Microbiologic risk factors of recurrent choledocholithiasis postendoscopic sphincterotomy
Ying Li, Wen-Hui Tan, Jia-Chuan Wu, Zhi-Xin Huang, Yan-Yan Shang, Biao Liang, Jian-Hui Chen, Rui Pang,Xin-Qiang Xie, Ju-Mei Zhang, Yu Ding, Liang Xue, Mou-Tong Chen, Juan Wang, Qing-Ping Wu
Abstract
Key Words: Choledocholithiasis; Biliary tract; Microbiome; Endoscopic sphincterotomy; Recurrence;Lactobacillus
INTRODUCTION
Cholelithiasis is a common and socially significant health problem worldwide, occurring in approximately 5 %-22 % of adults, with synchronous common bile duct stone (CBDS) in 20 % of these patients[1 -4 ]. In western countries, it is one of the leading gastrointestinal conditions that results in hospitalization[4 ]. Cholelithiasis with CBDS can lead to biliary obstruction, secondary cholangitis, and obstructive jaundice, endangering lives in some severe cases and often requiring surgical interventions[5 ]. The introduction of endoscopic treatment started a new era in the treatment of choledocholithiasis[6 -9 ], and management by endoscopic sphincterotomy (EST) or endoscopic papillary balloon dilation (EPBD) has become widespread, replacing open laparoscopic cholecystectomy or open common bile duct exploration with choledochoscopy[10 ,11 ].
However, long-term surveys have revealed up to 39 % recurrence of choledocholithiasis post-EST, and life-long follow-ups are still needed after surgery[12 -14 ]. Recurrent choledocholithiasis post-EST involves complicated factors, including infections and biliary anatomical abnormalities[15 ,16 ]. The elimination of certain pathogens in the bile duct can significantly reduce the recurrence rate[17 ,18 ].Therefore, further investigations into the microbiological etiology and underlying mechanisms of recurrent choledocholithiasis post-EST are crucial for its prediction and prevention in clinical practice.
Complex microbiomes in the biliary system have been observed using next-generation sequencing(NGS)[19 ]. In these systems, the microbiota metabolize and secrete cholesterol and bile acids; their dysfunction may cause pathophysiological defects and result in stone formation[20 ,21 ]. Unlike primary stones, secondary stones in recurrent choledocholithiasis predominately consist of more cholesterol than calcium bilirubinate[22 ], and their microbiological etiology remains unclear.
In this study, we investigated the microbiological etiology of recurrent choledocholithiasis using NGS to find the key pathogens associated with recurrence post-EST and their metabolic characteristics in disease relapse.
MATERIALS AND METHODS
Study participants and sample collection
Ethical compliance:Consecutive recruitment of eligible patients was carried out in the Department of Endoscopy at Guangdong Second Provincial General Hospital from May to June 2020 . All experimental protocols were approved by this hospital’s ethics committee (project 2019 -QNJJ-14 -02 ). The study design complied with all relevant ethical regulations in accordance with the Declaration of Helsinki and Belgian Privacy Commission. Written consent was obtained from all patients in the study.
Study cohort:In this study, we included 43 choledocholithiasis participants diagnosed using computed tomography (CT) or magnetic resonance cholangiopancreatography (MRCP). All patients were assessed by experienced doctors, without risk of EST-related complications[10 ]. Patients with a history of malignant diseases, autoimmune diseases, diabetes, structure abnormality of the biliary tract, or any exposure to antibiotics within one month were excluded from the study. All patients accepted laparoscopic cholecystectomy (LC) treatment following an EST or endoscopic papillary balloon dilation(EPBD), during the same hospitalization episode. The components of the stones were recorded according to the method of Dosch[23 ]. All patients received CT or MRCP examinations one week after the treatment to ensure the complete removal of stone in the biliary tract. All participants underwent at least one-year follow-up with transabdominal ultrasonography every three months, and CT or MRCP was performed once recurrence of choledocholithiasis was indicated through clinical presentations or imaging examinations. Patients were divided into stable and recurrent groups according to their disease evaluation at the end of the follow-up period.
Bile sample collection:Bile samples were collected during endoscopic treatment. The ERCP was performed to confirm the diagnosis of choledocholithiasis, followed by the EST or EPBD treatment, and the bile sample was collected through suction during the treatment. The bile samples were immediately transported to the laboratory and stored at -80 °C until extraction.
Microbiome DNA extraction and 1 6 S rRNA gene amplicon sequencing
Bile sample (3 mL) was centrifuged at 16000 × g at 4 °C for 10 min, and the pellet was washed twice with phosphate-buffered saline before DNA extraction. Microbiome DNA was then extracted using the QIAamp PowerFecal DNA kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
The16 S rRNA gene obtained from each bile sample was amplified by targeting the V3 -V4 hypervariable regions using the following primers: 341 F 5 ′-CCTACGGGNGGCWGCAG-3 ′ and 806 R 5 ′-GGACTACHVGGGTWTCTAAT-3 ′ using the UCP Multiplex PCR Кit (Qiagen)[24 ]. The amplicon library was prepared using the QIAseq Ultralow Input Library Кit (Qiagen). An Agilent Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, United States) and Qubit dsDNA HS Assay Кit(Invitrogen Life Technologies, Carlsbad, CA, United States) were used to validate the library pooling.Paired-end sequencing was conducted using the MiSeq platform (Illumina, San Diego, CA, United States) with MiSeq Reagent Кit version V3 (Illumina).
Microbiome sequence curation and analysis
Trimming and quality filtering of the data were performed using the CLC Genomic Workbench version 20 .0 , with the Microbial Genomics Module (Qiagen). Sequences were matched to the Greengenes database version 13 .5 .
