OsABT,a Rice WD40 Domain-Containing Protein,Is Involved in Abiotic Stress Tolerance
CHEN Eryong,SHEN Bo
(1College of Life and Environmental Sciences,Hangzhou Normal University,Hangzhou 311121,China;2Life School of Science and Technology,Henan Institute of Science and Technology,Xinxiang 453003,China)
Abstract:Plant growth and crop productivity are severely affected by abiotic stress on a global scale.WD40 repeat-containing proteins play a significant role in the development and environmental adaptation of eukaryotes.In this study,OsABT,a stress response gene,was cloned from rice (Oryza sativa L.cv.Nipponbare).OsABT encodes a protein containing seven WD40 domains.Expression analysis revealed that the OsABT gene was first up-regulated and then down-regulated following treatment with abscisic acid (ABA) and NaCl,but was down-regulated when treated with PEG8000.Subcellular localization results showed that OsABT was located in the cytoplasm and nucleus of Arabidopsis roots.OsABT transgenic Arabidopsis showed significantly increased tolerance to ABA and salt stress during plant seedling development.However,the transgenic lines were more sensitive to drought stress.Moreover,OsABT can interact with OsABI2,a component of ABA signaling pathway.These results showed that OsABT plays a positive regulatory role in response to salt stress and a negative role in response to drought stress in Arabidopsis.
Key words:abscisic acid;Arabidopsis;drought stress;rice;salt stress;WD40 domain-containing protein
Various abiotic stresses can severely affect the growth,development and yield of crop plants (Mishra et al,2012).Drought stress is a major limiting factor in agricultural production,leading to a decline in crop yields worldwide (Shinozaki and Yamaguchi-Shinozaki,2007).Therefore,plants have evolved diverse biological defense mechanisms to improve water use efficiency under drought conditions,including accumulation of dehydrin,maintenance of root water absorption and reduction of transpiration rate through leaf stomata (Shinozaki and Yamaguchi-Shinozaki,2007;Gong et al,2015;Li et al,2015).Salt stress,another key factor,has become a serious issue limiting plant growth and crop yields.Plant species have undergone diverse changes from physiological adaptation to gene expression changes in response to salt stress.Salt stress-responsive genes are broadly divided into two types:effector genes that directly play a protective role in plant cells against salt stress,such as Na+/H+antiporters,and regulator genes that control gene expression or stress signal transduction,such as transcription factors and protein kinases (Zhu,2001;Chinnusamy et al,2006).
WD40 proteins,also known as WD40 repeat-containing proteins,are junction proteins in eukaryotic proteomes.The WD40 domain provides a platform for mediating protein-protein or protein-DNA interactions,and it is involved in the scaffolding,assembly and regulation of active polyprotein complexes (Stirnimann et al,2010;Xu and Min,2011).There are multiple WD40 repeats in WD40 proteins,each of them contains 44-60 residue units as its main feature.This unit usually contains glycine-histidine (GH) dipeptide at N-terminus and tryptophan-aspartate (WD) dipeptide at C-terminus (van Nocker and Ludwig,2003).Moreover,each WD40 repeat includes a four-stranded anti-parallel β-sheet (Neer et al,1994).Generally,the WD40 domain typically exhibits 5-8 duplicates,mostly 7 duplicates,which form a stable β-propeller structure (Fülöp et al,1998;Juhász et al,2005).The WD40 proteins are implicated in diverse biological processes,such as signal transduction,transcriptional regulation,chromatin modification,damage response,ribosomal RNA biogenesis,cytoskeletal assembly,vesicle transport,cell cycle control and apoptosis (Neer et al,1994;Smith et al,1999;Wakasugi et al,2002;Xu and Min,2011).The WD40 proteins are also involved in plant stress tolerance.WDR5a,a WD40 protein ofArabidopsis,alters NOS-like activity,which plays a role in nitric oxide (NO) accumulation and stomatal closure under drought stress (Liu et al,2017).Another WD40 protein ofArabidopsis,HOS15,is involved in cold tolerance,and its mutant plants are hypersensitive to freezing temperatures (Zhu et al,2008).Transforming the wheat WD40 geneTaWD40DintoArabidopsisresults in transgenic plants with improved tolerance to abscisic acid (ABA),osmotic stress and salt stress (Kong et al,2015).In rice,SRWD,a WD40 protein subfamily,is predominately up-regulated under salt stress (Huang et al,2008).In light of the current research results,the various roles of WD40 proteins in plant stress tolerance are worthy of attention.
