Comparative transcriptome analysis provides insights into the mechanism of pear dwarfing
TANG Zi-kai ,SUN Man-yi ,Ll Jia-ming ,SONG Bo-bo ,LlU Yue-yuan ,TlAN Yi-ke ,WANG Cai-hong,WU Jun
1 College of Horticulture,State Key Laboratory of Crop Genetics and Germplasm Enhancement,Nanjing Agricultural University,Nanjing 210095,P.R.China
2 College of Horticulture,Qingdao Agricultural University,Qingdao 266109,P.R.China
Abstract Dwarfism is an important trait which is closely related to the efficiency of fruit orchard management and production.However,dwarfing cannot be widely applied in the cultivation of pears,especially Asian pears. Developing varieties with dwarf characteristics is a goal of paramount importance in pear breeding. In the present study,dwarf phenotype pears (DPPs) and arborescent phenotype pears (APPs) were obtained from the offspring of a cross between ‘Aiyuxiang’ and ‘Cuiguan’ pear cultivars,which exhibited dwarfed and arborescent statures,respectively. When compared with APPs,the heights of DPPs showed a 62.8% reduction,and the internode lengths were significantly shorter. Crossgrafting between DPPs and APPs demonstrated that the dwarfed phenotype of DPPs was primarily induced by the aerial portions of the plant,and independent of the root system. Observations of stem tissue sections showed that DPP cells were arranged chaotically with irregular shapes,and the average length was larger than that of the APP cells. A total of 1 401 differently expressed genes (DEGs) in shoot apices between DPPs and APPs were identified by RNA-sequencing (RNA-Seq),and these DEGs were mainly enriched in the ‘phytohormone-related pathways,cell wall metabolism and cell division’ categories. Moreover,101 DEGs were identified as transcription factors (TFs). In DPPs,several brassinosteroids (BR) signaling and cell cycle-related genes were significantly down-regulated,while genes involved in BR and GA degradation were up-regulated. Comprehensive analysis of RNA-Seq data and stem tissue sections suggested that the dwarfed phenotype of DPPs could be primarily attributed to deficiencies in cell division. Previous work using simple sequence repeat (SSR) markers narrowed the location of the gene responsible for the dwarf phenotype of ‘Le Nain Vert’. Through combined analysis of our transcriptomic data with the SSR results,we identified four genes as promising candidates for the dwarf phenotype,among which,a DELLA gene could be themost promising. The results presented in this study provide a sound foundation for further exploration into the genetic and molecular mechanisms underlying pear dwarfing.
Keywords:pear,dwarf phenotype,RNA-Seq,cell division,phytohormones
1.lntroduction
Dwarfism is a critical horticultural trait of many perennial fruit crops. As it makes overall orchard management easier and produces cost-saving benefits,dwarfing cultivation is widely applied in the production of many fruit species (Chenget al.2019;Ouet al.2019;Xuet al.2020). Pears are extensively cultivated worldwide and highly favored by consumers. Most pear cultivars have tall statures,which pose various challenges for orchard management. Meanwhile,few dwarfing pear rootstocks have been developed and widely produced,other than the Quince rootstocks which were used as a dwarfing rootstock for some European pears. Thus,the improvement of pear tree architecture has long been a point of interest for breeders (Qiet al.2020). Therefore,exploring the genetic and molecular mechanisms underlying dwarfism in pear is important for the advancement of breeding efforts to produce more dwarf pear cultivars and dwarfing rootstocks.
Stem elongation is affected by multiple factors. As the origin of all aerial organs of a plant,shoot apical meristem (SAM) development is tightly linked to plant height (Hanet al.2020). Stem elongation is severely inhibited when the homeostasis in SAMs is disrupted. For example,theKNOTTED-LIKE HOMEODOMAIN 4(Tkn4) gene is involved in maintaining SAM homeostasis,andTkn4overexpression in tomato produce developmentally defective SAMs that resulted in a dwarf phenotype (Yanet al.2015). Additionally,the ability of roots to absorb and transport water and nutrients also influences stem growth. TheSHORT-ROOT 1(SR1) gene is mostly expressed in the root system of rice plants,and plants harboring thesr1mutation experienced a reduction in water potential and exhibited a dwarfed phenotype (Xinget al.2020). In studies at the cellular level,plant dwarfism is typically attributed to suppressed cell division and/or cell expansion (Panget al.2019;Guo and Simmons 2011). The cell cycle,which is divided into the four phases of M,G1,S and G2,is a temporal regulator of cell division and expansion in proliferative cells (Joneset al.2017). Accurate control of cell cycle progression is essential for normal plant growth. In rice plants harboring the albinic leaf and growth retardation (alr) mutation,the cell cycle becomes arrested at the G1/S-phase due to a deficiency of deoxycytidine monophosphate deaminase (DCD),which results in a dwarfed stature (Niuet al.2017). The processes of both plant cell division and expansion involve cell wall remodeling (Speicheret al.2018),and defects in cell wall metabolism result in inhibited plant growth.OsPMEI28is a regulator of pectin methylesterification. Overexpression ofOsPME128in rice plants induced changes in the cell wall components,resulting in reductions in both plant height and culm diameter (Nguyenet al.2017).
