Uses of knockout,knockdown,and transgenic models in the studies of glucose transporter 4
Tian-Nan Wang,Xin-Ge Hu,Guo-Xun Chen
Tian-Nan Wang,Xin-Ge Hu,Guo-Xun Chen,Department of Nutrition,The University of Tennessee,Knoxville,TN 37996,United States
Abstract Currently,glucose transporter 4 (GLUT4) has been considered as the key player for the insulin-stimulated glucose transport in the muscle and adipose tissues.The development of recombinant DNA techniques allows the creations of genetically knockout,knockdown and transgenic animals and cells for the study of GLUT4’s physiological functions.Here,we have used key words to search the PubMed and summarized the methods used in Slc2a4 gene knockout,GLUT4 knockdown and overexpression in the whole body and tissue specific manner.The whole body GLUT4-null mice have growth retardation,but normal glucose tolerance and basal glucose turnover rates.Compared with whole body Slc2a4 knockout mice,adipose and muscle double knockout mice have impaired insulin tolerance and glucose intolerance.The results of GLUT4 knockdown in 3T3-L1 adipocytes have shown that its expression is needed for lipogenesis after,but not during,differentiation.Transgenic mice with the whole body GLUT4 overexpression have normal body weight and lowered blood glucose level.The adipose tissue specific overexpression of GLUT4 leads to increases in mouse body weight and adipose tissue weight.The insulin-stimulated GLUT4 translocation in the skeletal muscle contributes to the regulation of glucose homeostasis.Data from both transgenic overexpression and tissue specific Slc2a4 knockout indicate that GLUT4 probably plays a role in the glucose uptake in the fasting state.More studies are warranted to use advanced molecular biology tools to decipher the roles of GLUT4 in the control of glucose homeostasis.
Key Words: Glucose transporter 4;Knockout;Knockdown;Transgene;Overexpression;Insulin
INTRODUCTION
Genes in an organism are codes responsible for genetic traits.In most cases,genes usually exist in the form of nucleotide sequences.In a cell,the DNA sequence of a gene is first transcribed into mRNA,which serves as the template for protein translation.The newly synthesized proteins contribute to biological processes in an organism.To understand the biochemical,biophysical,and genetic functions of a given gene and its protein,recombinant DNA technologies have been developed and used extensively.Since 1970s,the discovery of restriction enzymes has facilitate the development of molecular cloning methods and allowed the manipulation of DNA sequences selectively and specifically to create novel recombinant molecules[1].DNA fragments are inserted into vectors to form recombinant genetic materials for their replication,studies of gene functions and productions of recombinant proteins.The recombinant DNA technology was first used to study gene functions when the genes responsible for metabolism of galactose in E.coli were fused into the SV40 vector in 1972[2].For the past few decades,procedures of molecular cloning have been simplified and standardized to construct recombinant DNA with various sizes for different purposes[3].All these have been applied to generate transgenic organisms and produce recombinant proteins for the use in variety of research and clinical settings.
Glucose enters cellsviaa family of proteins called glucose transporters (GLUTs),which have 14 known members.Glucose transporter 4 (GLUT4) encoded bySLC2A4gene in human genome andSlc2a4gene in others such as rodent genomes has 409 amino acid residues,and a Km value of 5 mmol/L for glucose[4].GLUT4 was first identified in a screen for the insulin-stimulated glucose transporter in cell membrane preparations of rat adipocytes using monoclonal antibodies against these membrane proteins[5].Subsequently,Slc2a4gene was cloned from rat adipose tissue,and is homologous with GLUT1,which is encoded bySlc2a1gene[6-8].GLUT4 is expressed in not only adipose and muscle cells,but also other tissues such as the heart and brain[9].The N- and C- termini of GLUT4 are located in the cytoplasm and responsible for the insulin-mediated translocation from the cytosol to the cell membrane[10].The current model is that insulin stimulates GLUT4 translocation from the intracellular locations to the plasma membrane,where it facilitates the glucose entry into cells[11].In addition,exercise also stimulates the expression ofSLC2A4mRNA in the skeletal muscle and improves insulin sensitivity in human patients[12],which may be mediated by GLUT4[13].Insulin-stimulated glucose transport is significantly impaired in the skeletal muscle of patients with type 2 diabetes[14].Therefore,understanding the role of GLUT4 in the regulation of glucose homeostasis is critical for the prevention and treatment of type 2 diabetes.