ABA signaling terminator(ABT),a gene encoding anArabidopsisWD40 repeat protein,can disintegrate ABA signaling and is critical for seed germination and seedling organization (Wang et al,2020).TransgenicABTArabidopsisseeds have higher seed germination rate and seedling greening than wild type seeds when being sown on Murashige and Skoog (MS) medium containing ABA.In contrast,Arabidopsisseeds withABTgene knockout have an opposite phenotype.However,the role ofABTin drought stress is still unclear (Wang et al,2020).These results highlight the importance ofABTin ABA signaling,seed germination and seedling development,but its role in rice stress tolerance should be further studied.
Despite growing evidence that WD40 proteins have function in abiotic stress tolerance in plants,research on the function of WD40 proteins in rice is still largely limited.Here,we identified OsABT,a rice WD40 protein,and elucidated its roles in plant stress tolerance.Our study found that theArabidopsistransgenic linesoverexpressingOsABTsignificantly increased tolerance to ABA and salt stresses,as well as sensitivity to drought stress.Moreover,the bimolecular fluorescent complimentary (BiFC) assay revealed that OsABT interacted with OsABI2,an important component in the ABA signaling pathway.Our results provided important insights in the role ofOsABTin ABA signaling pathway and its functions under diverse stresses,such as salt and drought stresses.
RESULTS
Identification and phylogenetic analysis of OsABT
To identify theOsABTgene from rice (Oryza sativa),the protein sequence ofArabidopsisABT (At1g49450) was used as a query to BLAST the rice RNAseq database ofOryza sativaJaponica Group (https:// archive.gramene.org/).TheABTsequence with higher similarity index was cloned and sequenced,and the homologous gene in rice was later named asOsABT(Os03g0738700).OsABT contains 488 amino acid residues and shares 47.95% identity with AtABT (Fig.1-A).According to the results of domain analysis performed using the SMART program,the deduced amino acid sequence possesses seven highly conserved WD40 domains (Fig.1-B).The results indicated that OsABT belongs to the WD40 protein family.
To understand the evolutionary relationships between OsABT and its homologs in different plant species,a BLASTP search was performed in the NCBI database.Then,a phylogenetic tree was constructed including OsABT,AtABT and 12 other homologous protein which sharing a high percentage of identity with OsABT.The phylogenetic tree showed that OsABT was clustered together with the proteins from monocots,but was far from the homologous proteins of dicots,especially from AtABT (Fig.1-C).These results suggested that the functions of OsABT and its homologs in monocots might be conserved,and that the functions of OsABT in rice might be different from those of AtABT inArabidopsis.
OsABT gene structure and protein analysis
From the database of Gramene (https://archive.gramene.org/),we found that the length ofOsABTis 2 175 bp,with the coding sequence of 1 470 bp,5′-UTR of 73 bp and 3′-UTR of 632 bp.To learn more about the gene,we analyzed the gene structure ofOsABTusing GSDS (http://gsds.cbi.pku.edu.cn/).TheOsABTgene sequence showed absence of intron,which was similar to theAtABTgene structure,although the lengths of their 5′-and 3′-UTRs were different (Fig.2-A).The results were consistent with the phylogenetic analysis results,showing similarities and differences in evolutionary process betweenOsABTandAtABT.