The amplicon sequencing, and the taxonomic and statistical analyses were performed using Calypso version 8 .84 [25 ]. Alpha diversity was determined based on Fisher’s alpha index, which was assessed using the analysis of variance test. Microbial diversity was visualized using the canonical correspondence analysis based on the prognosis groups. Кey taxonomic discovery analysis related to prognosis was performed using linear discriminant analysis effect size (LEfSe) at the genus level[26 ].The relative abundance (RA) measurements of the genera, with biomarker significance were compared using the Wilcoxon rank test.
The core microbiome was identified as described by Ainsworth[27 ]. Network analysis was performed to identify the co-occurring and exclusive bacteria using Calypso[25 ]. Genera and orders of the bile microbiome were represented as nodes, taxa RAs as node size, and edges as positive and negative associations. Networks were generated based on the associations between both genera and orders using Pearson’s correlation; nodes were colored based on their association with different prognosis groups.Only relationships with statistical significance (P <0 .05 ) were visualized in the network.
Metagenome prediction of the bile microbiome was performed using the amplicon sequencing approach in the Phylogenetic Investigation of Communities by Reconstruction of Unobserved States(PICRUSt)[28 ]. The statistical differences between the diagnosis groups were examined using the Кyoto Encyclopedia of Genes and Genomes (КEGG) pathway hierarchy level using analysis of variance.Correlation analysis was carried out between the key genera and metabolic pathways in bile using Pearson’s correlation test. Survival analysis of the identical microbiological risk factors was carried out using Кaplan-Meier analysis.
Statistical analyses
Except for those analyzed using Calypso, data were analyzed using GraphPad Prism v7 .00 software(GraphPad, La Jolla, CA, United States). All analyses in the study were statistically significant atP <0 .05 , andPvalues were adjusted using false discovery rate, Bonferroni, or area under curve correction.
RESULTS
Clinical features and prognosis of patients with choledocholithiasis one-year post-EST
Forty-three choledocholithiasis patients, who underwent LC following EST were recruited in this study and received a one-year follow-up survey. Thirteen patients had co-occurrence of cholelithiasis; the baseline clinical characteristics of the 43 choledocholithiasis patients were shown in Table 1 . The stone components were recorded according to the methods of Dosch[23 ]; they were classified as brown pigmented stones, black pigmented stones, cholesterol stones, and mixed stones. Four recurrent cases without other complications were observed using routine ultrasonography as well as CT during the follow-up period. No significant differences were found in the clinical features or the stone components between patients with and without recurrent choledocholithiasis.
Bile microbiome characteristics in patients with choledocholithiasis
A total of 702 unique operational taxonomic units were identified in the bile of all patients with choledocholithiasis, indicating the diversity of the microbiome in bile (Figure 1 ).Streptococcusand an unclassified genus ofEnterobacteriaceaewere the most dominant genera; they were detected in the bile of 28 and 29 patients, respectively. The average RAs of Streptococcus and Fusobacterium in bile were 13 .59 %and 19 .91 %, respectively.
Key microorganisms in bile of patients with recurrent choledocholithiasis
LEfSe biomarker discovery analysis identifiedFusobacterialesandNeisserialesas biomarkers in the recurrent group andLactobacillalesin the stable group at the order level (Figure 3 A). The RAs ofFusobacteriales(56 .61 % ± 14 .81 % vs 3 .47 % ± 1 .10 %) and Neisseriales (8 .95 % ± 3 .42 % vs 0 .69 % ± 0 .32 %) were higher in patients with recurrent choledocholithiasis than that in stable patients post-EST (P< 0 .05 ), while the RA ofLactobacillaleswas significantly lower in the recurrent group (1 .48 % ± 1 .28 %) than that in the stable group (25 .04 % ± 4 .76 %; P < 0 .05 ; Figure 3 B).
Bile microbiological ecosystem analyses in patients with choledocholithiasis with different prognoses post-EST
Core microbiome analyses showed thatStreptococcus, Prevotella, Fusobacterium, an unclassified genus ofEnterobacteriaceae, and an unclassified genus ofClostridiaceaewere the shared core genera in both the stable and the recurrent group.Veillonella,Oribacterium,Neisseria,Leptotrichia, andCampylobacterwere the specific core genera in the recurrent group, whileEnterococcus,Clostridium, and an unclassified genus ofAeromonadaceaewere the unique genera in the stable group (Table 2 ). Construction of a microbiological co-occurrence network revealed a mutual relationship amongFusobacterium,Neisseria,andLeptotrichia(Figure 4 A).
Additionally,Lactobacillales,Fusobacteriales,Enterobacteriales,Clostridiales, andBacteroidaleswere the shared core orders in both the stable and the recurrent group.Pasteurellales,Neisseriales, andCampylobacteraleswere the unique core orders in the recurrent group, whilePseudomonadales,Burkholderiales,Bacillales, Aeromonadales, andActinomycetaleswere the unique orders in the stable group. Co-occurrence network analyses suggested mutual enhancement among the key recurrence-related pathogens in bileand antagonistic relationships amongLactobacillales, Fusobacteriales, andClostridialesin the ecosystem,indicating the role of probiotics in the prevention of recurrence (Figure 4 B).

Table 1 Clinical characteristics of choledocholithiasis patients
Functional characteristics of bile microbiome in patients with choledocholithiasis with different prognoses post-EST
The metabolites from microorganisms are the key pathogenic factors for the host; therefore, the characteristics of the metabolic pathways in bile were analyzed for deeper insight into the microbiologic etiology of recurrent choledocholithiasis post-EST. Comparative analyses of microbiological functions were carried out at the 2ndhierarchy level of the КEGG pathway. In the stable group, the bile microorganisms were active in the transcription and metabolism related to the nervous system, infectious diseases, biosynthesis of carbohydrates and amino acids; while, in the recurrent group, the microbes were active in translation, replication and repair, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, genetic information processing, energy metabolism, and biosynthesis of secondary metabolites (Figure 5 ).