Here,we summarize the recombinant DNA technologies used to study expression profiles and functions of GLUT4 in tissues and cells.Key words as indicated in the following sections were used to search PubMed.The title and abstracts of the retrieved articles were read by authors.Only the articles that contained descriptions of knockout,transgenic overexpression and knockdown molecular techniques,and confirmed gene or protein expression levels were chosen for further reading.The methods used to manipulate the expression levels of GLUT4in vivoandin vitroand reported observations in retrieved studies were summarized here.This review may help researchers who are interested in the physiological functions of GLUT4 to have a clear understanding of the status.
THE COMMON MOLECULAR BIOLOGY TECHNIQUES TO STUDY GENE AND PROTEIN FUNCTIONS
The development of molecular cloning techniques allows isolation,generation,and production of DNA sequence independence of the species and organisms that carry the original sequences.DNA fragments isolated from genomes or createdviapolymerase chain reaction (PCR) are inserted into vectors that can replicate and express in the host cells,and in turn alter the genetic features of the host cells,tissues or organisms[15].PCR technique quickly produces large numbers of copies of a specific DNA fragment for sequencing analysis and molecular cloning.Cloning of a specific DNA sequence helps to explore the gene’s biological functions,and to create large amounts of protein,such as growth hormone,insulin and clotting factors for therapeutic purposes[16].In addition,a comparison of DNA sequences from different organisms can determine the evolutionary relationship within and between species,and functional domains of a gene.Recombinant DNA technologies can be used in gene therapies to treat diseases such as immunodeficiency diseases and metabolic disorders[17] and diagnosis of genetic diseases[18].Genetically modified organisms or genetically engineered organisms can be createdviaalterations of the genetic sequences of the chromosome or insertions of the foreign DNA fragments into the genome to alter the phenotypes of the offspring[19].
Genes in plants,animals and microorganisms have been deleted or their expression levels have been knocked down to investigate their functions or treat genetic diseases clinically[20].Methods are developed to silence or remove the target gene,such as gene silencing,conditional knockout,homologous recombination,and gene editing[20].Homologous recombination occurs when homologous recombinases (nucleases) recombine two linearized DNA fragments with the same terminal sequences to create a novel fragment for molecular manipulations[21].This makes accurate gene editing possible and becomes emerging tools in genetics[22].Zinc finger nucleases,transcription activator-like effector nucleases,and clustered regularly interspaced short palindromic repeat (CRISPR) are developed and shown differences in knockout efficiency,completion time,and off-target efficiency[20].Each of these techniques uses a nuclease to introduce DNA double-strand breaks at the targeted locations with the guidance of homologous binding proteins or RNA[23].Gene knockdown methods such as RNA-based RNA interference,small interfering RNA and short hairpin RNA (shRNA),and antisense oligonucleotides have been developed to inhibit protein expression[24].RNA interference (RNAi) is triggered by double-stranded RNA and causes the sequence-specific mRNA degradation of the single-stranded target RNA[25].Small non-coding RNA molecules can also act to inhibit RNA translation[26].
In addition to the change of gene expression,tagged proteins or fusion proteins with novel properties can be created using molecular biology tools[27].Fusion or tagged proteins with two or more domains from different proteins can be easily obtained and purified for their uses in research and clinical treatments,detection of the expression levels,and visualization of the intracellular locations of the expressed proteins[27].These have been used to create vaccines,multifunctional enzymes,targeted drugs,thrombolytics,antimicrobial peptides,etc[28].
MOLECULAR BIOLOGY TECHNIQUES USED IN THE STUDIES OF GLUT4
The identification of GLUT4 and cloning its gene[29] have facilitated the studies of its tissue distribution,functions,the mechanisms responsible for its translocation,and the regulations of its protein and mRNA expressions in different cells.The tagged or fluorescent GLUT4 fusion proteins are used to study its intracellular trafficking.GLUT4 overexpression and knockdown,andSlc2a4gene knockoutin vitroandin vivohave been developed to study the insulin-stimulated GLUT4 translocation and glucose homeostasis,which contribute significantly to our understanding of the role of GLUT4[29].To review the techniques of molecular biology in the study of GLUT4,"GLUT4,molecular biology" and "SLC2A4,molecular biology" as keywords were used to search the PubMed database to retrieve relevant articles.We have focused on the techniques used inSlc2a4knockout,knockdown and transgenic studies,and results associated with the genetic changesin vivoandin vitrowere analyzed and summarized here.As shown in Figure 1,Slc2a4genes have been knocked out and GLUT4 protein has been overexpressed in the whole body and in specific tissues and cells.In addition,GLUT4 protein has been knocked down using shRNA,and its translocation has been studied using fusion or tagged proteins.Various methods such as in situ hybridization,fluorescent microscopy,immunohistochemistry,Western blotting for protein and Northern blot and real-time PCR for mRNA used to determine endogenous or transgenic GLUT4 expressions are also summarized in this review.