To investigate the subcellular localization of the OsABT protein,theOsABTgene was fused with the green fluorescent protein (GFP) vector PEZR (K)-LC driven byCaMV35Spromoter,and transformed intoArabidopsisvia the floral-dip method (Clough and Bent,1998).GFP fluorescence was observed in the nucleus,peripheral membrane and cytoplasm ofArabidopsisroot cells expressing GFP-OsABT (Fig.2-B).These results suggested that the OsABT protein is distributed in multiple cellular locations.
Expression pattern of OsABT gene
AtABTis responsive to ABA and osmotic stress,as shown inArabidopsiseFP Brower (http://bbc.utoronto.ca.efp).However,there was no significant difference in phenotype betweenAtABTmutantArabidopsisand wild type (WT) plants under drought conditions (Wang et al,2020).To analyze whetherOsABTresponds to ABA and osmotic stress,we first intercepted the sequence of 1 947 bp upstream of the initiation codon ATG (Fig.S1),and then analyzed the promoter ofOsABTusing the New PLACE web (https://www.dna.affrc.go.jp/PLACE/?action=newplace).The results showed manycis-elements that respond to ABA and stress are in theOsABTpromoter,such as ABRELATERD1,MYB1AT,MYCCONSENSUSAT and DRECRTCOREAT (Table 1).Moreover,we aligned the promoters ofOsABTandAtABT,and the results revealed that they only shared 36.72% of identity (Fig.S2).These results suggested thatOsABTmight be a stress response gene,andOsABTandAtABThad different stress response patterns.
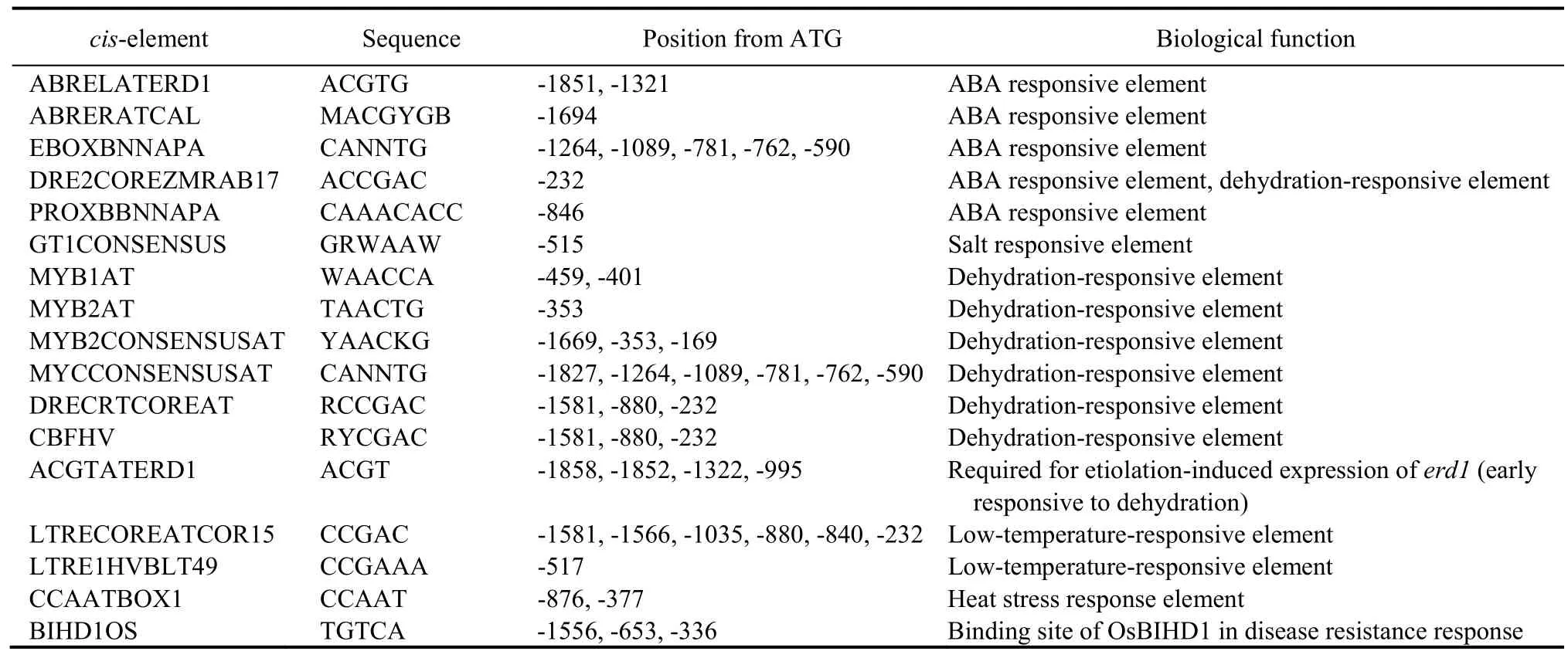
Table 1.cis-elements that respond to abscisic acid (ABA) and stress in OsABT promoter.