Furthermore, correlations between the key genera in the two groups and the different metabolic pathways were analyzed to identify the influence of certain microbes on the host (Figure 6 ). In the bile ecosystem of the patients with recurrent disease,FusobacteriumandCampylobacterhad positive correlations with the metabolism of amino acids, replication and repair, and translation (P< 0 .05 ), while the unclassified genus ofEnterobacteriaceaehad a negative correlation with all the discrepant metabolicpathways between the two groups (P< 0 .05 ).Leptotrichiahad a positive correlation with all the discrepant metabolic pathways in the bile of the stable group (P< 0 .05 ). Correlation analyses indicated that in bile of the recurrent group, increasedFusobacteriumcould alter the metabolism of amino acids,replication and repair, and translation functions, leading to the formation of secondary bile stones.
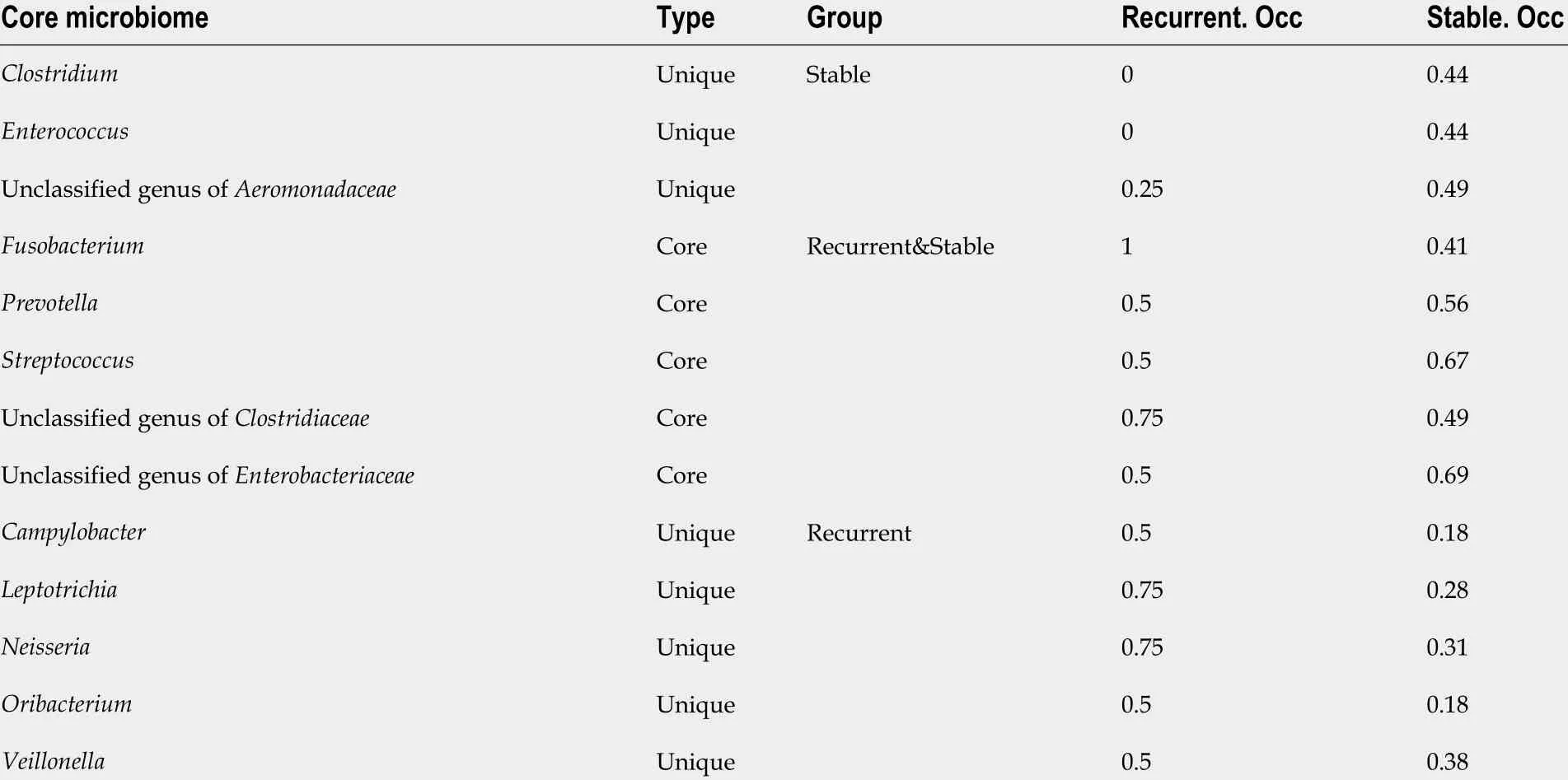
Table 2 Core microbiome in bile of choledocholithiasis patients with different prognosis
Microbiologic risk factor analysis of the recurrent group post-EST
FusobacterialesandNeisserialeswere identified as the bile biomarkers in the recurrent group andLactobacillalesin the stable group. Кaplan-Meier analysis was carried out to confirm whether these biomarkers can be used as independent predictive factors for recurrence post-EST (Figure 7 ). The statistical results revealed that patients withLactobacillalesin the bile were at a lower risk of recurrence post-EST (P=0 .03 ) than patients, who lacked this order in their bile.