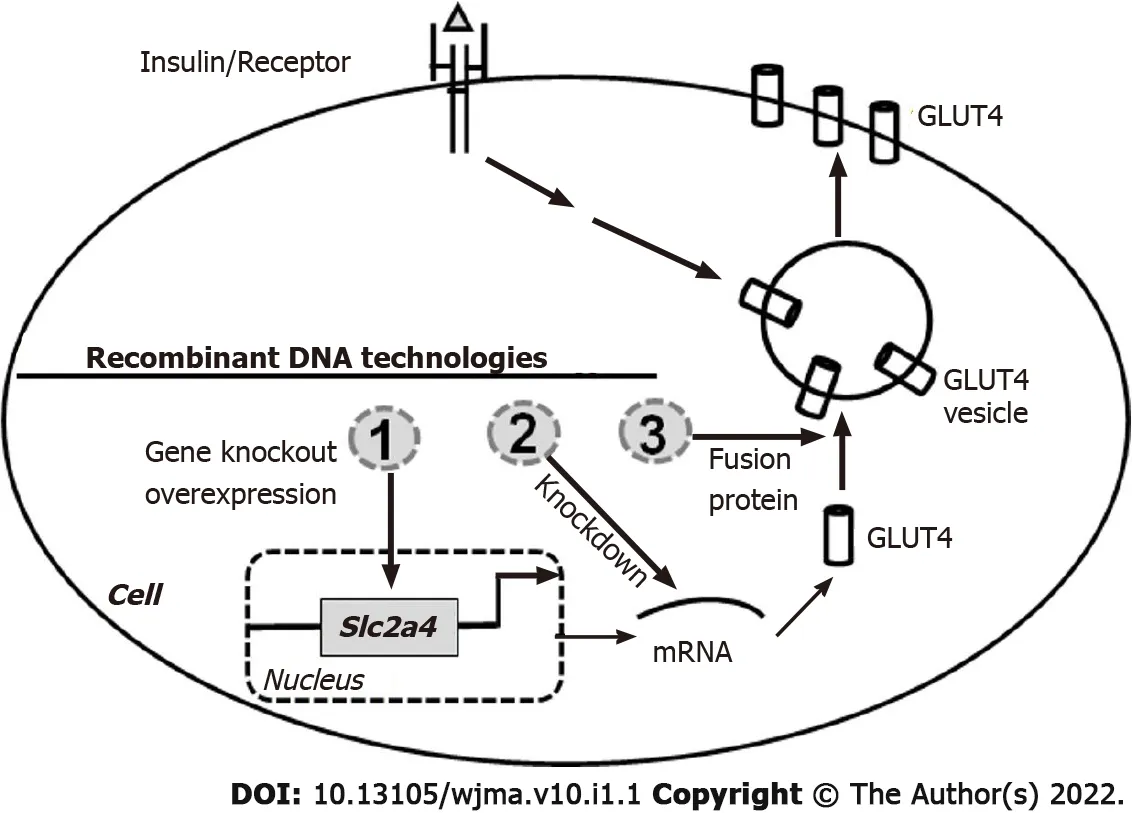
Figure 1 Recombinant DNA technologies used in the study of glucose transporter 4 functions and its translocation mechanism.
Whole-body and tissue specific Slc2a4 knockout studies
Slc2a4mRNA expression is detected not only in brown and white adipose tissue,skeletal and cardiac muscle,but also in other tissues such as neurons[30].To study GLUT4 functions,mice with theSlc2a4deletion in the whole body or specific tissues or cells have been created.We searched PubMed to retrieve the original articles that initially reported theSlc2a4deletions.Table 1 shows the techniques for creating knockouts,experimental animals,methods to confirm the gene deletion and expression,and observations.In the end,seven representative articles that the research groups generated a specific knockout model to study GLUT4 and clearly described the methods of GLUT4 deletion are summarized here as shown in Table 1.The animal models were also used by many other groups.