To analyze the expression patterns ofOsABT,rice seedlings of Nipponbare were treated with 150 mmol/L NaCl,50 μmol/L ABA or 10% PEG8000.The qRT-PCR results showed that the transcription level ofOsABTwas up-regulated,reaching a maximum level at 3 h after ABA treatment,and then gradually declined in the long-term treatment (Fig.3-A).The expression pattern ofOsABTunder the NaCl treatment was similar to that under the ABA treatment (Fig.3-B).In contrast,under the treatment with PEG8000,the transcription levelofOsABTwas down-regulated (Fig.3-C).These results suggested thatOsABTplays a vital role in drought and salt stresses,and that the function ofOsABTin stress responses is mediated by ABA.
OsABT transgenic Arabidopsis confers tolerance to ABA during greening
To investigate whetherOsABTis involved in ABA response,we cloned and transformedOsABTintoArabidopsis.Two homozygous35S::OsABTtransgenic lines (1-5 and 4-5) were chosen through RT-PCR and used for investigation of ABA response inArabidopsis(Fig.4-A).The seeds of Columbia-0 (Col-0) and two homozygousOsABTtransgenicArabidopsislines were sown on solid MS medium and MS medium containing ABA (1 μmol/L),and grown in a growth chamber.The greening rates of the three samples were calculated at 14 d after germination.The results showed that the greening rates were not different between the two transgenic lines and Col-0 grown on MS medium (Fig.4-B).On the contrary,the transgeniclines showed much better growth and higher greening rates than Col-0 under the ABA treatment (1 μmol/L) (Fig.4-B and -C),indicating thatOsABTis a negative regulator in the ABA signal transduction pathway.
OsABT improves greening rate of transgenic OsABT Arabidopsis under salt stress
To analyze whetherOsABTplays important roles in salt stress,the seeds of Col-0 and twoOsABTtransgenicArabidopsislines were sown on solid MS medium and MS medium containing NaCl (125 mmol/L).Four days later,the greening rates of the three samples were calculated.As shown in Fig.5-A,there was no significant difference between the two transgenic lines and Col-0 grown on MS medium.In contrast,under the NaCl treatment,the greening rates were significantly higher in the two transgeniclines than in Col-0 (Fig.5),suggesting thatOsABTpositively regulates plant resistance to salt stress.
OsABT transgenic Arabidopsis lines are sensitive to drought stress
To comprehend the molecular function ofOsABTin drought stress,Col-0 and the homozygous transgenic lines (1-5 and 4-5) were grown in four small pots and planted alternately.After transplanting for 2 weeks,all the seedlings were withheld water for 2 weeks and then rewatered.Upon watering,the transgenic lines showed higher sensitivity to drought stress and obviously lower survival rate than Col-0 (Fig.6),suggesting thatOsABTis a negative regulator under drought stress.
