SA) treatment,FvLOX3,FvLOX11 and FvLOX14 were up-regulated under methyl jasmonate (MeJA) treatment,FvLOX4 and FvLOX14 were up-regulated under abscisic acid (ABA) treatment. Promoter analysis showed that FvLOX genes were involved in plant growth and development and stress response. We analyzed and identified the whole genome of strawberry FvLOX family and characterized a variety of FvLOX candidate genes involved in abiotic stress response. This study laid a theoretical and empirical foundation for the response mechanism of strawberry to abiotic stress.
Keywords:lipoxygenase,gene family,strawberry,abiotic stress,expression pattern,cis-acting element
1.lntroduction
Cultivated strawberry (Fragaria×ananassa) is an octaploid plant (2n=8x=56) (Edgeret al.2019),which is an important soft fruit in the world. Strawberries have good taste and appearance,and are rich in nutrients. Plants often face a variety of abiotic stresses in the process of growth. Abiotic stress includes cold stress,heat stress,drought stress,salt stress and other factors. Under abiotic stress,the growth and nutrition of strawberries will be hindered,resulting in limited yield. According to the intensity,it will lead to nutritional imbalance,affect the metabolic function of cells,resulting in plant death. Therefore,increasing the tolerance to abiotic stress thereby improving the yield and quality of strawberry is one of the main challenges that need to be faced.
Oxolipids and their derivatives,such as jasmonic acid,divinyl ether and volatile aldehydes,play an important role in plant response to abiotic stress (Gouinguené and Turlings 2002;Nafieet al.2011). The synthesis of these oxygenated lipids is catalyzed by lipoxygenase.Lipoxygenase is a kind of non-heme,iron-containing dioxygenase,which exists widely in animals and plants (Andreouet al.2009). LOX can catalyze the oxidation of polyunsaturated fatty acids of carbon atom 9 and carbon atom 13,and their roles are 9-LOX and 13-LOX,respectively. Due to the difference of the specific motif of a specific binding site,the function of LOX may be different from 9-LOX to 13-LOX (Hornunget al.1999).According to the specificity of fatty acid oxidizing linoleic acid (LA) and linolenic acid (LEA),LOX proteins can be divided into two types:9-LOX and 13-LOX. In addition,13-LOX proteins can be divided into two subgroups:type I 13-LOX protein and type II 13-LOX protein. Among them,type I 13-LOX protein lacks chloroplast transport peptide,but the gene sequence has a high similarity,generally more than 75%,and is usually located in the cell. Type II 13-LOX protein has chloroplast transport peptide,but the gene sequence similarity is low,generally more than 35% (Shibata and Axelrod 1995;Brash 1999).
9s-Hydroxy peroxy octadecadienoic acid (9-HPOD) and 13s-hydroxy peroxy octadecadienoic acid (13-HPOD) produced by 9-LOX and 13-LOX can be further converted into various oxygen lipids by a variety of enzymes (Blée 2002). Related studies have shown that lipoxygenase and its derived oxygen lipids play an important role in seed germination (Baillyet al.2002),tuber development (Kolomietset al.2001),fruit ripening (Barry and Giovannoni 2007),sex determination (Chuck 2010) and immune response (Moran and Thompson 2001). More importantly,13-LOX-derived oxolipin jasmonic acid (JA) and its precursor (+)-12-oxo plant dienoic acid (OPDA) play an important role in plant growth and development and response to abiotic stress (Browse 2009).
Previous studies have shown that LOX gene has been well demonstrated in plant response to abiotic stress.InArabidopsisthaliana,the expression ofAtLOX1andAtLOX5is induced by stress hormones that affect immune response (Vellosilloet al.2007;Grebneret al.2013).AtLOX2is the main component of methyl jasmonate synthesis,and its expression is up-regulated under methyl jasmonate and stress treatment (wound and osmotic) (Bell and Mullet 1993).AtLOX3participates in the regulation of salt stress response (Huiet al.2016).AtLOX6is related to the rapid accumulation of JA and JA-lle in injured leaves (Grebneret al.2013). In pepper,CaLOX1plays an important role in regulating abiotic stress response by rapidly clearing reactive oxygen species (ROS) and activating defense-related marker genes (Limet al.2015). In persimmon,DkLOX3increases tolerance to senescence and salt stress by accumulating O2and H2O2and activating stress response genes (Houet al.2015).In cotton,GhLOX12andGhLOX13support its role in salt tolerance by regulating ROS (Shabanet al.2018).
At present,the LOX gene family has been studied in many plant species,such asArabidopsis(Bannenberget al.2009),rice (Umate 2011),radish (Wanget al.2019),pear (Liet al.2014),poplar (Chenet al.2015),and so on. However,so far,there is no information about genomewide identification and characterization of strawberry LOX gene family members. With the publication of the genome sequence of diploid woodland strawberry,it is possible to identify and characterize the whole genome of strawberry LOX gene family members,which is an indispensable step to study the function of strawberry LOX gene.
In this study,we conducted a genome-wide search for LOXs in diploid woodland strawberry (Fragariavesca) and identified 14FvLOXmembers. The expression level ofFvLOXsunder abiotic stress response (low temperature,high temperature,drought,salinity) and exogenous plant hormone treatment (salicylic acid,methyl jasmonate,abscisic acid) was studied. In addition,a series of bioinformatics analyses provide insights into the structure and function of genes,as well as evolution and function. To sum up,these results are helpful for further characterization of strawberry LOX gene.