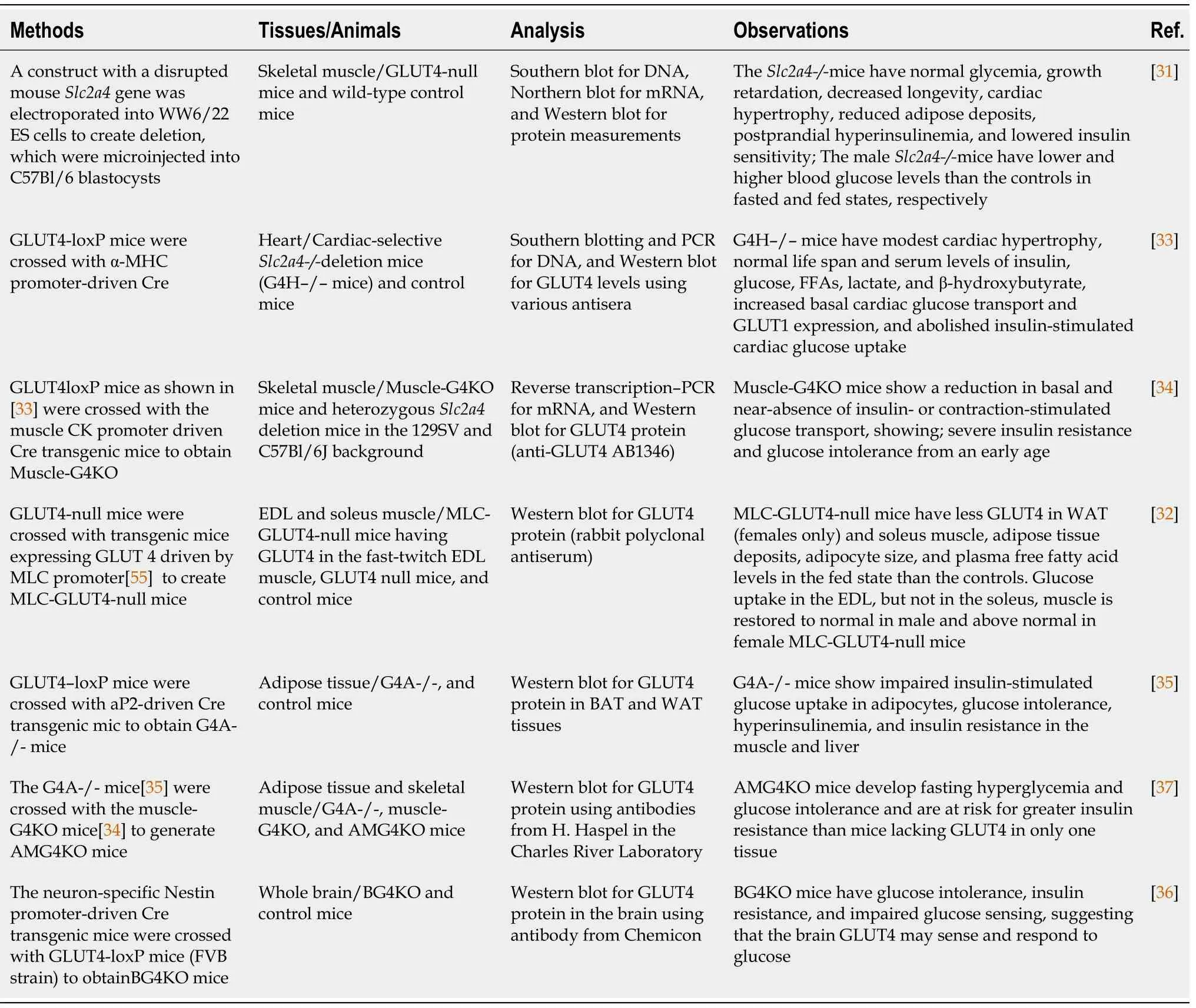
Table 1 Methods used to create whole body and tissue specific Slc2A4 knockout animals,tissues and animals studied,analytic methods included,and observations reported
In1995,the mouseSlc2a4locus was disrupted using homologous recombination in embryonic stem cells which generated mice without GLUT4 expression (GLUT4-null) in the whole body[31].The GLUT4-null mice showed growth retardation,enlarged hearts and complete lack of the white adipose tissue[31].GLUT4-null mice have normal glucose tolerance and basal glucose turnover rates.However,they are insulin intolerant,suggesting insulin resistance.Later on,the GLUT4-null mice[31] have been used to create mice expressing GLUT4 specifically in the extensor digitorum longus muscle[32].