OsABT interacts with OsABI2
WD40 proteins are usually regarded as a scaffolding platform for protein-protein interactions (Xu and Min,2011),and ABA is an important hormone for plant adaptation to abiotic stress (Hirayama and Shinozaki,2007).ABT is dependent on ABI2,which is a vital component in the ABA signaling transduction pathway to inhibit ABA signaling (Wang et al,2020).We speculated that OsABT interacted with OsABI2 in rice.To test this,full-length OsABT was fused within the N-terminus of yellow fluorescent protein (YFP) to generate OsABT-YFPN,and OsABI2 was fused to the C-terminus of YFP to generate OsABI2-YFPC.These constructs were co-induced into theleaf cellsofNicotiana benthamiana.As expected,there were strong YFP signals in both the cytoplasm and nucleus when OsABT-YFPNand OsABI2-YFPCwere co-induced intoleaf cellsofN.benthamiana(Fig.7).Nevertheless,no YFP signal was detected for OsABT-YFPNand YFPCor OsABI2-YFPCand YFPN(Fig.7),indicating that there is an interaction between OsABT and OsABI2 proteins.
DISCUSSION
OsABT is a typical protein of WD40 protein family in rice
WD40 proteins comprise the largest protein family in eukaryotes (Stirnimann et al,2010),which are characterized by the presence of multiple repeats of~40 amino acids named ‘WD40 repeats’ at the carboxyl termini,and ends with the sequence WD (Neer et al,1994).Generally,the WD40 domains contain seven repeats that form a highly stable β-propeller structure,and a minimum of four WD40 repeats are needed to form a higher-order and functional structure (Sondek et al,1996;Chothia et al,1997).
AtABT is a protein containing seven WD40 repeats inArabidopsis.In this study,we cloned the homologous gene ofAtABTin rice,namedOsABT.Protein sequence analysis revealed that OsABT possessed seven conserved WD40 repeats,the same as AtABT (Fig.1-A and -B).Phylogenetic analysis showed that OsABT had close evolutionary relationship with WD40 proteins inSorghum bicolor(Fig.1-C).These results indicated OsABT belongs to the WD40 protein family.The WD40 domain usually acts as a scaffold for protein-protein interactions and alter the process of molecular recognition (Xu and Min,2011).One well-studied example is a WD40-repeat protein HOS15 which can interact with histone H4 to regulate plant tolerance to cold stress (Zhu et al,2008).Here,BiFC analysis revealed that OsABT can interact with OsABI2 (Fig.7).This result provided another evidence to support that WD40 proteins are involved in protein interactions.
OsABT is involved in response to abiotic stress in rice
Although there are several studies on the functions of WD40 proteins,most of their functions in plants are still unclear.In recent years,many WD40 proteins have been found to participate in abiotic stress responses in the model plantArabidopsisand in crop plants.ArabidopsisWDR5a(WD40-REPEAT 5a) mutant,wdr5a,is more sensitive to drought stress than its WT plants,as they have reduced stomatal closure and decreased expression of drought-related genes (Liu et al,2017).WhenTaWD40D,a wheat WD40 protein,is exogenously overexpressed inArabidopsis,the transgenic plants show improved tolerance to ABA,salt and osmotic stresses during seed germination and seedling development.In addition,the increased tolerance of transgenic lines may be due to changes in the expression patterns of genes in salt overly sensitive (SOS) pathway,ABA-dependent pathway and ABA-independent pathway (Kong et al,2015).Furthermore,OsRACK1A,a WD40 protein in rice,negatively affects the salt tolerance phenotype in rice.OsRACK1A-suppressed transgenic rice can significantly accumulate more ABA and more transcripts of ABA-and stress-inducible genes compared with the WT plants (Zhang et al,2018).