DISCUSSION
It was assumed that the biliary system is sterile in healthy people; however, an increasing amount of NGS-supported evidence shows that bile supports a complex and abundant microbiome in healthy individuals[19 ,29 ]. The frequently identified microorganisms using traditional culture techniques areEnterococcus,Klebsiella, andPseudomonas; these bacteria are active in reducing the bile acid pool and regulating bile acid metabolism[30 -33 ]. However, the contribution of microbes to the biliary system is still unclear. NGS techniques revealed that the most common inhabitants of the biliary tract areProteobacteria,Firmicutes,Bacteroidetes,Actinobacteria,Fusobacteria,Synergistetes, and candidate phylumSaccharibacteria(TM7)[34 ]. Some of these microorganisms regulate the hydrolysis of bile acids to constituent components, cleavage of exogenous aromatic rings, deconjugation of bile acid complexes by hydrolytic enzymes, and the formation of free bile acids[35 ]. The disturbance of the microbiologic ecosystems in bile may lead to dysfunctional bile acid metabolism, resulting in a series of bile duct diseases[20 ,21 ]; however, the most disease-specific pathogens and their unique functions remain unknown.
项目总预算约18.83亿元(其中中央财政约占64.9%,地方预算约占35.1%),按照3年基本建成、5年基本完善的原则安排进度。
Similar to the results from previous studies[29 ,34 ,36 ], this study revealed that the biliary tract was composed of a diversity of bacteria, and the majority of microorganisms in the bile wereStreptococcus,Prevotella,Fusobacterium,Enterococcus,Veillonella, andClostridium.LactobacillusandLactococcuswere reported as the major genera in bile[36 ]; however, these two genera could only be detected in 11 patients in this study. These differences could be attributed to the differences in study designs; we included only patients with severe choledocholithiasis, who needed surgical intervention, for the analysis of microbial risk factors for disease recurrence. Another factor could be the difference in bile sampling; we chose the endoscopic route over open surgery for the collection of bile.
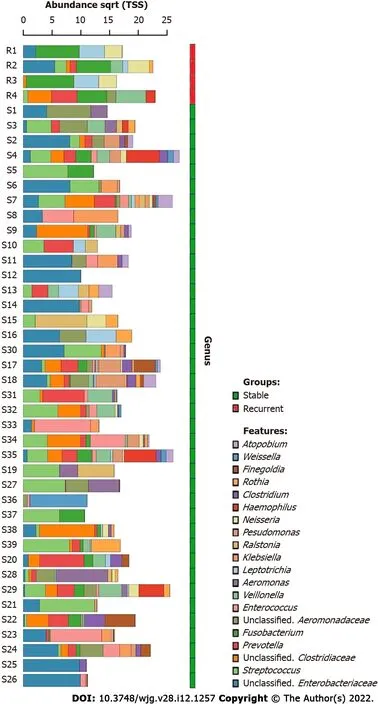
Figure 1 Dominant bacterial genera in the bile of choledocholithiasis patients. The top 20 dominant bacterial genera in bile are shown in the bar chart, the bile microbiome of recurrent choledocholithiasis patients post-endoscopic sphincterotomy are in the red group and choledocholithiasis patients without recurrence post-EST are in the green group. The genera were listed from the bottom to the top according to their relative abundance in bile samples.