Phytohormones are known as the center of the plant growth-regulatory network (Deepikaet al.2020).Many plant dwarfing mechanisms are associated with various phytohormones,as revealed by previous studies (Sazukaet al.2009;Wanget al.2014;Nemotoet al.2017;Taoet al.2020). Among all of the known phytohormones,gibberellin (GA) and brassinosteroids (BRs) are the ones predominantly associated with plant height. Changes in expression of the genes involved in GA and BR metabolism have been found to be responsible for dwarfing. InArabidopsis,Gibberellin2-oxidases 6(GA2ox6) andCYP72C1participate in the degradation of bioactive GA and BR,respectively,and upregulation of these genes results in reductions in plant height (Takahashiet al.2005;Huet al.2017). Similar to the results observed by reducing GA and BR contents,suppression of their respective signaling pathways also yields a dwarfed phenotype. DELLA proteins (DELLAs) are key transcriptional repressors involved in GA signaling,and GA is known to promote plant growth by inducing the degradation of DELLAs. Previous studies found that a single-base substitution in the coding sequence ofGA receptor1 c(GID1c) abolishes its role in mediating DELLA degradation,resulting in a dwarfed phenotype in peaches (Chenget al.2019). In theBrachypodiumloss-of-functionBRI1-RNAi mutant,the expression of genes involved in BR signaling,such asBdBZR1,BdBES1andBdBLE2,was down-regulated which also resulted in a dwarfed phenotype (Fenget al.2015).
Many transcription factor (TF) families have also been found to participate in plant height regulation. Many of these TFs are related to phytohormones. They generally act upstream of phytohormone metabolism pathways or downstream of phytohormone signaling pathways,and further participate in phytohormone-regulated plant growth. Overexpression ofGmMYB14reduced the height of soybean by activating the expression ofBEN1to decrease BR levels (Chenet al.2020).OsEATB,which encodes an ERF/AP2 protein,was identified as a suppressor of GA biosynthesis,and its overexpression in rice plants reduced overall plant height and panicle length (Qiet al.2011). OFP1,a member of the Ovate Family Protein class of plant growth regulators,was strongly induced by BR and contributed to the inhibition of plant growthviaincreased BR accumulation or enhanced BR signaling (Xiaoet al.2017).
In pears,several dwarf cultivars and dwarfing rootstocks have been characterized,such as ‘Nain Vert’ (Wanget al.2011) and ‘Zhongai 1’ (Ouet al.2019),however,the molecular mechanisms underlying these dwarfed phenotypes in pears are still unclear. A previous report has identified the gene contributing to the dwarf phenotype of ‘Le Nain Vert’ asPcDwusing simple sequence repeat (SSR) markers (Wanget al.2011,2016). Subsequently,several genes have been identified as candidates forPcDw(Xiaoet al.2019),however,no further information is available on this topic. In this study,a trait segregant population for plant height was obtained by crossing ‘Aiyuxiang’ (Pyrus communis×Pyrus bretschneideri) which inherited the dwarf character of ‘Le Nain Vert’ (P.communis) and ‘Cuiguan’ (P.bretschneideri) which exhibits an arborescent stature. The dwarf phenotype pear (DPP) represents hybrid individuals with dwarfed stature,and arborescent phenotype pear (APP) represents hybrid individuals with arborescent architecture. Cytological observations were used to confirm the cellular basis of variations in the DPPs. We further cross-grafted DPP and APP to investigate which tissue (the aerial parts or roots) was responsible for inducing the dwarf phenotype of DPPs. Furthermore,expression differences in the genes associated with plant growth were identified by RNA sequencing (RNASeq). Our study provides new insights into the genetic and molecular mechanisms underlying dwarfing in pear plants.
2.Materials and methods
2.1.Plant materials
The dwarf pear cultivar ‘Aiyuxiang’ (female parent) was crossed with ‘Cuiguan’ (arborescent phenotype;male parent) and a hybrid pear population was obtained in Qingdao Agricultural University,China. The population was grown in the pear germplasm resource nursery under normal field management conditions. This population was divided into two groups according to plant height and designated as DPP and APP,respectively. The shoot apices of DPP and APP plants were collected in the middle of June of that year,which is the rapid development stage of shoots and the plants show the difference between the dwarfing and arborescent phenotypes. To ensure the accuracy of results,three plants in each group were randomly chosen as three biological replicates. All the samples were stored at -80°C.
2.2.Paraffin section preparation and analysis
The first internodes (including the shoot apex) of three plants from DPP and APP were randomly collected as the material for paraffin sectioning. The internodes were immersed in formalin-acetic acid-alcohol (FAA) fixative and sent to the Servicebio Company (Wuhan,China) for paraffin section preparation and scanning. The cell length in each paraffin section image was measured using a Pannoramic Viewer 1.15.3 (3DHISTECH Ltd.). Then,the total number of cells in a single row was estimatedviadividing the plant height by the average cell length,according to the method used in previous research (Ripettiet al.2008),with slight modifications.