Tissue specific GLUT4 knockout mice have been created by crossing mice carrying aSlc2a4allele with exon 10 flanked by loxP sites with those carryingCregene expression driven by tissue specific promoters[33-36].The various phenotypes of these knockout mice help us to understand the roles of GLUT4 in different tissues and glucose metabolism.For example,the muscle-specific GLUT4 knockout mice (muscle-G4KO) were created by breeding mice carrying theSlc2a4exon 10 flanked by loxP sites with mice carrying a transgene encoding Cre recombinase under the control of the muscle creatine kinase promoter[34].Compared with GLUT4-null mice,muscle-G4KO mice have normal body weight and fat pad weight at least before 6 mo of age[34].The skeletal muscle mass is also normal.The increase in heart weight is consistent with GLUT4-null mice[31] and cardiac-G4KO mice[33].Compared with the shortened lifespan of GLUT4-null mice,the life span of muscle-G4KO mice is normal.In contrast to GLUT4-null mice and cardiac-G4KO mice,adipose-G4KO mice[35] are similar to muscle-G4KO mice.However,adipose-G4KO mice have glucose intolerance[35].Interestingly,unlike other GLUT4 knockout mice,the heart weight of adipose-G4KO mice is normal.
In addition,adipose and muscleSlc2a4double knockout (AMG4KO) mice are also created by crossing the respective tissue knockout mice[37].Interestingly,these AMG4KO mice develop hyperglycemia in the fasting state[37].It appears that GLUT4 also plays a role in a physiological condition that does not need the insulin-stimulated glucose uptake.
GLUT4 knockdown studies
The deletion of a gene completely stops the genetic information flow.Another way to block the protein expression is to knockdown a gene’s expression,which temporarily stops or reduces the expression of the targeted gene.Unlike knockout,gene knockdown involves the methods interfering with RNA molecules (mRNA or non-coding RNA) that bridges DNA and proteins."GLUT4,knockdown" and "SLC2A4,knockdown" as keywords were used to search the PubMed database to retrieve relevant articles.Table 2 summarizes the methods of knockdown and confirmation,cells used,observations of GLUT4 knockdown studies.
Recently,RNAi has emerged as a powerful tool for the study of gene function in mammalian cells[38].After transfection,the shRNAs molecules are transcribed under promoters in constructs that drive the RNA synthesis within the targeted cells.Oligo nucleotides with sequences of shRNAs may be transfected directly into the cells[38].In all studies summarized in Table 2,shRNAs method is used to achieve GLUT4 knockdown[39-41],which is deliveredviarecombinant retroviruses.Two of three studies investigated the roles of GLUT4 in 3T3-L1 adipocytes.It appears that GLUT4 expression is needed for the lipogenesis after differentiation in 3T3-L1 cells,but not necessary for lipogenesis during differentiation[41].In addition,the insulin-regulated aminopeptidase trafficking is not always associated with the GLUT4 movement[40].
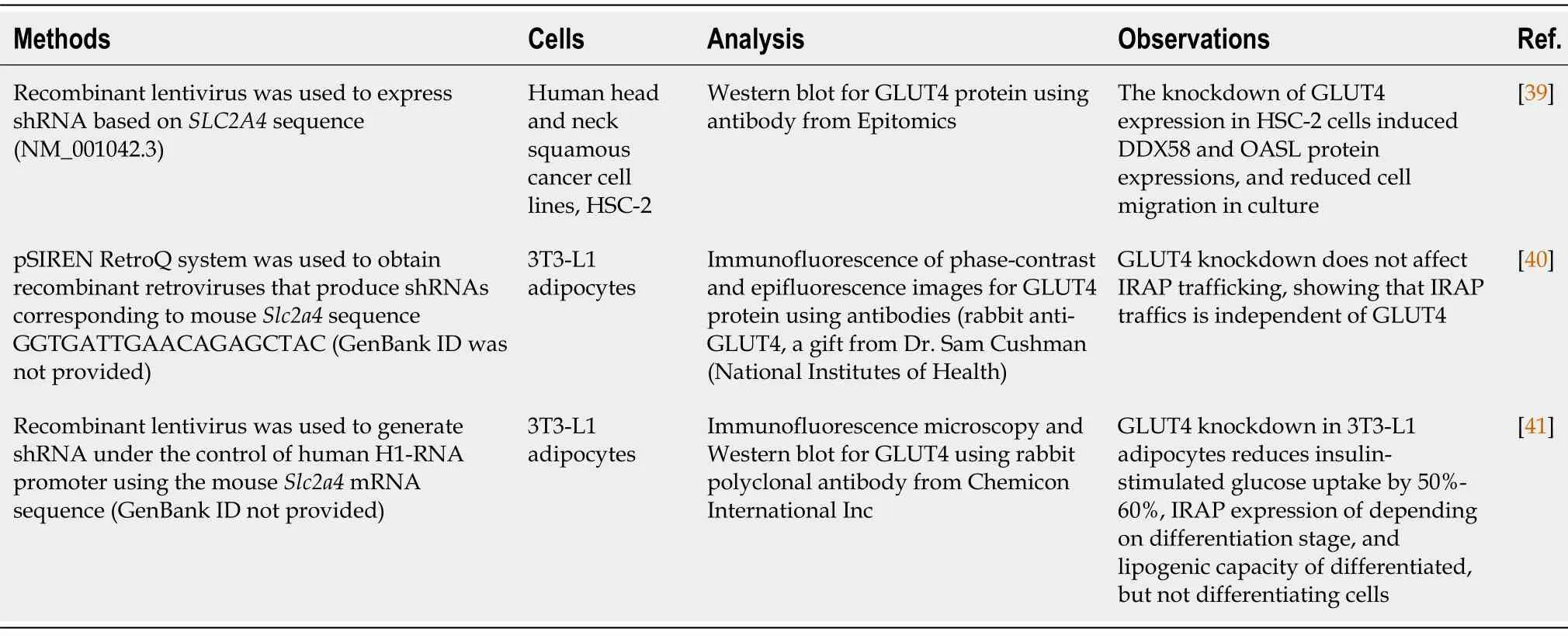
Table 2 Methods used to knockdown glucose transporter 4 and analyze its expression in cell lines,and reported observations
Transgenic studies