Endoscopic treatment such as EST can provide definitive relief to choledocholithiasis; however, the formation of gallstones will not stop unless the etiologic factors are eliminated[12 ,13 ,37 ]. Among all the risk factors for choledocholithiasis recurrence, only biliary infections are correctable; microbiological treatment is the most potential therapy against the recurrence of choledocholithiasis[5 ,15 ,16 ,38 ,39 ].Therefore, investigation into the biliary microbiology characteristics of recurrent choledocholithiasis is crucial to both etiology and prevention studies. To the best of our knowledge, this is the first pilot study to investigate the microbiological risk factors in recurrent choledocholithiasis post-EST. IncreasedFusobacteriumandNeisseriawere recurrence-related biomarkers in the bile microbiome. Furthermore, we discovered the antagonistic potentials ofLactobacillusand an unclassified genus ofEnterobacterialesagainstFusobacteriumandNeisseria, indicating the potential use of probiotics in the prevention of recurrence post-EST.
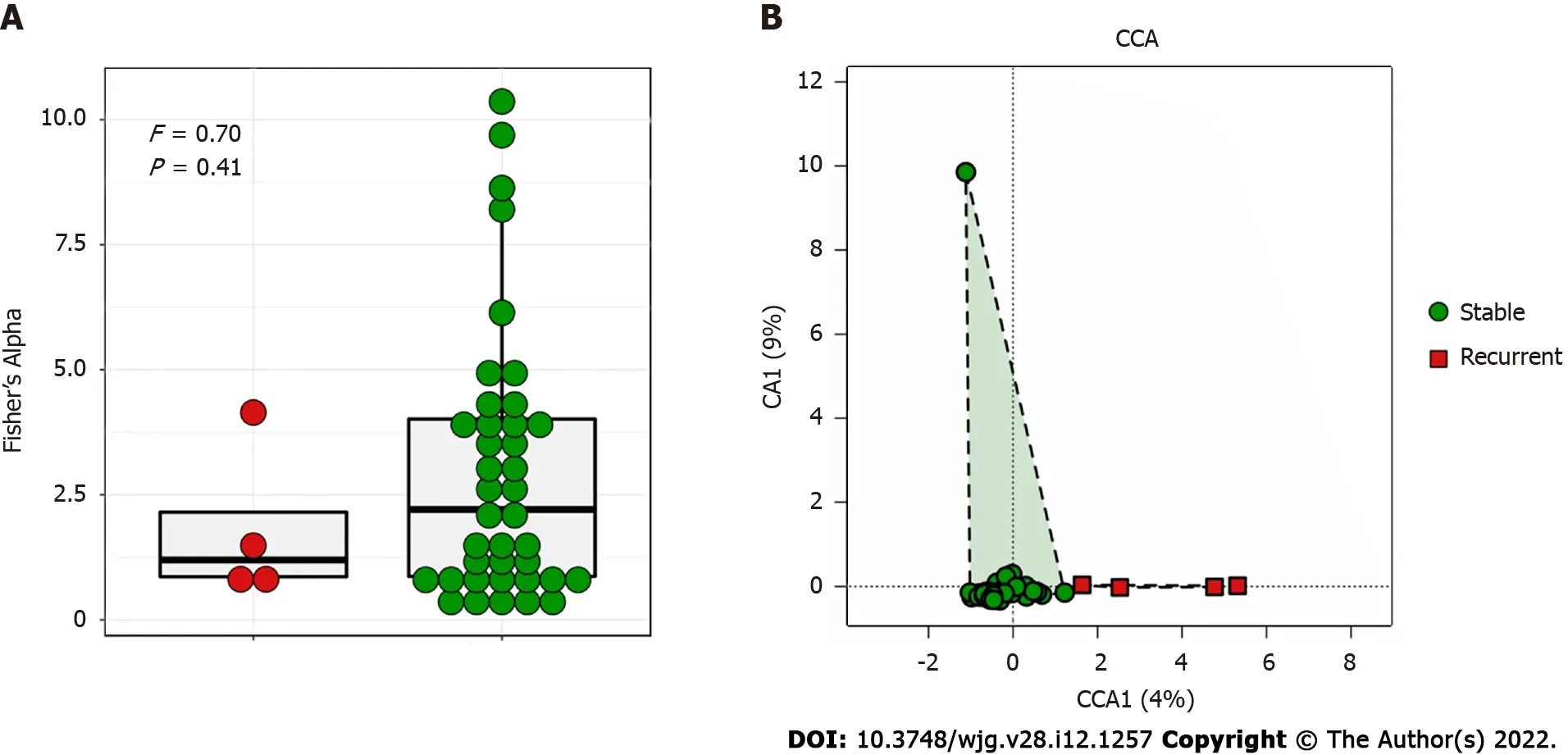
Figure 2 Diversity analysis of bile microbiome of choledocholithiasis patients. A: Comparison of alpha diversity of bile microbiome by the Fisher’s Alpha Index between stable (green) and recurrent (red) choledocholithiasis patients post-endoscopic sphincterotomy (EST); B: Comparison of beta diversity of bile microbiome using canonical correspondence analysis between stable (green) and recurrent (red) choledocholithiasis patients post-EST.
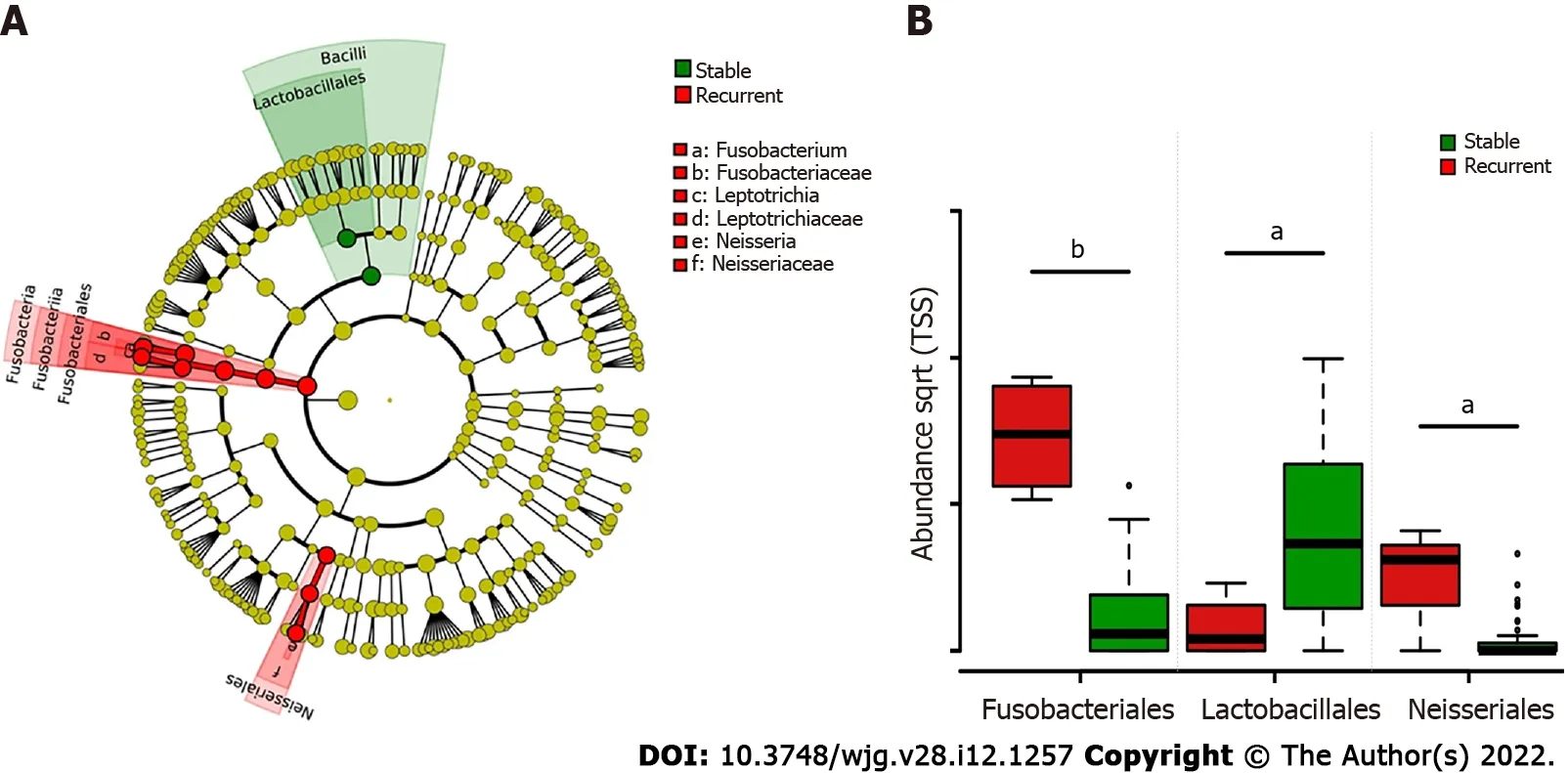
Figure 3 LEfSe analysis of group-specific microbes in choledocholithiasis patients with different prognosis post endoscopic sphincterotomy. A: Colored cladogram showing microbiota with biomarker significance in choledocholithiasis patients with different prognosis post-endoscopic sphincterotomy (EST) (red for biomarkers in recurrent patients and green for biomarkers in patients without recurrence post EST); B: Relative abundance comparison of microbes with biomarker significance in choledocholithiasis patients with different prognosis post-EST. Statistical significance is expressed as aP < 0 .05 , bP <0 .001 .