2.Materials and methods
2.1.Plant materials,growth conditions,and stress treatments
In this study,diploid woodland strawberry,namely ‘Yellow Wonder’,were used as wild type. Seeds of wild diploid woodland strawberry were obtained from the strawberry germplasm resource nursery of the College of Horticulture,Fujian Agriculture and Forestry University (26°10´N,119°23´E),Fuzhou,China. The seeds were sown on Murashige and Skoog medium and then transferred to soil for subsequent growth after germination. The growth environment was 22°C,relative humidity 75%,and a photoperiod of 13 h/11 h (day/night). As materials for gene expression analysis in different organs of woodland strawberry plants,samples of the roots,stems,leaves,flowers (at anthesis),and fruits (at the red fruit stage) were collected.
Four-month-old seedlings of uniform growth were selected for abiotic stress treatment. Among them,three strawberries were used as control,three strawberries were treated with low temperature,high temperature,drought and salt stress,and six strawberries were treated with salicylic acid (SA),methyl jasmonate (MeJA),and abscisic acid (ABA). A solution of 400 mmol L-1NaCl was sprayed onto potted plants to induce salt stress for 12 h. Potted plants were transferred to a growth room maintained at either 4 or 42°C for 12 h for treatment with low-temperature and high-temperature stress,respectively. Plants were uplifted from the soil and placed on dry filter paper for 12 h of drought treatment. In addition,leaves were evenly sprayed with 40 μmol L-1SA,40 μmol L-1MeJA,and 40 μmol L-1ABA solution.The leaves of treated plants were sampled at 0,3,6,and 12 h after initiation of low temperature,high temperature,drought,and salt stress. The leaves of plants treated with SA,MeJA and ABA were sampled at 0,4,8,12,16,20,24,and 48 h after treatment. Six leaves from three individual seedlings were selected and combined as one sample (Weiet al.2016). All treatments were evaluated in triplicate. All collected samples were immediately wrapped in tin foil,frozen in liquid nitrogen,and stored at −80°C until use.
2.2.Genome-wide identification of LOX genes
The gene files of diploid woodland strawberry (FragariavescaGenome v4.0.a1),rose (RosachinensisWhole Genome v1.0),and peach (PrunuspersicaGenome v2.0.a1) were downloaded from Rosaceae genome database (https://www.rosaceae.org). The gene files of pear,Chinese plum,Arabidopsis,and apple were downloaded from following databases,respectively (http://peargenome.njau.edu.cn/,http://prunusmumegenome.bjfu.edu.cn/,http://www.arabidopsis.org/,respecitvely). The hidden Markov model of LOX protein characteristic domain (PF00305) was downloaded from Pfam (Finnet al.2016) (http://pfam.xfam.org/) database.HMMER Software was used to identify the whole genome ofArabidopsis,strawberry,Chinese plum,rose,peach,pear and apple under the condition of E-value<1×10-10and Score>100. The published amino acid sequences of LOX family members ofArabidopsiswere compared with the strawberry,Chinese plum,rose,peach,pear and apple genome database by Blastp. The protein domains were verified by CD-search (https://www.ncbi.nlm.nih.gov)) and the deleted genes in the [His-(X)4-His-(X)4-His-(X)17-His-(X)8-His] domain were deleted (Appendix A) The subcellular localization of LOX genes inA.thalianaand six species of Rosaceae were predicted by online bioinformatics software Wolf PSORT (https://wolfpsort.hgc.jp/) (Appendix B).
2.3.Phylogenetic analysis
The multiple sequences of LOX gene protein inArabidopsis(A.thaliana),strawberry (F.vesca),Chinese plum (P.mume),rose(R.chinensis),peach (P.persica),pear (P.bretschneideri) and apple (M.domestica) were compared by MEGA6.0 Software (Tamuraet al.2013). A phylogenetic tree was constructed using the maximum likelihood method with MEGA6.0 Software. Support for the topology was assessed by performing a bootstrap analysis with 1 000 replicates. The phylogenetic tree was visualized and edited using the iTOL (Letunic and Bork 2016) online tool (https://itol.embl.de/itol.cgi).
2.4.Gene structure and conserved sequence analysis
The GSDS 2.0 online server (Guoet al.2007) (https://gsds.cbi.pku.edu.cn/) was used to visualize the exon and intron structure of each gene. The amino acid sequence of each LOX protein of strawberry was submitted to the MEME online analysis website (Baileyet al.2015) (http://meme-suite.org/tools/meme) to identify conserved protein motifs. The optimized MEME parameters were as follows:minimum pattern width 6;maximum pattern width 100;and use the maximum number of programs,including up to 16. The conserved sequences were visualized using Adobe Illustrator 2020 Software,USA.
2.5.Synteny analysis
Based on the method used in the Plant Genome Duplication Database (Leeet al.2013),we use an improved method for synteny analysis. First,a BLASTP search was performed on the entire genome to identify candidate homologous gene pairs (E-value<1×10−5,the first five matches). The candidate genes were analyzed with MCScanX (Wanget al.2012) for detection of syntenic blocks using the default parameters. MCScanX was used to distinguish singleton genes,whole-genome duplication (WGD)/segmental duplication,dispersed duplication,proximal duplication,and tandem duplication events in the strawberry LOX family (Appendix C).
