Genetic Improvement of Rice for Bacterial Blight Resistance: Present Status and Future Prospects
R. Abdul Fiyaz, D. Shivani, K. Chaithanya, K. Mounika, M. Chiranjeevi, G. S. Laha, B. C. Viraktamath, L. V. Subba Rao, R. M. Sundaram
Review
Genetic Improvement of Rice for Bacterial Blight Resistance: Present Status and Future Prospects
R. Abdul Fiyaz1, D. Shivani2, K. Chaithanya1, K. Mounika2, M. Chiranjeevi1, G. S. Laha1, B. C. Viraktamath1, L. V. Subba Rao1, R. M. Sundaram1
()
The production and productivity of rice has been challenged due to biotic and abiotic factors.Bacterial blight (BB) disease, caused bypv., is one of the important biotic stress factors, which reduces rice production by 20%–50%.The deployment of host plant resistance is the most preferred strategy for management of BB disease, and breeding disease resistant varieties remains a very economical and effective option. However, it is difficult to develop rice varieties with durable broad-spectrum resistance against BB using conventional approaches alone. Modern biotechnological tools, particularly the deployment of molecular markers, have facilitated the cloning, characterization and introgression of BB resistance genes into elite varieties. At least 46 BB resistance genes have been identified and mapped from diverse sources till date. Among these, 11 genes have been cloned and characterized. Marker-assisted breeding remains the most efficient approach to improve BB resistance by introducing two or more resistance genes into target varieties. Among the identified genes,,andare being widely used in marker-assisted breeding and more than 70 rice varieties or hybrid rice parental lines have been improved for their BB resistance alone or in combination with genes/QTLs conferring tolerance to other stress. We review the developments related to identification and utilization of various resistance genes to develop BB resistant rice varieties through marker-assisted breeding.
rice; bacterial blight; resistance gene; marker-assisted breeding; gene pyramiding
Rice (L.) is one of the most important food crops grown in various agro-climatic conditions throughout the world. More than 90% of the world’s rice is produced and consumed in Southeast Asia and tropical Latin America. In India, rice is grown on about 4.4 × 107hm2field and provides food for more than 75% of the population. To meet the growing demand of nearly 5.0 billion consumers worldwide, with annual average population growth rate of ~1.5%, the demand for rice is expected to increase by 40% by 2030 (Khush, 2005). This demand can be met only if there was increase in production and productivity. However, the production of rice is being adversely affected by two factors, viz., biotic stress factors and abiotic stress factors. The biotic stress factors include fungi, bacteria, viruses, weeds, nematodes, insects, parasites, rodents and birds present in the environment. Among the various bacterial diseases affecting the rice crop, bacterial blight (BB) is one of the major diseases, caused bypv.(), resulting in yield losses ranging from 20% to 50% and is a potential threat to rice cultivation (Singh et al, 2011). BB was reported for the first time in 1884 by farmers of Southern region of Japan (Tagami and Mizukami, 1962). In 1908, Takaishi found bacterial masses in dew drops of rice leaves but the organism could not be named (Mizukami and Wakimoto, 1969). In 1911, Bokura first isolated a bacterium from bacterial oozes on the leaves, and after study of its morphology and physiology, the bacterium was named asHori et Bokura (Mizukami and Wakimoto, 1969). Ishiyama (1922) further studied the disease and renamed the bacterium asIyeda and Ishiyama according to Migula’s taxonomic system. Later, it was transferred toand subsequently to(the name used for the last 40 years). During 1990’s, the taxonomy of plant pathogenic bacteria has been extensively revised and as per revised International Code of Nomenclature of Bacteria (ICNB), the Committee of International Society of Plant Pathology adopted the namepv.Dye. The pathogen was elevated to a species status in 1990 and was named aspv.(). Infection byhas been reported in all major rice growing countries in Asia, Africa, Australia and America.