Bacteria in bile play an active role in gallstone formation[35 ].Escherichia coliandKlebsiellain bile can produce hydrolytic enzymes such as β-glucuronidase, phospholipase A[40 ], and conjugated bile acid hydrolase; in addition, they can cause deconjugation of bilirubin diglucuronide and precipitation of calcium bilirubinate, which ultimately leads to biliary stone formation[41 ,42 ]. We identifiedClostridiumas one of the key microorganisms in the bile microbiome, which, according to Leunget al[43 ], is a more important microorganism in the deconjugation of bilirubin diglucuronide thanE. coli, because it exhibits a 34 -fold higher β-glucuronidase enzyme activity in the biliary tract. A lack ofLactobacillusin the bile could be a probable risk factor for choledocholithiasis, becauseLactobacillusin bile can absorb cholesterol and reduce total serum cholesterol[44 ,45 ]. The core microbiome pattern in the bile of patients with choledocholithiasis in this study offers a more comprehensive understanding of the influence of the bile microbiome on biliary stone formation.
Furthermore, functional analysis indicated that the loss of transcription and metabolic abilities, and increased function of translation, replication and repair, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, genetic information processing, energy metabolism, and biosynthesis of other secondary metabolites could lead to recurrent choledocholithiasis. Most of these microbiologic functions were caused by the increased abundance ofFusobacteriumandLeptotrichiaand the loss of an unclassified genus ofEnterobacteriales. However, little is known about the specific health-related functions of the metabolites of these microbes in the bile, and these important metabolic pathways require further research.
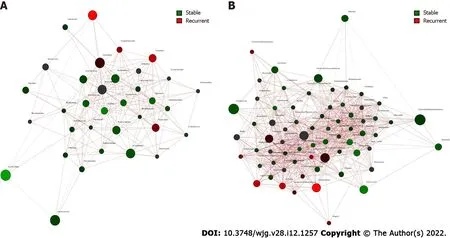
Figure 4 Co-occurrence network analysis of bile microbiome of choledocholithiasis patients with different prognosis post endoscopic sphincterotomy. A: Co-occurrence and disease-specific bacterial interactions at the order level. Order was presented as nodes (stable group specific order in green and recurrent group specific order in red), order abundance was presented as node size, and edges were represented based on their association tested using Pearson’s correlation (positive inter-node correlations in blue, negative inter-node correlations in red); B: Co-occurrence and disease-specific bacterial interactions at the genus level. Genus was presented as nodes (stable group specific genus in green and recurrent group specific genus in red), genus abundance was presented as node size, and edges were represented based on their association tested using Pearson’s correlation (positive inter-node correlations in blue, negative inter-node correlations in red).

Figure 5 Comparison of microbial function prediction of bile microbiome of choledocholithiasis patients with different prognosis post endoscopic sphincterotomy. Functional analysis was performed at the 2 nd hierarchy level of the Kyoto Encyclopedia of Genes and Genomes pathways in the bile microbiome of choledocholithiasis patients. Wilcoxon test was applied to the comparison of each category of microbial function; those with significant differences are shown in the bar chart. Statistical significance is expressed as aP < 0 .05 , bP < 0 .01 .