2.6.RNA extraction and gene expression analysis
To evaluate the expression levels of LOX genes in strawberry,total RNA was extracted from collected plant samples or treated leaves using the RNAprep Pure Plant Plus Kit (Tiangen Biotech.,Beijing,China) in accordance with the manufacturer’s recommendations. Before reverse transcription,the RNA was treated with DNase I (Tiangen Biotech) to remove residual DNA contamination. According to the RNA concentration of the sample extract,1-7 μg total RNA was reverse-transcribed into the first-strand cDNA using the FastKing gDNA Dispelling RT SuperMix (Tiangen Biotech). The reaction mixture contained 4 μL of 2× FastKing-RT SuperMix,4 μL total RNA,and 12 μL RNase-free ddH2O in a total reaction volume of 20 μL. The DNA polymerization temperature was 42°C for 15 min and reverse transcriptase was deactivated at 95°C for 3 min. Primer5 Software was used to design gene-specific primers for eachFvLOXgene (Appendix D).
Eyes rolling up in my head, I replied, Tom, you don t understand. You don t just go out and casually4 throw around an all-leather, NFL regulation, 1963 Chicago bears-inscribed football. I told you before; it s special.
Quantitative reverse transcription-PCR (qRT-PCR) reactions (Tiangen Biotech) were performed in a 10 μL volume containing 5 μL of 2× Super Real Pre Mix Plus,0.3 μL forward primers,0.3 μL reverse primers,1 μL cDNA template,and 3.4 μL RNase-free ddH2O.Each amplification was conducted under the following conditions:pre-denaturation of 1 cycle for 15 min at 95°C,followed by 40 cycles of denaturation for 10 s at 95°C and annealing/extension for 32 s at 60°C. The temperature was gradually increased by 0.5°C every 10 s to analyze the melting curve.FvPDB(Hoffmannet al.2006) was used as an internal reference gene to normalize expression data. For each sample,the determination comprised three technical replicates and three biological replicates. Each replication included the internal reference gene. The relative expression level of each gene was calculated using the 2−ΔΔCtmethod,and the standard deviation was calculated from the three biological replicates and three biological replicates (Schmittgen and Livak 2008).
2.7.Analysis of cis-acting regulatory elements in the promoter
Using the woodland strawberry reference genome,the sequence 1 500 bp upstream of the start codon for eachFvLOXgene was selected for analysis ofcisacting elements in the promoter using TBtools Software (Chenet al.2020),and thecis-acting elements were predicted using PlantCARE (Lescotet al.2002) (http://bionformatics.psb.ugent.be/webtools/plantcare/html). The motifs that may be involved in plant growth and development,plant hormone responses,and abiotic and biotic stress responses were summarized (Appendix E).
2.8.Statistical analysis
Statistical significance was determined using Student’st-test as implemented in Graphpadprism7.0 Software.The average±standard deviation of at least three repeated samples was calculated. The significance of differences compared with the control were expressed as*P<0.05 and**P<0.01.
3.Results
3.1.ldentification and phylogenetic analysis of LOX gene in Rosaceae
In this study,14,17,19,14,22,and 24 LOX genes were identified from diploid woodland strawberry (F.vesca),Chinese plum (P.mume),rose (R.chinensis),peach (P.persica),pear (P.betulifolia) and apple (M.domestica),respectively,namedFvLOX,PmLOX,RcLOX,PpLOX,PbLOXandMdLOX(Table 1). In addition,in order to explore the evolutionary relationship betweenA.thalianaand six species of Rosaceae,we constructed a phylogenetic tree based on multiple sequence alignment of LOXs genes from sixArabidopsisLOXs (AtLOX) and 110 Rosaceae plants using MEGA6.0. According to the classification ofArabidopsisAtLOX,the LOX family is divided into two subgroups:9-LOX and 13-LOX. We found that both 9-LOX and 13-LOX contain genes fromArabidopsisand six Rosaceae species,indicating thatArabidopsisLOX gene and Rosaceae LOX gene evolved from a common ancestor. On the basis of this classification,according to the phylogenetic relationship,9-LOX was further divided into Group 9a-9d,and 13-LOX into Group 13a-13g. We found that independent evolutionary events occurred among Rosaceae plants in the subbranches of Group 9a,Group 9b,Grou13a,Group 13b,Group 13c,Group 13d,Group 13e,and Group 13g. Interestingly,Group 13f contains only the LOX gene of roses and strawberries. In addition,the number of pear and apple genes in Group 9c increased sharply,suggesting the potential function of LOX in pears and apples (Fig.1).

Fig.1 Phylogenetic analysis of LOX proteins from Arabidopsis,strawberry,Chinese plum,rosa,peach,pear and apple. The full-length amino acid sequences of LOX proteins from Arabidopsis (AtLOX),strawberry (FvLOX),Chinese plum (PmLOX),rosa (RcLOX),peach (PpLOX),pear (PbLOX),and apple (MdLOX) were aligned by ClustalX,and the phylogenetic tree was constructed using the Maximum Likelihood method with 1 000 bootstrap replicates by MEGA 6.0. The branched lines of the subtrees are colored to indicate different LOX subgroups.