In this study,the expression ofOsABTinitially increased until reaching a peak,and then gradually decreased after 3 h of treatment with ABA or NaCl (Fig.3-A and -B).In contrast,the expression ofOsABTwas down-regulated after the PEG8000 treatment (Fig.3-C).These results indicated that theOsABTgene may be involved in salt and drought stresses.WhenOsABTwas transformedintoArabidopsis,the resulting transgenic lines had higher greening rates than Col-0 under the NaCl treatment (Fig.5).On the contrary,the transgenic lines had lower survival rates under drought conditions and were more sensitive to drought stress (Fig.6).These results indicated thatOsABThas a positive regulatory effect on salt stress and a negative regulatory effect on drought stress inArabidopsis.Under the drought conditions,bothArabidopsislines withAtABToverexpression orAtABTknockout have similar drought phenotype to Col-0 plants (Wang et al,2020).This result was inconsistent with our finding on the function ofOsABTin drought tolerance.There were two reasons whyOsABTandAtABThad different functions under drought stress.The first one may be that they shared low percentages of identity (47.95%) with each other,which resulted in different protein structures (Fig.1-A),and the other one may be that they shared low percentages of promoter identity (36.72%),which resulted in different expression patterns (Fig.S1).
ABA is a vital hormone for plant adaptation to stress.Many components in the ABA signaling pathway are involved in plant stress tolerance (Miller et al,2007;Cutler et al,2010;Hubbard et al,2010).PP2Cs (type 2C protein phosphatases),includingArabidopsisABI1,ABI2,HAB1,AtPP2CA and RAB18,are important components and negative regulators of ABA signal transduction (Leung et al,1997;Sheen,1998;Gosti et al,1999;Kuhn et al,2006).Previously,ZmPP2Coverexpression inArabidopsisresults in decreased tolerance of transgenic plants to salt and drought stresses.ABI2-dependent ABA signaling can control HrpN-induced drought tolerance inArabidopsis(Dong et al,2005).SOS2,a protein kinase and a key component of the SOS pathway,can physically interact with ABI2 (Rodriguez et al,1998;Qiu et al,2002;Ohta et al,2003).These results suggested thatABI2is a key factor for plant adaptation in response to salt and drought stresses.BiFC assay confirmed that ABT can interact with ABI2 (Wang et al,2020).Here,we also found a similar result that OsABT can interact with OsABI2 (Fig.7).Moreover,the transgenicOsABTArabidopsislines showed a higher resistant to salt stress than Col-0 plants (Fig.5).These results suggested thatOsABTregulates salt tolerance through the ABA and SOS signaling pathways mediated byOsABI2.TheOsABTtransgenicArabidopsislines were insensitive to ABA and sensitive to drought stress,compared with Col-0 plants (Figs.4-B,4-C and 6).Furthermore,ABT switches off the ABA signaling pathway by obstructing the interaction between PYR1/ PYL4 and ABI1/ABI2 (Wang et al,2020).All these results implied that OsABT inhibits the ABA signaling pathway and decreases plant responses to drought stress via interaction with OsABI2.Although we have analyzed the roles ofOsABTin abiotic stress conditions inArabidopsis,the roles and the regulatory mechanisms ofOsABTin rice under abiotic stress conditions still need further research.
METHODS
Plant materials and growth conditions
Rice (O.sativaL.cv.Nipponbare) was used as an experimental material.Rice seeds were cultivated in Hoagland’s solution (Hoagland and Arnon,1950) in 96-well plates with cut bottom.Rice leaves were harvested at the 3-leaf stage for RNA and DNA extraction.The rice seedlings were grown in a growth room at temperatures of 28 °C day/25 °C night with a 16-h light/8-h dark photoperiod under 50% relative humidity (Kawasaki et al,2001).
A.thalianaCol-0 was used.Col-0 seeds were surface-sterilized using 15% sodium hypochlorite,washed four times with sterile water,sown on solid MS medium and vernalized for 2 d at 4 °C in dark.Next,the MS plates were placed in a growth chamber.After 11 d,the Col-0 seedlings were transplanted into a mixture which contains two thirds nutrient soil and one third vermiculite,and grown at 22 °C in a growth room under a 16-h light/8-h dark photoperiod.One month later,the adultArabidopsisplants were used for transformation studies.