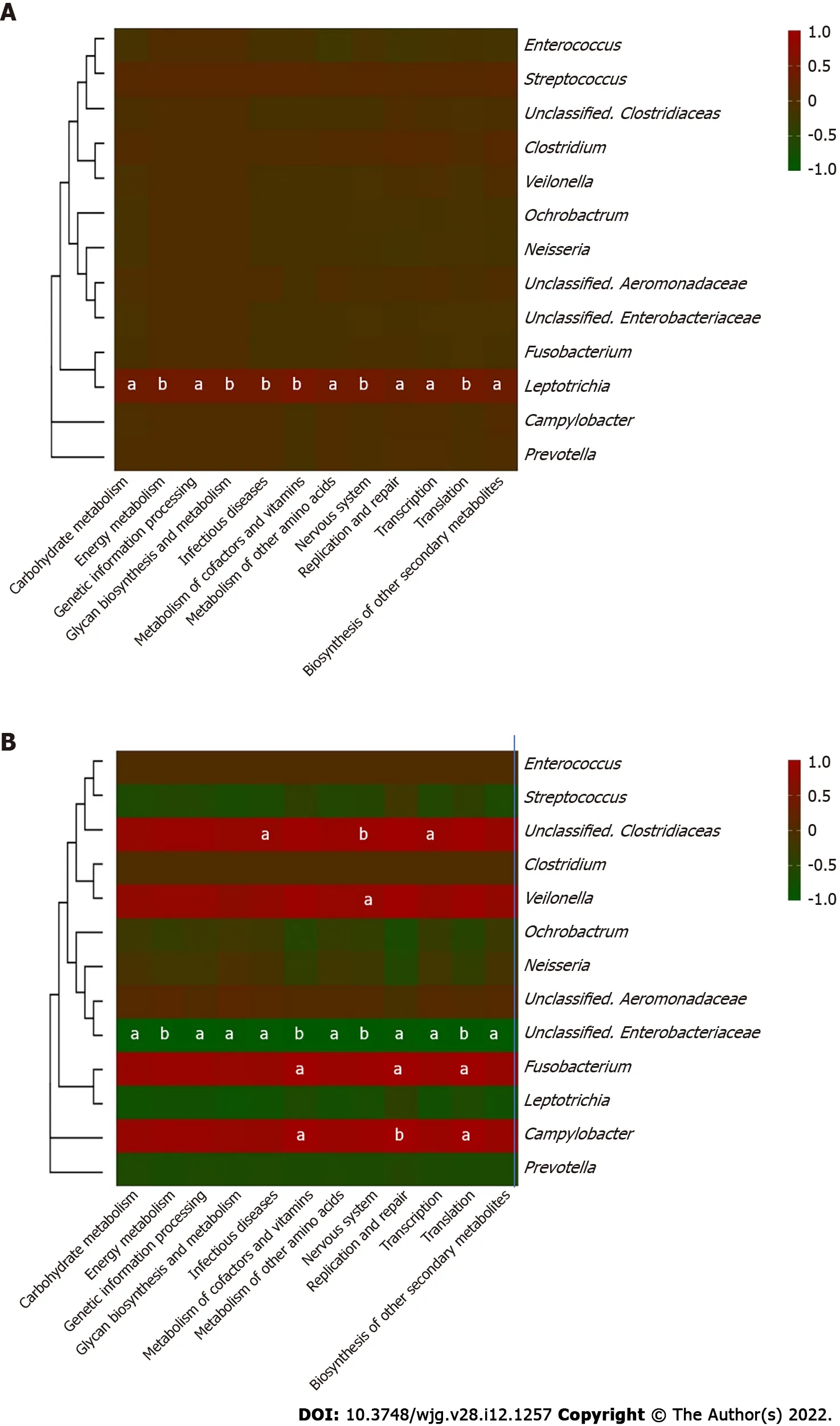
Figure 6 Heatmap of correlation between the core microbiome and key metabolic pathway in choledocholithiasis patients. Thirteen core genera of bile microbiome and their correlations with the twelve discrepant metabolite pathways in different prognosis groups were analyzed using Pearson correlation analysis. The Pearson correlation coefficient between the genus and the metabolic pathway was calculated and shown in colored matrix; red represents a positive correlation, while green represents a negative correlation. A: Matrix heatmap shows the correlations between different genera and metabolite pathways in choledocholithiasis patients without recurrence post-endoscopic sphincterotomy (EST); B: Matrix heatmap shows the correlations between different genera and metabolite pathways in recurrent choledocholithiasis patients post-EST. Statistical significance is expressed as aP < 0 .05 , bP < 0 .01 .

Figure 7 Kaplan-Meier analysis of recurrent time post endoscopic sphincterotomy with different microbiologic risk factors. A: Kaplan-Meier analysis of recurrent time post-endoscopic sphincterotomy (EST) between choledocholithiasis patients with (red) and without (green) Fusobacteriales in bile; B:Kaplan-Meier analysis of recurrent time post-EST between choledocholithiasis patients with (red) and without (green) Neisseriales in bile; C: Kaplan-Meier analysis of recurrent time post-EST between choledocholithiasis patients with (red) and without (green) Lactobacillales in bile.
Certain microorganisms in bile could predict the time taken before disease recurrence post-EST, and this was evaluated. The existence ofLactobacillalesis crucial for predicting recurrence time post-EST,because patients withLactobacillalesin their bile had a longer progression-free time post-EST than patients withoutLactobacillales. Therefore, the examination ofLactobacillalesexistence in bile at the time of endoscopic examination could help doctors identify high-risk patients, who are likely to have early choledocholithiasis recurrence post-EST.
The limited number of early recurrent choledocholithiasis patients could have introduced a bias in statistical analysis and could have limited the generalizability of the prediction model in this study.Furthermore, the diagnosis of choledocholithiasis recurrence relied mainly on the imaging examinations; we could have missed some stones which were invisible in the CT, underestimating the rate of choledocholithiasis recurrence. The molecular mechanisms of microorganisms underlying the recurrence post-EST was based on the PICRUSt model. Verification experiments such as analyzing the correlation between the bile microbiome and the stone composition, and animal experiments to ascertain the preventive effects ofLactobacillusin choledocholithiasis recurrence are warranted.
CONCLUSION
The microbiological characteristics of bile from patients with recurrent choledocholithiasis post-EST indicated that an increase inFusobacteriumandNeisseriaare potential biomarkers for the identification of high-risk patients in the first EST. It elucidated the role of microbial metabolites in the underlying etiology of choledocholithiasis. A co-occurrent network of the biliary bacterial community was constructed. Potential preventive therapy against recurrent choledocholithiasis through supplementation withLactobacillusand maintenance of the balance of the microbial systems could be promising. These findings could help doctors better understand the etiology of recurrent choledocholithiasis and develop better monitoring and treatment strategies against recurrence post-EST.
ARTICLE HIGHLIGHTS
Research background
Choledocholithiasis is a common and socially significant health problem worldwide, and endoscopic sphincterotomy (EST) has become widespread in treating choledocholithiasis; however, recurrence post-EST is relatively common. The bile microbiome has a profound influence on the recurrence of choledocholithiasis; however, the key pathogens and their functions are not fully elucidated.