We have used key words “GLUT4 transgenic” (314 hits) and “GLUT4 overexpression” (609 hit) to search PubMed to retrieve GLUT4 transgenic studies.After going through the titles or abstracts containing “GLUT4 overexpression and GLUT4 transgenic”,we found 15 papers that have described their original methods or clearly cited the methods used by them,confirmed the GLUT4 overexpression in mice and provided results.Table 3 summarizes the techniques used to overexpress GLUT4,the methods to confirm the expression and results observed in those animals.
In 1992,a 2.4-kb fragment of 5’ flanking DNA of humanSLC2A4promoter fused with the bacterial chloramphenicol acetyltransferase (CAT) as a reporter construct was developed and used to show theSLC2A4expression profile in mice[42].In 1993,an 11.5-kbSLC2A4mini gene in pHSS6 vector was created and used to overexpress human GLUT4 in the whole body of mice[43].As shown in Table 3,the mouse line containing the 11.5-kb mini gene has been used in seven out of eight papers testing the effects of overexpression of human GLUT4 in the mouse whole body on metabolism[43-49].CAT activity,SLC2A4mRNA and/or GLUT4 protein level in adipose tissue and skeletal muscle and other tissues have been analyzed to confirm the success of transgenic expression[43,44].In general,the wholebody human GLUT4 overexpression reduces blood glucose in both fasting and fed states and increased glucose uptake in mice,but affects the blood insulin level in wild type mice and diabetic mice differentially[44].Whole body GLUT4 overexpression does not alter body weight,but can reduce blood glucose level and affect serum insulin level.
GLUT4 has been overexpressed in a tissue specific manner as shows in Table 4.Majority of the studies have been focused on adipose tissues[50-54].There is one for the skeletal muscle[55] and one for adipocytes[56].A 6.3-kb genomic DNA fragment of humanSLC2A4gene driven by the mouseap2[50] promoter has been used to overexpress GLUT4 in mouse adipose tissues.This method was first published in 1993[50] and was used in many other studies in the genetic settings of wild-type and diabetic mice[51-54].Like the papers summarized in Table 3,a humanSLC2A4gene under a tissuespecific promoter was used to overexpress GLUT4.HumanSLC2A4cDNA was also used to overexpress GLUT4 in adipocytes[56],but mouseSlc2a4gene was used to express GLUT4 in the hindlimb muscle[55].All adipose tissue specific GLUT4 overexpression studies[50-55] tested theSLC2A4mRNA or GLUT4 protein level to confirm GLUT4 expression,which is found to be expressed in both brown and white adipose tissues.In conclusion,adipose tissue specific GLUT4 overexpression in mice can cause increases in body and adipose tissue weights[50,51].The adipose tissue specific GLUT4 transgenic mice also have higher glucose disposal rate,may be caused by increased basal and insulin-stimulated glucose transport rate.It is interesting to find out that the elevated expression of GLUT4 in the adipose tissue only can increase glucose transport rate and adipose tissue weight,which is associated with the significant increase in body weight,suggesting the importance of GLUT4 expression in adipose tissue.