2.3.Grafting
A total of 10 healthy leaf buds with 1 cm of stem below the buds were randomly collected from 10 DPP plants and 10 APP plants. The stem below the buds was cut to fit the shape of a two-sided wedge. Then,10 APP plants and 10 DPP plants exhibiting similar growth conditions were randomly selected and truncated to the same height to form the rootstocks. A longitudinal incision was made on each rootstock,and the wedge-shaped scions were inserted in the incisions of the rootstock. Plastic film was used to wrap the graft junction to reduce water loss and secure the scions.
2.4.RNA extraction and reverse transcription
The total RNA of six independent samples were extracted using the Plant Total RNA Isolation Kit (FORGENE Co.,Ltd.,Chengdu,China) according to the instructions. The concentration and purity of products were detected using the Thermo NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific,USA). High-quality RNA from each sample (500 ng) was used for reverse transcription. The HiScript 1st Strand cDNA Synthesis Kit (Vazyme Biotech Co.,Ltd.,Nanjing,China) was used to perform reverse transcription.
2.5.Library construction and lllumina sequencing
mRNA was enriched using Oligo(dT) magnetic beads (Thermo Fisher Scientific,USA) and then broken using fragmentation buffer (Ambion,USA). The mRNA fragments were used as the reference sequence,and random hexamers were used for synthesizing singlestranded cDNA. Double-stranded cDNA was synthesized,and Poly-A tail and sequencing adaptors were added. The final cDNA library was constructed by PCR amplification,and then sequenced on the Illumina HiSeq™ 2500 platform (Novogene Corporation,Tianjing,China).
2.6.Analysis of transcriptome data
The raw reads of six samples were filtered to remove the adaptor sequences and obtain high quality reads through Trimmomatic (version:0.39) Software (Bolgeret al.2014),and the clean reads were mapped to the Chinese white pear genome using HISAT2 Software (version:2.1) (Kimet al.2019). SAMtools (version:1.9) (https://github.com/samtools/samtools) Software was used for converting and sorting the Sequence Alignment/Map file. Quantification of gene expression levels was estimated by Reads Per Kilobase per Million mapped reads (RPKM) using the FeatureCounts (Liaoet al.2014) (version:1.6.3) Software and an in-house Python script.
To evaluate sample repeatability,the RPKM values from six samples were used for correlation analysis by using the ‘rcorr’ function of the Hmisc/R package (https://cran.r-project.org/web/packages/Hmisc/index.html),and clustered and visualized by using the Pheatmap/R package (https://cran.r-project.org/web/packages/pheatmap/index.html). PCA was performed using the prcomp/R function (https://www.r-project.org/),and the ggplot2/R package (https://ggplot2.tidyverse.org/) was used for visualization.
2.7.Differential expression,function annotation,and enrichment analysis
To compare the differential expression of genes between DPP and APP plants,the DESeq2/R package (http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) was used for filtering and detecting differentially expressed genes (DEGs). First,genes with read counts lower than 20 were filtered in all samples. Then,differential expression analysis was performed using the ‘DEseq’ function. Finally,genes with an adjustedP-value≤0.05 and a |log2(fold-change)|≥1 were identified as DEGs for further function annotation and enrichment analysis.
For analyzing the potential functions of the DEGs,Gene Ontology (GO),BioCyc and Kyoto Encyclopedia of Genes and Genomes (KEGG) function annotations were performed. Furthermore,the enrichment analysis was performed using the website of KOBAS3.0 (http://kobas.cbi.pku.edu.cn/kobas3/genelist/) with Fisher’s exact test and Benjamini and Hochberg correction methods. The results were displayed using the ggplot2/R package (https://github.com/tidyverse/ggplot2).
2.8.Collinear analysis between scaffold00074 and the ‘Dangshansuli’ genome
The protein sequence and annotation files of scaffold00074 fromP.communisandP.bretschneiderigenome were downloaded from the Genome Database of Rosaceae (https://www.rosaceae.org/) and the Pear Genome Project (http://peargenome.njau.edu.cn/). BLASTP was used to make a BLAST search with e-value<1E-10 and the top five matches. The BLAST output and the annotation file were imported into MCScanX Software (Wang Yet al.2012) to identify collinear blocks with default parameters (Tanget al.2008).
2.9.RT-qPCR validation of RNA-Seq data
Specific primers of 15 genes were designed using Primer 5.0 Software,andPyrus UBQwas used as an internal reference (Appendix A). The 10 μL RT-qPCR reaction consisted of the following:5 μL of LightCycler 480 SYBR Green I Master,1.25 μL of each primer (0.5 mmol L-1),2.4 μL of ddH2O,and 0.1 μL of cDNA. All reactions were conducted on the LightCycler 480 II (Roche,USA) using the follow cycling conditions:95°C pre-denaturation for 5 min,45 cycles of 95°C denaturation for 3 s,60°C annealing for 10 s,and 72°C extension for 30 s,followed by a 72°C final extension for 5 min. Finally,fluorescence acquisition was performed at 60°C for 30s. The average threshold cycle (Ct) was calculated at the end of the program. The 2-ΔΔCtmethod was used to calculate the relative expression levels.