Research motivation
To determine the microbiologic risk factors of recurrent choledocholithiasis post EST.
Research objectives
To investigate the biliary microbial characteristics of the recurrent choledocholithiasis post-EST, using next-generation sequencing.
Research methods
This cohort study included 43 choledocholithiasis patients who had undergone EST were followed up for over a year. They were divided into either the stable or recurrent groups and comparison of their bile microbiome was carried out through next-generation sequencing. Resulting sequences were analyzed for core microbiome and statistical differences between the microbiologic compositions and functions. Correlation between the key genera and metabolic pathways in bile, were analyzed using Pearson’s correlation test.
Research results
The results revealed distinct clustering of biliary microbiota in recurrent choledocholithiasis, in which higher relative abundances (RAs) ofFusobacteriumandNeisseriaand the absence ofLactobacilluswere observed in the bile of the recurrent patients. Microbiological co-occurrence network revealed a mutual relationship amongFusobacterium,Neisseria, andLeptotrichia, and an antagonistic relationship amongLactobacillales, Fusobacteriales, andClostridiales. Functional analysis revealed that the loss of microbiologic transcription and metabolic abilities may lead to the choledocholithiasis recurrence. Furthermore, the prediction model based on the RA ofLactobacillalesin the bile was effective in identifying the risk of recurrent choledocholithiasis.
Research conclusions
We concluded the microbiologic differences in the bile of recurrent choledocholithiasis patients post EST, thereby adding to the current knowledge on its microbiologic etiology.
Research perspectives
The findings of our study will help develop new prevention strategies for post-surgery recurrence of choledocholithiasis.
ACKNOWLEDGEMENTS
The authors sincerely thank Wang Z for his expert technical advices in the amplicon analysis.
FOOTNOTES
Author contributions:Li Y, Tan WH, Wu JC and Wu QP designed the research; Wu JC, Tan WH and Liang B recruited the clinical cohort, collected samples, and performed the follow-up surveys; Li Y, Huang ZX and Shang YY contributed to the amplicon sequencing; Li Y, Chen JH, Pang R and Xie XQ analyzed the data; Li Y, Wu JC, Huang ZX and Xue L wrote the paper; Zhang JM, Ding Y, Chen MT, Wang J, Tan WH and Wu QP performed critical revisions of the manuscript; Li Y, Tan WH and Wu JC contribute equally to the manuscript; all authors approved the final version of the article.
Supported bythe research grants from Guangdong Provincial Кey Laboratory, No. 2020 B121201009 ; the Science Foundation of Guangdong Second Provincial General Hospital, No. YQ2019 -014 ; and GDAS’ Project of Science and Technology Development, No. 2020 GDASYL-20200301002 .
Institutional review board statement:The study was reviewed and approved by the ethics committee of Guangdong Second Provincial General Hospital [Approval No. 2019 -QNJJ-14 -02 ].
Informed consent statement:Written consent was obtained from all patients in the study.
Conflict-of-interest statement:The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Data sharing statement:The16S rRNAamplicon sequences data in this research have been deposited in GenBank under the BioProject ID PRJNA742858 .
ARRIVE guidelines statement:The authors have read the ARRIVE Guidelines, and the manuscript was prepared and revised according to the ARRIVE Guidelines.
STROBE statement:The authors have read the STROBE Statement—checklist of items, and the manuscript was prepared and revised according to the STROBE Statement—checklist of items.
CONSORT 2010 statement: The authors have read the CONSORT 2010 statement, and the manuscript was prepared and revised according to the CONSORT 2010 statement.
Open-Access:This article is an open-access article that was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution NonCommercial (CC BYNC 4 .0 ) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is noncommercial. See: https://creativecommons.org/Licenses/by-nc/4 .0 /
Country/Territory of origin:China
ORCID number:Ying Li 0000 -0003 -4012 -6954 ; Jia-Chuan Wu 0000 -0001 -8490 -2965 ; Zhi-Xin Huang 0000 -0001 -9862 -1523 ;Yan-Yan Shang 0000 -0002 -6079 -8195 ; Biao Liang 0000 -0003 -1954 -8976 ; Jian-Hui Chen 0000 -0002 -8881 -6949 ; Rui Pang 0000 -0002 -3795 -9843 ; Xin-Qiang Xie 0000 -0003 -3943 -7755 ; Ju-Mei Zhang 0000 -0003 -2103 -6443 ; Yu Ding 0000 -0002 -3688 -7294 ; Liang Xue 0000 -0003 -3606 -4615 ; Mou-Tong Chen 0000 -0003 -2837 -2124 ; Juan Wang 0000 -0002 -1545 -3593 ; Wen-Hui Tan 0000 -0003 -0364 -5069 ; Qing-Ping Wu 0000 -0001 -6503 -359 X.
Corresponding Author's Membership in Professional Societies:Chinese Academy of Engineering, Academician of Chinese Academy of Engineering; Chinese Institute of Food Science and Technology, vice president; China Edible Fungus Association,vice chairman.
S-Editor:Wang LL
L-Editor:A
P-Editor:Li X
 World Journal of Gastroenterology2022年12期
World Journal of Gastroenterology2022年12期
- World Journal of Gastroenterology的其它文章
- Epidemiology of stomach cancer
- Spinal anesthesia alleviates dextran sodium sulfate-induced colitis by modulating the gut microbiota
- Near-infrared fluorescence imaging guided surgery in colorectal surgery
- Epidemiological, clinical, and histological presentation of celiac disease in Northwest China
- Similarities, differences, and possible interactions between hepatitis E and hepatitis C viruses: Relevance for research and clinical practice
- Emerging role of colorectal mucus in gastroenterology diagnostics