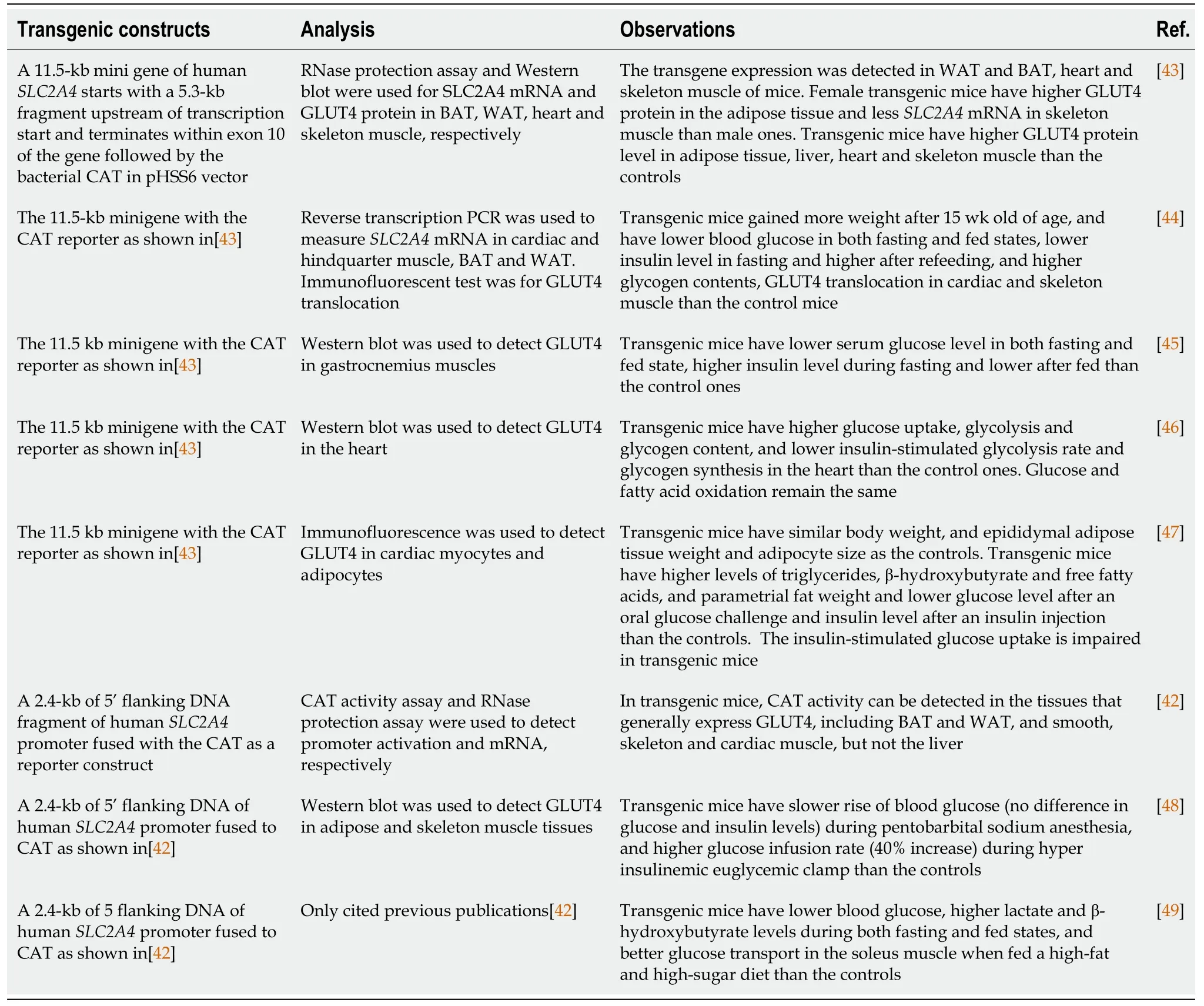
Table 3 The transgenic studies using the SLC2A4 mini gene and its promoter for the whole-body expression in mice

Table 4 Recombinant DNA techniques to create tissue specific glucose transporter 4 overexpression in animals and cells,analysis performed,and observations reported
CONCLUSION
Conclusion and future perspectives
As summarized in this review,methods such as whole body and tissue specific gene knockout,recombinant viruses,real-time PCR,immunofluorescence,stable cell line and transgenic animals have been used to study GLUT4 system and insulin action in different target cells and tissues.The advantages of using multiple molecular biology methods allow us to confirm the functions of GLUT4 for insulin-stimulated glucose transport in different cells and tissues,and in the regulation of wholebody glucose homeostasis.Interestingly,GLUT4-null mice which do not have a functionalSlc2a4gene in the whole genome have normal glucose tolerance and basal glucose turnover rates,but they are insulin-intolerant which suggests insulin resistance[31].AMG4KO mice (adipose and muscle double knockout) have reduced whole body glucose uptake and hyperglycemia[37].Compared with GLUT4-null mice,AMG4KO mice have more severe glucose homeostasis defects.Although the explanation for this difference is not clear,differences in genetic background and differences in developmental stages,where GLUT4 is deleted have been proposed.More importantly,the hyperglycemia in these double knockout mice develops in the fasting state,rather than fed state[37].This phenomenon appears to indicate that GLUT4 plays an important role in the control of glucose homeostasis during fasting,a state that insulin level is low.The translational value of these observations is that GLUT4’s physiological role from the integrated homeostatic point of view may be extended beyond the insulin-stimulated glucose uptake.Of course,more studies are warranted on this line of research.