3.Results
3.1.Phenotypic analysis of the hybrid offspring of ‘Aiyuxiang’ and ‘Cuiguan’ pear cultivars
Dwarf phenotype pears and arborescent phenotype pears were obtained by crossing the ‘Aiyuxiang’ and ‘Cuiguan’ cultivars. Significant differences in plant architecture were observed between DPPs and APPs. The DPPs had thick upright main stems with few lateral branches (Fig.1-A). Under the same growth conditions,the heights of DPPs were significantly (P=3.61E-38,t-test) shorter than those of APPs,reaching only 37.2% the average height of the latter (Fig.1-B). In addition,the average internode length of each DPP was less than 1 cm,which was significantly (P=9.23E-28,t-test) lower than the average of the APPs at 2.2 cm (Fig.1-C and E). These results suggest that the differences in plant height observed between DPP and APP may be caused by reduced internode length in DPP.
To identify the plant organs responsible for inducing the dwarfing phenotype,the two types (DPP and APP) of pears were cross-grafted. The leaf buds of ten APP were grafted onto ten DPP plants (APP scions+DPP rootstocks,labeled Graft A),andvice versa(DPP scions+APP rootstocks,labeled Graft B). After one-year of growth under routine field management conditions,six trees from the ‘Graft A’ group and eight from the ‘Graft B’ group (Fig.1-D) survived. The morphology of all ‘Graft A’ plants was consistent with APP,while all of the ‘Graft B’ plants exhibited dwarfing architecture. The plant heights and internode lengths of Graft A were still significantly (P=9.69E-10,t-test) larger than those of ‘Graft B’ (Fig.1-E-G). These results indicate that the dwarfing phenotype is controlled by the aerial plant parts of the F1hybrid population of the ‘Aiyuxiang’ and ‘Cuiguan’ cross.

Fig.1 Phenotypic characterization of dwarf phenotype pear (DPP),arborescent phenotype pear (APP) and their cross-grafted plants. A,morphotypes of DPP and APP plants. B,heights of APP and DPP plants. C,average internode lengths of APP and DPP plants. D,morphotypes of cross-grafted pear plants. Graft A,scion of APP grafted onto rootstock of DPP;Graft B,scion of DPP grafted onto rootstock of APP. E,comparison of internodes. F,average internode lengths of grafted plants. G,overall heights of grafted plants. Bars are SD (for Graft A,n=6;Graft B,n=8;APP,n=30;DPP,n=30). **,P<0.01.
Cell division and cell expansion are directly responsible for stem elongation. To investigate the cellular basis of the dwarfing phenotype,the first internodes from the tops of DPPs and APPs were evaluated using microscopic observations of longitudinal paraffin sections. Compared with APPs,the cells of DPPs were irregularly and loosely arranged (Fig.2-A and B),and the cortex of DPP plants was much thicker than that of APP plants (Fig.2-B).Notably,the average cell length of DPPs (15.85 μm) was significantly (P=4.56E-3,t-test) longer than APPs (10.77 μm) (Fig.2-C). The number of cells in the DPP plants was only 31.67% of that in APP plants (Fig.2-D).These results indicate that the dwarfed growth of DPP might be caused by deficient cell division,rather than repressed cell expansion.

Fig.2 The anatomical structures of stems from arborescent phenotype pear (APP) and dwarf phenotype pear (DPP) plants. A,cells in the pith of APPs and DPPs. B,cells in the cortex of APPs and DPPs. C,cell lengths of APPs and DPPs. Bars are SD (n=50). D,estimated cell numbers of APPs and DPPs. Bars are SD (n=30). **,P<0.01.
3.2.ldentification of DEGs between shoot apices of DPP and APP plants
To further investigate the molecular mechanisms underlying dwarfing of DPP plants,shoot apical tissues were collected from three DPPs and three APPs,and used for RNA-Seq to identify key genes that might be involved in regulating the dwarfing phenotype. Approximately 256 million raw reads were obtained with an average of 42.67 million reads per sample. After adaptor sequences,and lowquality and N-containing reads were removed,an average of 40.75 million reads remained for each sample,and the average clean-reads rate reached 95.46%. Clean reads were then mapped to the ‘Dangshansuli’ pear genome (P.bretschneideri) (Wuet al.2013). The mapping rates of all samples were over 78%,and the average paired mapping rate exceeded 70% (Table 1). High correlations between the three biological repeats were observed in both groups (r>0.92). Cluster analysis and principal component analysis (PCA) revealed that six samples were differentiated into two clusters (Appendix B). These results illustrated the similarities between biological replicates.