Table 1 Summary of LOX gene family in six species
3.2.Gene structure and conserved motif analysis
We studied the gene structure and intron number ofFvLOXfamily (Fig.2). The results showed that the LOX gene ofA.thalianaand six species of Rosaceae contained introns,and the number of introns ranged from 1 to 11. Most of the introns of LOX gene in 9a,9b,9c,13a,13b,13d,13e,13f,and 13g subfamilies are 8-10. On the whole,most of the LOXs genes in the same subgroup have similar exon-intron structure,and the relative positions and relative positions of introns are highly conserved in each subgroup.
A total of 16 conserved motifs were identified in the LOX amino acid sequence. Fig.2 shows the motif distribution corresponding to the phylogenetic tree of the LOXs gene family. In addition,we looked at the number of motifs in each subgroup. All the LOXs genes in Group 9a did not contain motif 16,eight motifs were identified inMdLOX08,and motif 4 was missing inPbLOX20. All the LOXs in Group 9b contained 16 motifs. In Group 9c,except forPbLOX13lacking motif 4,other LOXs genes were contained 16 motifs. In Group 9d,AtLOX1lacked motif 7,11 motifs were identified inPmLOX17,13 motifs were identified inPmLOX05,and 16 motifs were found in other LOXs. In Group 13a,RcLOX12lacked motif 2 and identified 12 motifs inPmLOX07,while other LOXs contained 16 motifs. In Group 13b,10 motifs were identified inMdLOX12,11 motifs were identified inPbLOX10,and 16 motifs were identified in other LOXs.In Group 13c,AtLOX3lacked motif 13 and identified 11 motifs inRcLOX02,4 motifs inRcLOX01,and 16 motifs in other LOXs. All the LOXs genes in Group 13d were contained 16 motifs. In Group 13e,nine motifs were identified inFvLOX11andFvLOX13,13 motifs were identified inPmLOX13,and 16 motifs were identified in other LOXs. In Group 13f,12 motifs were identified inFvLOX07,13 motifs were identified inRcLOX18,and 16 motifs were identified in other LOXs. In Group 13g,motif 10 and motif 13 were missing inPmLOX11,nine motifs were identified inMdLOX10,motif 12 was missing inMdLOX13,and 16 motifs were found in other LOXs.
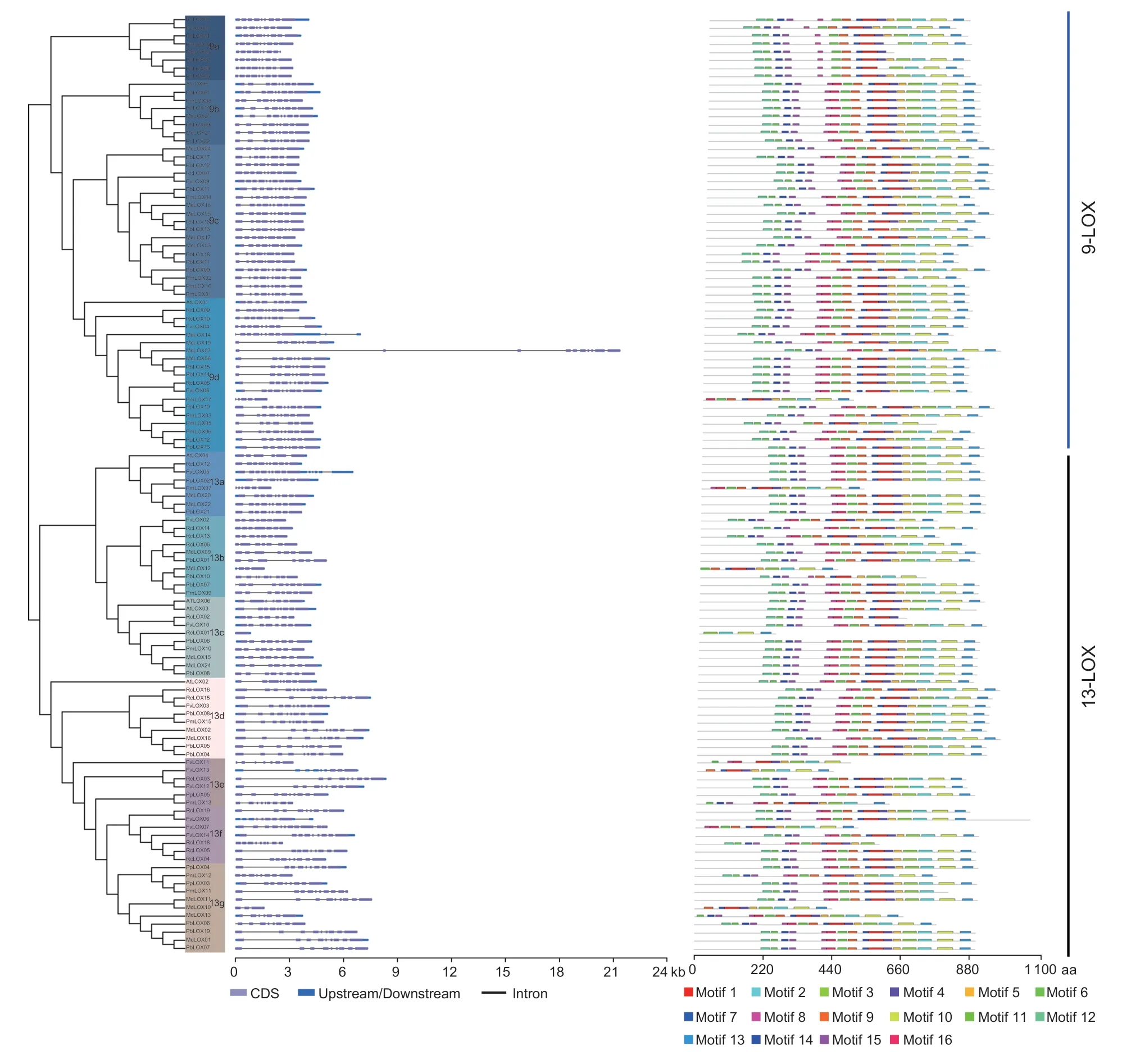
Fig.2 Analysis of gene structure and conserved motif in each LOX group of strawberry. The exon/intron structure of strawberry LOX gene is shown on the left. The blue-gray box represents the intron. The blue box represents upstream/downsteam region. The black lines connecting two exons represent introns. The one on the right represents the distribution of conserved motifs in each LOX gene. Boxes of different colors represent different conservative patterns and show their relative positions.