On the other hand,the GLUT4 knockdown studies used the shRNAs method and have been done in cell lines to reduce GLUT4 expression.This may be helpful for us to understand the GLUT4 functions and the underlying mechanisms in particular cells.It appears that GLUT4 expression is not necessary for lipogenesis during 3T3-L1 cells differentiation.Apparently,it will be helpful when more GLUT4 knockdown studies are done in animals.
The GLUT4 overexpression in transgenic mice at wholebody level reduces blood glucose in both fasting and fed states and increased glucose uptake,glycolysis and glycogen level[44].Compared to the control mice,overexpression of GLUT4 in adipose tissue in mice leads to lowered blood glucose in the fasting state,and increase in body weight and adipose tissue weight[50].The expression of GLUT4 in adipose tissue and skeleton muscle affects the rate of whole-body glucose disposal,which may be caused by increased basal and insulin-stimulated glucose transport rates.This lowered blood glucose level in the transgenic mice also indicates that GLUT4 probably plays a role in the basal glucose uptake.
For the tissue specific GLUT4 knockout,Cre-loxP-mediated gene recombination under the control of promoters has been the main method to deleteSlc2a4gene.Since the development of CRISPR technology,it has not been used to knockoutSlc2a4in whole body or tissues,which is a limitation in the field.We have used “GLUT4” and “CRISPR”,and “SLC2A4” and “CRISPR” as key words to search PubMed,and retrieved eight and two published articles,respectively.However,none of the published articles used the CRISPR methods to knockoutSLC2A4orSlc2A4gene in cells or animals.All of them used CRISPR methods to study the components in the exocytosis process of GLUT4 translocation.As CRISPR has been developed and used widely,GLUT4 knockout/knockdown through this system may be worth to be done.This may provide us another tool to manipulate the GLUT4 expression in the whole body or in tissue specific manners.
In addition,results of glucose tolerance are different between mice with whole body and tissue specific GLUT4 knockout.Therefore,whether the loss of GLUT4 in a specific tissue (muscle or fat) or the expression of GLUT4 in other tissues without gene deletion plays a role in this difference is worth to be investigated.It is safe to say that more research works are anticipated in the future to precisely define the role of GLUT4 in the control of glucose homeostasis at whole body and tissue levels.In so doing,we develop effective ways to prevent and treat type 2 diabetes mellitus.
FOOTNOTES
Author contributions:Chen GX was responsible for the design of the topics;Wang TN and Hu XG were responsible for PubMed search and information collection;Wang TN,Hu XG and Chen GX were responsible for writing.
Conflict-of-interest statement:All authors confirmed that there is no conflict-of-interest to be reported.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers.It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4.0) license,which permits others to distribute,remix,adapt,build upon this work non-commercially,and license their derivative works on different terms,provided the original work is properly cited and the use is noncommercial.See: https://creativecommons.org/Licenses/by-nc/4.0/
Country/Territory of origin:United States
ORCID number:Tian-Nan Wang 0000-0001-5584-3628;Xin-Ge Hu 0000-0002-3253-7537;Guo-Xun Chen 0000-0001-6226-4050.
S-Editor: Liu JH
L-Editor: A
P-Editor: Liu JH
 World Journal of Meta-Analysis2022年1期
World Journal of Meta-Analysis2022年1期
- World Journal of Meta-Analysis的其它文章
- Hepatitis C virus among blood donors and general population in Middle East and North Africa:Meta-analyses and meta-regressions
- Effects of dexmedetomidine on cardioprotection and other postoperative complications in elderly patients after cardiac and non-cardiac surgerie
- Leptin levels in women with unexplained infertility:A systematic review and meta-analysis