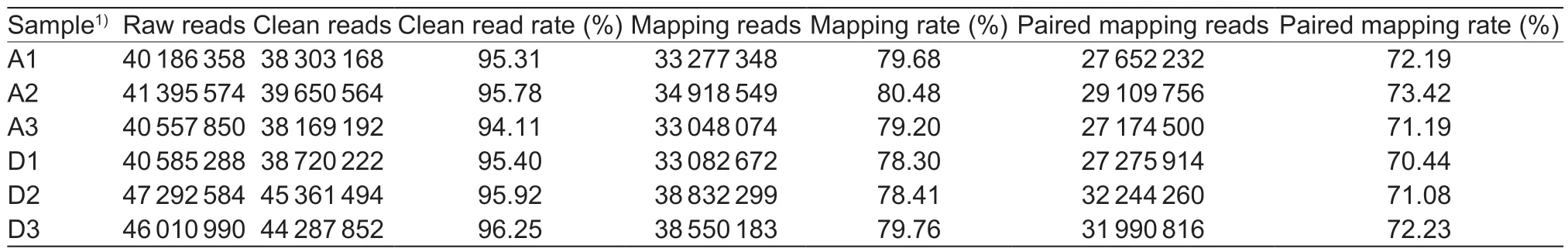
Table 1 Summary of RNA-Seq data
From the two groups of samples,the expression data for 34 582 genes was obtained,excluding genes with RPKM value=0 in all samples (Appendix C). A total of 1 401 genes were found to be differentially expressed (Appendix D). Compared with APP plant gene expression levels,714 genes in DPP were up-regulated and 687 genes were down-regulated (Appendix E).
3.3.Functional enrichment analyses of DEGs
To understand the potential functions of DEGs,Gene Ontology analysis (GO) was performed. The 1 401 DEGs were classified into three GO categories:biological process,cellular component,and molecular function. In the molecular function category,protein binding (220),mRNA binding (108),and DNA-binding transcription activity (89) were the most abundant terms. Signaling receptor activity (P=5.20E-9) and UDP-glycosyltransferase activity (P=1.25E-4) were also significantly enriched. For the cellular component category,chloroplast (466),cytosol (254) and cytoplasm (251) were the predominant subcategories. Additionally,the cell wall (P=4.76E-6) and DNA replication factor A complex (P=3.79E-3) categories were also significantly enriched,with 38 and five genes,respectively. For the biological process category,regulation of transcription (76),response to abscisic acid (39) and oxidationreduction process (47) were the top three returned terms,and five subcategories were involved in stress responses such as light,wounding,water deprivation,cold and bacterial (Fig.3-A;Appendix F). These results indicate that many transcription factors (TF),signaling receptors,cell division-related genes,chloroplast-related genes and stress-response factors might participate in the regulation of DPP plant height.
To further identify and characterize the critical pathways and genes which may be involved in the dwarfing trait in pears,KEGG pathway analysis was also carried out based on the 1401 DEGs. A total of 103 KEGG pathways were enriched (Appendix G),and the top 20 pathways are shown in Fig.3-B. Metabolic pathways,photosynthesis,and biosynthesis of secondary metabolites were the three most significantly enriched pathways. It is noteworthy that many plant hormone-related pathways are significantly enriched,including terpenoid biosynthesis,zeatin biosynthesis,MAPK signaling pathway-plant,brassinosteroid biosynthesis,carotenoid biosynthesis,and plant hormone signal transduction,and phenylpropanoid biosynthesis. Furthermore,Bio Cyc database enrichment analysis showed that the metabolic pathways of multiple plant hormones were also enriched,including gibberellin (GA),brassinosteroid (BR),ethylene (ETH),abscisic acid (ABA),auxin,jasmonic acid (JA),and cytokinin (CTK) (Fig.3-C;Appendix H). These results support the idea that plant hormones may play an important role in the dwarfing phenotype.

Fig.3 Functional enrichment of differently expressed genes (DEGs). A,GO classification. B and C,bubble diagrams of KEGG pathway and Bio Cyc pathway,respectively. The size of each bubble indicates the number of genes enriched to that KEGG pathway. The colors represent the significance,where red represents a higher -log10(P-value) and thus higher significance,and blue represents a lower -log10(P-value) and thus lower significance.
3.4.Expression differences of phytohormone-related genes
According to the results of GO,KEGG and Bio Cyc pathway enrichment analyses,a total of 45 genes involved in seven plant hormone metabolism and signal transduction pathways were identified. Eight DEGs were involved in GA-related pathways:Pbr041942.1andPbr006571.1encoding GIBBERELLIN INSENSITIVE DWARF1(GID1),had higher expression levels in DPP;two gibberellin regulated proteins (Pbr007305.1andPbr032532.1) were down-regulated in DPP;and two DELLA proteins (Pbr014064.1andPbr012085.1) had diametrically opposed expression trends between DPPs and APPs. Interestingly,twoCYP714A1(Pbr001398.1andPbr001399.1) genes,the products of which can degrade active gibberellin (Nomuraet al.2013),had significantly higher expression levels in DPPs than APPs. In BR-related pathways,theBZR1/BES1genes (Pbr022869.1,Pbr000539.1,andPbr016089.1) with important roles in BR signal transduction were downregulated in DPPs. The same trend also occurred for a BR synthetic gene,CYP90A1/CPD(Pbr009519.1).However,Pbr006166.1annotated asCYP734A1,showed the opposite expression trend,which makes it appear to be a negative regulator of BR activity. The other 23 genes were involved in ABA,auxin,CTK,ETH,and JA-related pathways. These results indicate that genes involved in plant hormone metabolism and signaling pathways had significantly different gene expression profiles,and may further affect the growth and development of DPPs (Table 2).