3.3.Different replication events control the amplification of LOX gene in Rosaceae plants

Fig.3 Comparison of different modes of gene duplication in six species.
3.4.Expression patterns of FvLOX genes in different organs
To study the expression patterns of LOX genes in different organs of diploid woodland strawberry,qRTPCR was used to quantify the expression level ofFvLOXgenes in the root,stem,leaf,flower,and fruit.CertainFvLOXgenes were highly expressed in specific organs,whereas otherFvLOXgenes showed similar expression patterns in different organs (Fig.4),which may be indicative of functional differences of the genes in strawberry growth and development. For example,FvLOX1,FvLOX6andFvLOX11are more expressed in roots than in other tissues,and they may play a role in root growth. The expression ofFvLOX3,FvLOX7andFvLOX10in leaves and flowers was higher than that in other tissues. In particular,FvLOX14showed a very high level of relative expression in leaves,suggesting that it may play a role in the growth of strawberry leaves.FvLOX4,FvLOX8,FvLOX9,FvLOX12,andFvLOX13showed relatively low expression levels in different tissues. It is worth noting that the relative expression levels of mostFvLOXsgenes in stems and fruits are relatively low (Fig.4).

Fig.4 Quantitative RT-PCR analysis of 14 FvLOX genes in different organs of woodland strawberry. Expression levels were normalized to that of FvPDB. The mean and SD were calculated from three biological and three technical replicate samples.
3.5.Expression pattern of FvLOX genes under stress treatments
To explore the potential role ofFvLOXgenes in plant responses to various environmental stresses,the expression level of the 14FvLOXgenes under low temperature,high temperature,drought,and salt stress was determined by qRT-PCR analysis. Generally,theFvLOXgenes differed in the degree of response to low temperature,high temperature,drought,and salt stress. The expression ofFvLOX1andFvLOX8were up-regulated under low-temperature stress,FvLOX3andFvLOX7were up-regulated under drought stress,FvLOX6andFvLOX9were up-regulated under salt stress,and the other genes were down-regulated to varying degrees (Fig.5).
The plant hormones SA,MeJA and ABA play important roles in plant stress signal response and in plant growth and development. To evaluate ifFvLOXgene expression was induced in response to plant hormone treatment,the expression patterns of theFvLOXgenes in the leaf in response to exogenous SA,MeJA and ABA treatment were analyzed by qRT-PCR. In response to SA treatment,the expression levels ofFvLOX2,FvLOX3,andFvLOX6were significantly increased to a high level from 4 h after treatment,and were maintained at a high expression level during the entire experimental period,whereas the other genes were down-regulated in certain periods (Fig.6). Under MeJA treatment,the expression ofFvLOX3,FvLOX11,andFvLOX14were up-regulated,whileFvLOX7andFvLOX8were significantly down-regulated. Under ABA treatment,FvLOX4andFvLOX14were up-regulated to varying degrees,whereasFvLOX3,FvLOX6,FvLOX12,andFvLOX13were significantly down-regulated. It is notable thatFvLOX3was up-regulated in response to treatment with SA and MeJA,FvLOX14was up-regulated in response to treatment with MeJA and ABA (Fig.5).

Fig.5 Hierarchical clustering of the expression profiles of 14 LOX genes in diploid woodland strawberry (Fragaria vesca) in response to different treatments. Expression accumulation profiles of 10 woodland strawberry LOX genes in response to low temperature (4°C),high temperature (42°C),drought,NaCl,salicylic acid (SA),methyl jasmonate (MeJA),and abscisic acid (ABA) treatments as determined by quantitative RT-PCR analysis and displayed in the form of heat maps. The color scale indicates the change in expression level,where red indicates an increase and green indicates a decrease,relative to that at 0 h. The experiment was repeated three times and the results were consistent. Genes were hierarchically clustered using the the average Pearson distance index and the“average link”method.