To analyze the function ofOsABTin ABA and salt stresses,the seeds of Col-0 and homozygousOsABTtransgenicArabidopsiswere surface-sterilized,sown on solid MS medium or MS medium containing ABA (1 μmol/L) or NaCl (125 mmol/L),and vernalized in dark at 4 °C for 2 d.Next,the samples were moved to a growth chamber with a constant temperature of 22 °C under a 16-h light/8-h dark photoperiod.The greening rates were calculated after 14 d of culture on MS medium containing ABA,and after 5,6,7 and 8 d of culture on MS medium containing NaCl.
For drought assay,the seeds of Col-0 and homozygousOsABTtransgenicArabidopsiswere sown on solid MS medium after surface-sterilization and moved to a growth chamber for 10 d.Next,the seedlings were transplanted into four small pots and alternately planted with four replicates.After transplantation for 2 weeks,the seedlings were grown with no water supply for 2 weeks.Next,the seedlings were rewatered and grown in a growth room at 22 °C under a 16-h light/8-h dark photoperiod.After 1 week of rewatering,the survival rates were calculated,and the phenotypes were recorded by taking photos.
Vector construction and Arabidopsis transformation
To generate35S::OsABTtransgenicArabidopsislines,full-lengthOsABTcoding sequence was amplified from rice using specific set of primers and cloned into a pMD18-T simple vector.The pMD18-T-OsABTvector was sequenced to test the correctness ofOsABTsequence at Wuhan GeneCreate Biological Engineering Co.,Ltd.(Wuhan,Hubei,China).The pMD18-T-OsABTvector with correctOsABTsequence was digested withSpeI andSacI (TaKaRa,Dalian,China),and the full-length coding sequence ofOsABTwas cloned into theSpeI andSacI sites of a p6MYC vector to generate p6MYC-OsABTconstruct.Then,the correctness of the p6MYC-OsABTwas identified by enzyme digestion withSpeI andSacI and sequencing.The correct recombinant vector p6MYC-OsABTwas used to obtain the transgenicArabidopsisofOsABT.The primers used for cloningOsABTwere OsABT-OV-F and OsABT-OV-R,as shown in Table S1.
To obtain the transgenicOsABTArabidopsislines,theAgrobacteriumGV3101 containing the recombinant vector p6MYC-OsABTwas suspended,and then transformed into the buds ofArabidopsisusing the floral-dip method (Clough and Bent,1998).
Phylogenetic and domain analyses,gene structure analysis,and protein sequence alignment
TheOsABTsequence was obtained from Gramene (https:// archive.gramene.org/) BLAST program.OsABT homologous protein was searched in NCBI (http://www.ncbi.nlm.nih.gov/) using BLASTP.The neighbor-joining method was used for phylogenetic tree construction in the MEGA software (version 6.0).The SMART program (http://smart.embl-heidelberg.de/) was used to analyze conserved domains in the OsABT protein.
To analyze the gene structure ofOsABT,the gene sequences ofOsABTandAtABTwere obtained and the gene structure was constructed via GSDS (http://gsds.cbi.pku.edu.cn/).To analyze the sequence identity between OsABT and AtABT,the protein sequences of OsABT and AtABT were aligned using DNAMAN 7.0.
Subcellular localization analysis
To analyze the subcellular localization of OsABT,the coding sequence ofOsABTwas amplified through PCR and then cloned into a binary vector PEZR (K)-LC betweenEcoRI andXbaI restriction sites.The construct was transformed intoA.thalianathrough the floral-dip method (Clough and Bent,1998),whereas the pEZR(K)-LC vector was used as a control.Fluorescence imaging was performed by a confocal laser scanning microscopy (Leica SP8;Leica Microsystems,Wetzlar,Germany).The primer set used for cloningOsABTconsisted of OsABT-GFP-F and OsABT-GFP-R,as shown in Table S1.