Table 2 Differential expression of phytohormone metabolism and signaling-related genes
3.5.Differentially expressed TFs
To identify TFs associated with the dwarfed pear phenotype,the 1 401 DEGs were scanned using iTAK,with default parameters (Zhenget al.2016). A total of 101 DEGs were identified as transcription factors,and further divided into 38 categories (Appendix I). Among these genes,67 were up-regulated and 34 were downregulated in DPP plants.AP2/ERFwas the most enriched TF family with a total of 14,and most of these genes were highly expressed in DPPs. Four out of sevenbZIPs1)DPP,dwarf phenotype pear;APP,arborescent phenotype pear.were expressed at low levels,while five of sevenbHLHs were expressed at a higher level in DPPs than APPs.Interestingly,all nineMYBs,fourTifys and sixNACs identified from DEGs were expressed at higher levels in DPP plants. These differentially expressed TFs may participate in regulating the dwarfed phenotype in pears.
3.6.The cell cycle and cell wall-related genes
A total of 26 DEGs involved in the cell cycle and cell wall metabolism were differentially expressed between DPP and APP plants.CYCA-2(Pbr022014.1),CYCD3-1(Pbr019683.1) andCYCU4-1 (Pbr002851.1),annotated as cyclins which promote the cell cycle process,were down-regulated in DPP. Two genes (Pbr032386.3andPbr009388.1) annotated asCELL CYCLE SWITCH 52B,targeting mitotic cyclins for degradation and stimulating the conversion of mitotic cycles to endocycles,respectively (Xiaoet al.2020),exhibited lower expression levels in DPPs (Fig.4-A),while a cell cycle inhibitor (Pbr025495.1) was highly expressed (Takatsukaet al.2015). According to GO analysis,17 DEGs associated with cell wall loosening,organization,biogenesis and degradation were identified. Among these,four genes (Pbr034745.1,Pbr032622.1,Pbr039073.1,andPbr009385.1) annotated asexpansinwere downregulated in the DPPs.Pbr017885.1andPbr036871.1,both involved in cell wall biogenesis,were expressed at a lower level in DPP plants. However,four genes encoding galactosidases showed irregular expression patterns,in which two were up-regulated and the other two were down-regulated in DPPs (Fig.4-B).

Fig.4 Expression changes of cell cycle and cell wall-related genes. A,cell cycle-related genes. B,cell wall-related genes. A1-A3,three biological replicates from arborescent phenotype pears (APPs);D1-D3,three biological replicates from dwarf phenotype pears (DPPs). Red indicates higher expression and blue indicates lower expression.
3.7.Validation of RNA-Seq data by RT-qPCR
To validate the reliability of our RNA-Seq data,the expression levels of 11 randomly selected genes related to phytohormones,the cell cycle and TFs,and four specifically selected genes located in scaffold00074 identified as DEGs,were confirmed by quantitative RTPCR (RT-qPCR). Subsequently,the Pearson correlation coefficients (PCC) between RT-qPCR and RNA-Seq data for each gene were calculated in R using the Pearson function. The results showed that high correlations between transcriptome and RNA-Seq data were exhibited for each gene,i.e.,the average Pearson correlation coefficient was more than 0.86. These results reflect the validity of the RNA-Seq data (Fig.5).

Fig.5 RT-qPCR validation of RNA-Seq data. A1-A3,three biological replicates from arborescent phenotype pears (APPs);D1-D3,three biological replicates from dwarf phenotype pear (DPPs). Bars are SD (n=3).
3.8.DEGs within the candidate interval of PcDw
To further explore key genes associated with the dwarfing phenotype,we mapped the RNA-Seq data to the scaffold00074 (P.communis),which harbors the SSR markers linked to the dwarf phenotype. Four synteny blocks were identified through collinearity analysis between the scaffold00074 (P.communis) and theP.bretschneiderigenome. A total of 81 genes were identified in these synteny blocks,four of which (Pbr012061.2,Pbr012068.1,Pbr012069.1,andPbr012085.1) showed expression differences greater than two-fold between DPPs and APPs (Fig.6). ExcludingPbr012068.1,which lacked functional annotation and was down-regulated,the other three genes were highly expressed in DPP plants. Pbr012085.1,annotated as GA1-3 repressor protein 1 (RGA1),belongs to the DELLA protein family which,as previously mentioned,plays significant roles in the GA signal pathway.Pbr012069.1andPbr012061.2were annotated as major latex proteinlike gene and Sulphate anion transporter,respectively.These results suggest thatPbr012061.2,Pbr012068.1,Pbr012069.1,andPbr012085.1might play important roles in pear plant height regulation.