Fig.6 Quantitative RT-PCR analysis of woodland strawberry LOX gene expression in response to low temperature (4°C),drought,NaCl,salicylic acid (SA),methyl jasmonate (MeJA),and abscisic acid (ABA) treatments. The expression levels of the FvLOX genes revealed the epigenetic patterns in response to the SA,MeJA and ABA treatments. The expression levels were normalized to that of FvPDB. The mean and SD were calculated from three biological and three technical replicate samples. Asterisks indicate that the gene was significantly up-regulated or down-regulated after treatment (*,P<0.05;**,P<0.01;Student’s t-test).
3.6.ldentification of cis-acting regulatory elements in the promoter of FvLOX genes
To explore the regulation of strawberry LOX family members,analysis of thecis-acting regulatory elements in the 1.5 kb region upstream of the initiation codon of allFvLOXfamily members was conducted using the PlantCARE online portal. The GCN4 motif,acisacting regulatory element associated with endosperm expression,was detected inFvLOX6(Fig.7). It was noteworthy that the element involved in the regulation of circadian rhythms were detected inFvLOX3,FvLOX10andFvLOX14.FvLOX1,FvLOX3andFvLOX6were contained an element involved in mechanical injury response (WUN motif). In addition,the gliadin metabolic regulatory element (O2-site),meristem expression and specific activation elements (CAT-box and CCGTCCbox),and seed-specific regulatory element (RY-element) were identified in the promoter ofFvLOXgenes. With regard to hormone-relatedcis-acting elements,the SA response element (TCA-element),methyl jasmonate response elements (CGTCA-motif and TGACG-motif),and ABA response element (ABRE) were identified in the promoters of nine,eight,and sevenFvLOXgenes,respectively. Gibberellin response elements (GAREmotif,P-box and TATC-box) and auxin response elements (TGA-element and AuxRR-core) were observed in eleven and fiveFvLOXgenes,respectively. A large number of elements associated with hormone response were identified in theFvLOXpromoter sequence,which indicated that plant hormones may play a crucial role in regulating the functions ofFvLOXgenes in plant growth and development (Fig.7-B). In addition,severalcisacting elements associated with response to stresses (such as drought,extreme temperature,and salinity) were observed in the promoter region ofFvLOXgenes.
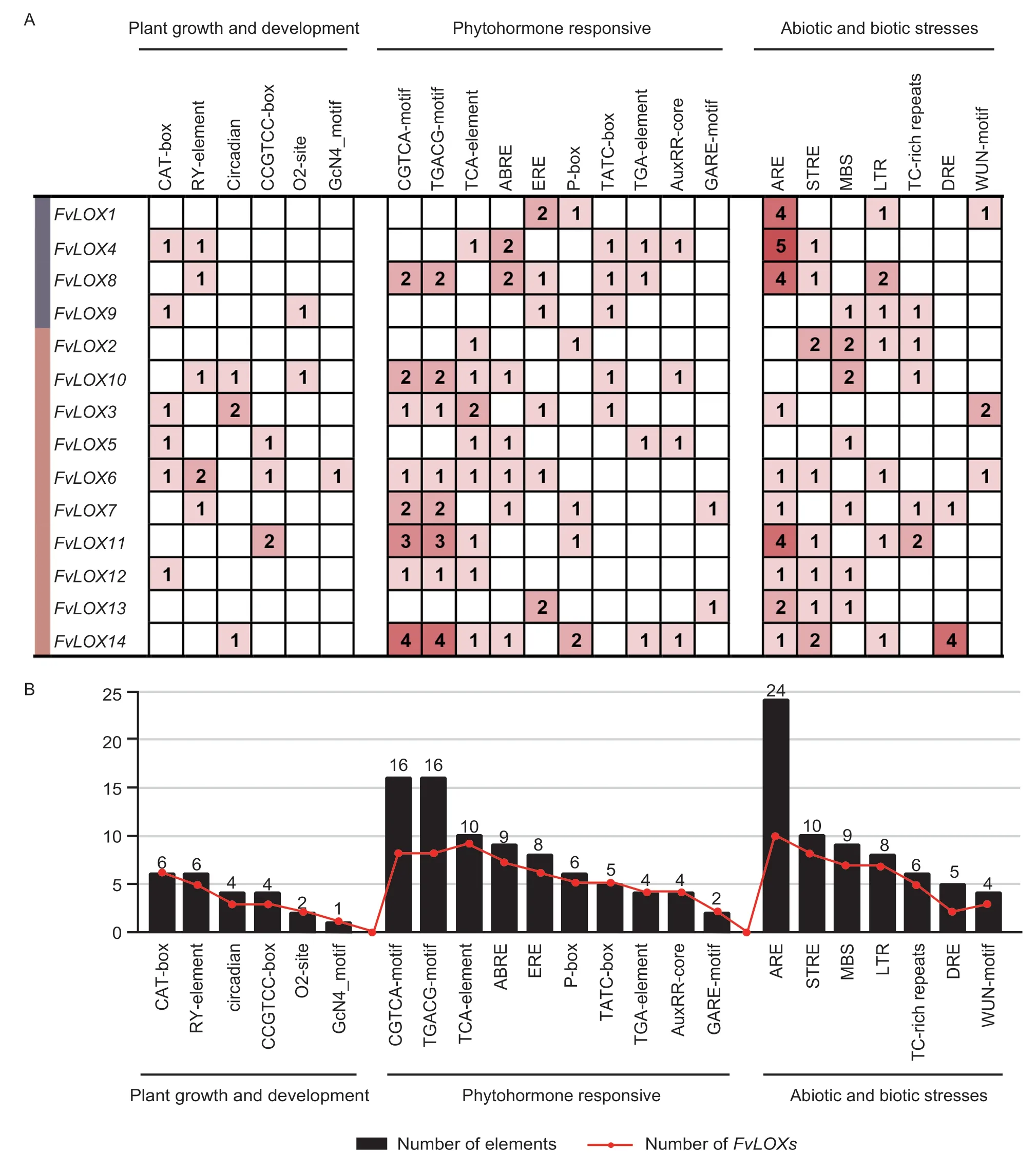
Fig.7 Analysis of cis-acting regulatory elements in the promoter of woodland strawberry LOX genes. A,number of each cis-acting element in the promoter region (1.5 kb upstream of the translation start site) of FvLOX genes. B,statistics for the total number of FvLOX genes,including the corresponding cis-acting element (red dot) and the total number of cis-acting elements in the FvLOX gene family (black box). On the basis of the functional annotation,the cis-acting elements were classified into three major classes:plant growth and development,phytohormone response,and abiotic and biotic stress response.