RNA extraction,RT-PCR and qRT-PCR analyses
Total RNA was extracted from transgenic and control plantlet leaves using TRIzol reagent according to the manufacturer’s instructions (Invitrogen,Shanghai,China).To detectOsABToverexpression inArabidopsis,we synthesized first-strand cDNA using the total RNA extracted from transgenic and Col-0 plants.2× Es Taq master mix (Dye) (CWBIO,China) was used for PCR amplification.The sequences of specific primers OsABT-F and OsABT-R are shown in Table S1.RT-PCR was performed using a heated lid thermal cycler (Bio-Rad,USA) under the following conditions:pre-denaturation at 94 °C for 4 min,denaturation at 94 °C for 30 s,annealing at 56 °C for 30 s,and extension at 72 °C for 60 s,for 28 cycles.AtUBQ10(At4g05320) was used as an internal control.The primers used were based on those described by Chen et al (2017).
To investigate the transcript patterns ofOsABTunder the ABA,salt and drought stress conditions.The rice seedlings were treated with 50 μmol/L ABA,150 mmol/L NaCl or 10 g/mL PEG8000.The rice roots were collected at the time point of 0 (control),1,3,6,12 and 24 h after treatment and frozen in liquid nitrogen immediately for RNA isolation.cDNAs were synthesized from the total RNA extracted from the rice roots.qRT-PCR was performed using a SYBR®premix Ex Taq™ (Tli RNaseH Plus) (TaKaRa,Dalian,China) in an ABI Prism 7000 system (Thermo Fisher Scientific,USA).Each PCR was conducted with three biological and technical replicates.OseEF-1αwas used as an internal reference.The standardized 2-ΔΔCTmethod was used to normalize the obtained results.The primer sets used for qRT-PCR are given in Table S2.
BiFC assay
The binary vectors used for BiFC assay were pEarleyagate201-YN and pEarleygate202-YC vectors,in accordance with Wang et al (2015).TheOsABTcoding sequence was cloned into pEarleyagte201-YN to obtain OsABT-YFPN,whereas the coding sequence ofOsABI2was cloned into pEarleygate202-YC to yield OsABI2-YFPC.The reconstructed vectors were transformed intoA.tumefaciensstrain GV3101 and introduced intoN.benthamianaepidermal cells.At 3 d after injection,YFP fluorescence was imaged through a confocal laser scanned microscopy (Leica SP8;Leica Microsystems,Wetzlar,Germany).
ACKNOWLEDGEMENTS
This study was supported by the Major Program of the Zhejiang Province for Food Crop Breeding (Grant No.2016C02050-6) and the Key Program of Hangzhou Agricultural Scientific Research (Grant No.20191203B08).We also thank LI Xia and WANG Zhijuan from Huazhong Agricultural University,for their help in experimental ideas and methods.
SUPPLEMENTAL DATA
The following materials are available in the online version of this article at http://www.sciencedirect.com/journal/rice-science;http://www.ricescience.org.
Fig.S1.Promoter sequence ofOsABT.
Fig.S2.Multiple sequence alignment ofOsABTandAtABTpromoters.
Table S1.Primers used in this study.
Table S2.qRT-PCR primers used for analyzing expression patterns ofOsABTunder ABA,salt and drought stresses.
- Rice Science的其它文章
- Effect of Roasting Temperature on Production of Functional Wild-Rice Tea and Factors Influencing Purchase Intention Towards Wild-Rice Tea
- Rice Germination and Its Impact on Technological and Nutritional Properties:A Review
- Nucleus-Encoded Thylakoid Protein,OsY3IP1,Confers Enhanced Tolerance to Saline and Alkaline Stresses in Rice
- NRL3 Interacts with OsK4 to Regulate Heading Date in Rice
- Conservation and Utilization of Genetic Resources of Wild Rice in China
- Poaceae Orthologs of Rice OsSGL, DUF1645 Domain-Containing Genes,Positively Regulate Drought Tolerance,Grain Length and Weight in Rice