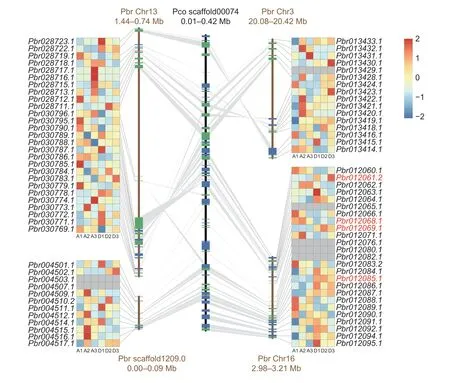
Fig.6 Expression levels of genes within scaffold00074. The vertical black line and brown lines represent synteny blocks between scaffold00074 (Pyrus communis) and the Pyrus bretschneideri genome;Pbr and Pco indicate P.bretschneideri and P.communis,respectively. Genes within blocks are shown as horizontal lines,with colors indicating the gene transcriptional orientation (green lines are forward,blue lines are reverse). The grey area in the heatmap indicates that the genes were not expressed in all six samples. The red font indicates the genes that were differently expressed between arborescent phenotype pear (APP;A1-A3) and dwarf phenotype pear (DPP;D1-D3) plants.
4.Discussion
4.1.Exploration of the potential core genes regulating the dwarfing trait in pear plants
Since dwarfing cultivation is the potential trend of orchard production,research on dwarfing traits of fruit trees has attracted more and more attention. In apple,several dwarfing rootstocks with good compatibility have been identified and widely used in production,such as M9 and M26 (Ganet al.2018;Zhenget al.2018). In the last century,researchers have identified three recessive single genes,nameddw,dw2,anddw3,which control three dwarf traits in peach,and applied them to develop new varieties (Hollenderet al.2016;Cantinet al.2018).However,compared with other fruit crops,dwarfing germplasm resources of pear are scarce,and our understanding of the pear dwarfing mechanism is also relatively limited. In previous studies,thePcDwgene,which responsible for the dwarf trait of the ‘Le Nain Vert’ pear cultivar,was localized between two SSR markers in scaffold00074 of the pear (P.communis) genome. Six genes have been identified within the candidate interval,but none have been shown to impart the dwarf phenotype of pear (Wanget al.2016). This is likely due to the imperfect nature of the existing pear reference genome,which degraded the accuracy of thePcDwlocus (Xiaoet al.2019). Therefore,we expanded the candidate gene interval to include the entire scaffold00074. In doing so,we identified four differentially expressed genes between APP and DPP in this interval.Pbr012069.1andPbr012061.2were annotated as major latex protein-like gene and sulphate anion transporter,respectively. Both of them were proposed to be involved in the plant resistance response,and no report to date has shown their functions to be associated with plant dwarfing (Chen and Dai 2010;Chen Zet al.2019). Given thatPbr012068.1has an unknown function,these three genes might not be the primary subjects in our further investigations.
However,Pbr012085.1was predicted to encode a DELLA protein,which is proposed as a primary inhibitor of GA signal transduction pathways. As one of the most important phytohormones in determining plant height,GA promotes the degradation of DELLA proteins and releases its inhibitory effect on downstream growth-related genes,thereby promoting stem elongation (Dillet al.2001).When DELLAs fail to be degraded,plants exhibited a dwarf stature. In apple,a GA-insensitive DELLA gene,Mhgai,reduced the stature of transgenic plants when ectopically expressed in tomatoes (Wang S Set al.2012). Likewise,with reductions in bioactive gibberellin levels,DELLAs gradually accumulate and eventually inhibit plant growth (Achardet al.2008). Previous studies revealed that DELLAs can effectively suppress cell division. InArabidopsis,DELLA activates the expression ofKRP2,a cell cycle inhibitor,resulting in restricted cell division in SAMs (Serrano-Mislataet al.2017). In addition,SLR1 (DELLA) also indirectly inhibited the transcription of rice cell cycle-related genes (cycOs1andcycOs2) in rice plants by up-regulating the expression ofmiR396,ultimately producing a dwarfed phenotype (Luet al.2020). Our RNA-Seq data showed that twoCYP714A1genes (Pbr001398.1andPbr001399.1) which function in bioactive GA degradation (Nomuraet al.2013) and theDELLAgene (Pbr012085.1) were significantly more highly expressed in DPPs. These findings provide conducive conditions for the accumulation of DELLA proteins in DPPs. Furthermore,acting downstream of DELLA,Pbr025495.1,a homologous gene of theKRP2gene,was up-regulated while several cyclins (Pbr022014.1,Pbr019683.1,andPbr002851.1) were down-regulated.This could further lead to deficient cell division in DPPs.Subsequently,our statistical analysis of cell numbers verified this inference to some extent. Based on this reasoning,we suspectPbr012085.1is a promising candidate gene for the dwarfed phenotype,while genesPbr001398.1andPbr001399.1may also play important roles in regulating the dwarf phenotype of DPP.
4.2.Changes in BR-related genes are associated with the dwarfing phenotype in pear plants
Besides GA,BR is another phytohormone which is closely associated with plant cell division (Fabianet al.2000;Sunet al.2015). BR promotes cell division and early fruit development in cucumber,and the expression of cell cycle-related genes (CycD3;1andCycD3;2) were upregulated by the application of EBR (exogenous BRs) and suppressed by the application of BRZ (BR biosynthesis inhibitor) (Fuet al.2008). In our RNA-Seq data,CYP90A1(Pbr009519.1) was down-regulated and the expression level ofCYP734A1(Pbr006166.1) increased over 9-fold in DPPs (Table 2). CYP90A1/CPD is a critical enzyme involved in BR biosynthesis,as CPD loss-offunctionArabidopsismutants exhibited extremely low levels of BR and a severely dwarfed phenotype (Ohnishiet al.2012). In contrast,CYP734A1was shown to play a negative regulatory role in BR activity,and overexpression ofCYP734Ain rice plants resulted in a significant reduction in plant height (Sakamotoet al.2011). These results indicate that the BR content in DPPs may be reduced and may further contribute to the low expression levels of cell cycle-related genes.