4.Discussion
LOX gene family plays an important role in plant growth and development and abiotic stress response. However,the LOX gene family has not been studied in strawberries. Therefore,this study hopes to identify candidate genes involved in stress regulation from strawberry LOX gene. Based on the diploid woodland strawberry genome (V4a version),14FvLOXgenes were identified in this study.Decentralized replication is the main driving force for the expansion of strawberry LOX gene family. Strawberry LOX gene was expressed in different tissues and responded to low temperature,high temperature,drought,salinity,SA,MeJA and ABA in varying degrees,indicating thatFvLOXgene has a potential regulatory function in abiotic stress. These results laid a foundation for further study on the role of strawberry LOX gene in abiotic stress.
For gene family evolution,one of the main mechanisms for producing new evolutionary innovation patterns is gene replication,such as tandem replication and WGD/segmental replication. According to the genomic data of six Rosaceae plants,they all experienced a WGD event of a Rosaceae ancestor (Jiaoet al.2011). Recent WGD events can be traced back to apples (Velascoet al.2010) and pears (Wuet al.2013) about 40 million years ago,rather than in woodland strawberries (Shulaevet al.2010),roses (Raymondet al.2018),peaches (IPGIet al.2013) and Chinese plum (Zhanget al.2012). Through the analysis of gene replication events,it was found that the tandem repeat frequency of LOX gene in Chinese plum was higher than that in strawberry and peach,which showed an increase in the number of LOX gene in Chinese plum. In pears and apples,WGD/segmental replication is another major driving force for the expansion of the LOX gene family. As a result,more LOX genes were found in pears and apples,possibly because the two species experienced two WGD events. In addition,the LOX gene of Chinese plum only experienced tandem replication,dispered replication and proximal replication,and there was no WGD/segmental replication. This shows that the replication of LOX gene family in these Rosaceae plants is diversified. Previous studies have shown that a gene family may have a common non-random origin pattern and conservative replication patterns in different species (Rodgers-Melnicket al.2012). However,in this study,it is found that the main replication patterns of LOX gene families in six species of Rosaceae are not always strictly conservative,and non-random patterns from different sources are common.
Previous studies have shown that some LOX genes have unique expression patterns,which may provide important clues for understanding their physiological functions. InA.thaliana,AtLOX1andAtLOX5can control the emergence of lateral roots through the production of 9-HOT,thus acting as regulators of root development (Vellosilloet al.2007). In this study,it was found thatFvLOX1andFvLOX8were highly expressed in roots,andFvLOX1,FvLOX8,AtLOX1,andAtLOX5all belonged to the 9-LOX subfamily,suggesting that they may have similar functions in roots. Compared with the control plants,the expression ofAtLOX2in leaves and flowers of transgenic plants was up-regulated,indicating thatAtLOX2plays an important role in the development of leaves and flowers (Bellet al.1995). We found thatFvLOX3andFvLOX10are highly expressed in leaves and flowers,andFvLOX3,FvLOX10andAtLOX2all belong to the 13-LOX subfamily,suggesting that they may have similar functions in leaves and flowers. It is worth noting that mostFvLOXgenes have low expression levels in fruits,and similar results have been found in peaches (Zhanget al.2006),grapes (Andriyet al.2010),watermelons (Liuet al.2020) and tomatoes (Upadhyay and Mattoo 2018). The low expression of LOX gene in fruits of these plants indicates that plants may need LOX activity for cell division and fruit expansion in the early stage of development.