4.3.Differentially expressed TFs involved in the dwarfing phenotype in pear plants
TFs act as a critical bridge which connects phytohormone signaling and plant-growth-related gene expression. Our transcriptome analysis found that 101 TFs involved in 38 gene families were differentially expressed between DPP and APP. Notably,three genes annotated asBZR1/BES1were down-regulated in DPP. As a main TF in the BR signal transduction pathway,BZR1/BES1regulates the expression of numerous genes to mediate BR-regulated plant growth (Ohet al.2014). InArabidopsis,bzrhextuple mutants exhibited a dwarfing phenotype similar to BRinsensitive mutants (Chen L Get al.2019). Moreover,previous studies revealed intimate signal crosstalk between BR and GA (Unterholzneret al.2015). DELLAs interacted with BZR1/BES1 and suppressed its DNAbinding activity (Zentellaet al.2017). The low transcript levels ofBZR1/BES1genes and the functional inhibition of DELLA proteins may further suppress the expression of downstream growth-related genes in DPP plants.
BRAVOis a R2R3 MYB transcription factor,which directly inhibits the expression of cell cycle genes (CYCB1;2,CYCD3;3,andCYCD2;2) and cell division of the quiescent center (QC) inArabidopsis(Vilarrasa-Blasiet al.2015). Further investigation found that BZR1/BES1 negatively regulated the activity of BRAVO through direct interaction with BRAVO to directly repress its transcription (Vilarrasa-Blasiet al.2015). In our study,a total of nine MYBs were highly expressed in DPP,including a homologous gene ofAtBRAVO(Pbr016851.1). These MYBs may play a negative regulatory role in controlling plant height.
AP2/ERF family genes widely participate in regulating plant growth by influencing the signaling or metabolism of phytohormones (Liuet al.2018). When overexpressed in rice plants,OsRPH1,a member of the AP2/ERF gene family,suppresses GA biosynthesis,triggering its degradation and eventually leading to reductions in plant height (Maet al.2020). Another study found that AtERF11 positively regulated GA biosynthesis and interacted with DELLA to antagonize its function,promoting hypocotyl elongation (Zhouet al.2016). In our results,14 AP2/ ERF genes were differentially expressed between the two types of pears studied,10 of them were up-regulated and the other four were down-regulated in DPPs. This suggests that AP2/ERF genes may play an important role in GA mediated regulation of height in pear plants.
5.Conclusion
In the present study,we confirmed that the dwarf phenotype of DPP is controlled by plant organs located in the scions,independent of the rootstock. Stem tissue analysis revealed that the reduced height in DPP plants can be primarily attributed to deficient cell division. Subsequently,1 401 DEGs between DPP and APP plants were identified by RNA-Seq. Among these,101 genes were predicted to function as TFs. Further functional analysis of the DEGs suggested that changes in the expression of genes involved in GA and BR degradation and signal transduction could potentially explain the reduced numbers of cells observed in DPP plants. Furthermore,four candidate genes were identified through combined analysis of our transcriptome data and molecular markers provided by a previous study. Among these genes,Pbr012085.1was the most important one related to the formation of this dwarf trait in pear.These findings provide new insights for further investigation into the genetic and molecular mechanisms underlying dwarfing in pear plants.
Acknowledgements
This work was supported by the National Key Research and Development Program of China (2018YFD1000200),the Earmarked Fund for Jiangsu Agricultural Industry Technology System,China (JATS [2020]401),the National Science Foundation of China (31801835),the China Agriculture Research System of MOF and MARA (CARS-28),the Natural Science Foundation of Jiangsu Province,China (BK20180516),and the Agricultural Variety Improvement Project of Shandong Province,China (2019LZGC008).
Declaration of competing interest
The authors declare that they have no conflict of interest.
Appendicesassociated with this paper are available on http://www.ChinaAgriSci.com/V2/En/appendix.htm.
 Journal of Integrative Agriculture2022年7期
Journal of Integrative Agriculture2022年7期
- Journal of Integrative Agriculture的其它文章
- Roles of mushroom polysaccharides in chronic disease management
- lnfluence of high-molecular-weight glutenin subunit deletions at the Glu-A1 and Glu-D1 loci on protein body development,protein components and dough properties of wheat (Triticum aestivum L.)
- Variations in the quality parameters and gluten proteins in synthetic hexaploid wheats solely expressing the Glu-D1 locus
- ldentification of candidate genes related to soluble sugar contents in soybean seeds using multiple genetic analyses
- ldentification and characterization of long-lnDels through whole genome resequencing to facilitate fine-mapping of a QTL for plant height in soybean (Glycine max L.Merr.)
- Photosynthetic properties of the mid-vein and leaf lamina of fieldgrown,high-yield hybrid rice during senescence