Abscisic acid (ABA) plays an important role in the response of higher plants to environmental stress during plant growth and development. One of the responses mediated by ABA is to induce gene expression,which is regulated by abscisic acid response element (ABRE).Previous studies have shown that single-copy ABRE elements are not enough to give ABA the ability to respond to promoters,while multi-copy ABRE elements give ABA the ability to respond to promoters (Hoboet al.1999). We found two ABRE elements in the promoter ofFvLOX4,which may be the reason for the upregulation ofFvLOX4expression. Previous studies have shown thatAtLOX1washighly expressed under ABA treatment. In this study,it was found thatFvLOX4was up-regulated by 15.92 times during the treatment of ABA,and bothFvLOX4andAtLOX1belonged to the 9-LOX subgroup,suggesting that they may have similar functions. Related reports show that the response ofrd29Agene to ABA expression is due to the presence of DRE element and ABRE element in the promoter. DRE element acts as a coupling element of ABRE in ABA-dependent gene expression (Narusakaet al.2003). We found an ABRE element and a DRE element in theFvLOX14promoter,which may be the reason whyFvLOX14responds to the upregulation of ABA expression. SA plays an important role in regulating plant response to abiotic stress. Under SA treatment,the expression ofFvLOX2,FvLOX3andFvLOX6was up-regulated. Promoter analysis showed that they all contained TCA-element,and speculated that they might be the cause of their upregulated gene expression. Similarly,methyl jasmonate plays an important role in regulating plant growth and environmental response. Under MeJA treatment,the expression of MeJA was up-regulated in response toFvLOX3,FvLOX11,andFvLOX14. CGTCA-motif and TGACG-motif elements were found in their promoters,which may be the reason for their up-regulation.
Temperature is one of the main environmental factors that restrict plant growth and development and affect plant geographical distribution. Low temperature and high temperature stress will lead to plant stunting and reduce crop yield. Therefore,it is very important to improve the ability of plants to resist low temperature and high temperature stress. Previous studies have found that barleyblt4.9gene promoter contains CCGAAA sequence. Through the mutation analysis of CCGAAAmotif,it is found that the deletion of this motif reduces the basic level of response to low temperature,indicating that CCGAAA-motif plays an important role in barley low temperature stress response (Dunnet al.1998). In this study,CCGAAA-motif elements were found inFvLOX1andFvLOX8promoters,suggesting that they may be the reason for their up-regulation under low temperature stress. In particular,this study found that there was no upregulation ofFvLOXgene expression under high temperature stress. Previous studies have shown that heat shock response element (HSE) exists in heat shock protein gene. Heat shock factor (HSF) stimulates the expression of heat shock proteins by binding to HSE (Storozhenkoet al.1998). We did not find HSE elements in the promoter ofFvLOXgene,which may be the reason why there is no up-regulated expression ofFvLOXgene under high temperature stress.
Drought and high salt stress have great effects on plant growth and productivity. These stresses induce the expression of many genes in different plants. Previous studies have shown that GT-1 components directly control the up-regulation ofOsRAV2reaction in high salt (Duanet al.2015). In this study,GT-1 (GAAAAA) elements were also found in the upstream of the promoters ofFvLOX6andFvLOX9. It is speculated that this element may be the reason for the up-regulation of gene expression under salt stress. InA.thaliana,AtLOX3was significantly induced by salt treatment (Huiet al.2016). Phylogenetic analysis showed thatFvLOX6andAtLOX3belong to 13-LOX subgroup,suggesting that they may have similar functions in salt stress. Under drought stress,the expression ofFvLOX3andFvLOX7was up-regulated. Previous studies have found that ABRE and DRE elements are important causes ofOsDhn1expression (Kumaret al.2014). We also found ABRE and DRE elements inFvLOX7,which may be an important reason for the upregulation ofFvLOX7expression. In addition,FvLOX3is also up-regulated in response to drought stress,but there is a lack of relevant reports and there is no more basis for description. We speculate that the upregulation ofFvLOX3expression may be due to the existence of some new motifs in the promoter,which may be the key elements of their response to drought stress.
Different environmental factors affect gene expression,and gene expression also requires the coordination of induciblecis-acting regulatory elements and transcription factors in the promoter of environment-responsive genes. The type,number,and location of these elements may affect the level of gene expression. Therefore,further research is needed to gain an improved understanding of transcriptional regulatory mechanisms in strawberry,including transcription factors and their specificcis-acting regulatory elements.
5.Conclusion
We identified 14FvLOXfamily members in the genome of woodland strawberry. Dispered duplication is indicated to have been the primary driving force for expansion of the strawberry LOX family. TheFvLOXgenes were differentially expressed in vegetative and reproductive organs of strawberry. The genes showed varying degrees of response to low temperature,high temperature,drought,and salinity stress,and to exogenous SA,MeJA and ABA treatment. The present results provide a basis for investigation of the transcriptional regulatory mechanisms of growth and development,and the functional identification of stress-resistance genes in strawberry.
Acknowledgements
This work was supported by the Science and Technology Innovation Fund of Fujian Agriculture and Forestry University (KHF200005). The authors would like to thank anonymous reviewers for comments on this manuscript.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm
 Journal of Integrative Agriculture2022年7期
Journal of Integrative Agriculture2022年7期
- Journal of Integrative Agriculture的其它文章
- Roles of mushroom polysaccharides in chronic disease management
- lnfluence of high-molecular-weight glutenin subunit deletions at the Glu-A1 and Glu-D1 loci on protein body development,protein components and dough properties of wheat (Triticum aestivum L.)
- Variations in the quality parameters and gluten proteins in synthetic hexaploid wheats solely expressing the Glu-D1 locus
- ldentification of candidate genes related to soluble sugar contents in soybean seeds using multiple genetic analyses
- ldentification and characterization of long-lnDels through whole genome resequencing to facilitate fine-mapping of a QTL for plant height in soybean (Glycine max L.Merr.)
- Photosynthetic properties of the mid-vein and leaf lamina of fieldgrown,high-yield hybrid rice during senescence