Bacterial leaf blight in India is a serious problem in South-West monsoon season (Kharif or wet season). It is a vascular disease and the symptoms can be visualized in terms of drying and yellowing of leaves which start from the leaf tips and proceed downward. The favorable temperature for the development of disease is 25 ºC–34ºC, with relative humidity above 70%. The BB pathogen colonizes the xylem vessels by invading the host through natural openings in leaves, viz. hydathodes and wounds. Under field conditions, symptoms start at the tillering stage and disease incidence increases with plant growth, peaking at the flowering stage. The kresek is the severe form of disease that occurs at the seedling stage, resulting in a partial or total crop failure.
The high degree of breakdown of resistance is due to the evolution of new pathotypes because of considerable genetic variation in the pathogen. Several researchers have reported that under epidemic situations, cultural and chemical means of managing the disease are not very effective and are often very costly. Among the available options for successful management of the BB disease, host plant resistance offers the best solution, as it is not only ecologically safe, but also very cost effective. Till date, as many as 46 genes conferring resistance against different strains of the BB pathogen are available for use, and 22 pathotypes of the pathogen have been characterized in India (Yugander et al, 2018).
Both the conventional and molecular breeding approaches for BB resistance aim at improving the genetic underpinnings of rice cultivars and the BB pathogen for host plant resistance (Li et al, 2020). In the present situation, it is very difficult to meet the growing challenges only with the application of conventional plant breeding techniques and tools. It would be very difficult to select rice lines possessing multiple resistance genes using the conventional approach alone, because of the several limitations in conventional breeding (Sundaram et al, 2014). Molecular tools of biotechnology can be helpful to meet the rice production and productivity targets by improving resistance against several diseases. This review highlights the bacterial blight resistance genes, genetics of resistance and the recent progress made in application of molecular tools in breeding for BB resistant rice varieties.
Genetics of pathogenicity in Xoo
is a gram-negative proteobacteria. The pathogen colonizes the xylem vessels with bacterial cells and extracellular polysaccharides by invading the host through natural openings in leaves, including wounds or hydathodes. Several races ofare present, all of which secrete race-specific effectors into the xylem to trigger individualized response and cause infection. Factors are also released by bacteria which bind and activate transcription of genes that activate resistance response, known as resistance genes (Rgenes) (Kumar et al, 2020). Recently, genes related to virulence of pathogen were studied by different research groups (Ryan et al, 2011) and the interactions between the bacteria and plants have been critically analyzed (White and Yang, 2009).requiresgene associated with thiamine biosynthesis for its virulence (Yu et al, 2015). By carrying out extensive BLAST searches and by studying the genome data base available forstrain PXO99A (GenBank accession number NC_010717) using the respective homologous enzymes from other organisms, open reading frames (ORFs) of ThiC (, YP_001915417), ThiD (, YP_001912869), ThiE (, YP_001915211), ThiG (, YP_001914750) and ThiL (, YP_001912199) have been discovered. Virulence is not only as a result of the formation of cell-cell aggregates, but also related to xanthan synthesis inand also quorum sensing (Yu et al, 2015).proteins coordinate quorum sensing-controlled virulence associated functions by affecting cyclic di-GMP synthesis and degradation (Sundaram et al, 2014). There are also many genes associated with thiamine biosynthesis reported in other studies. The factor responsible for virulence is,which isidentified by evaluating a Tn-5 tagged library of(Zou et al, 2011). Theencoded protein serves as an essential enzyme in synthesis of thiazole moiety.
At present, 46 major BB Rgenes are identified in rice (Table 1). Many of these genes are tagged with markers (DNA based), while some have been cloned and characterized (Kumar et al, 2020). Marker-assisted introgression of disease resistance genes into a single genetic background might be expected to give more durable disease resistance, as more resistance genes are incorporated into single genotype (Koide et al, 2010).

Table 1. Summary of resistance genes to bacterial blight in rice.
Characterization of DNA marker for allele mining of BB resistance genes
Important genes/QTLs encoding disease resistance and morphological traits have been obtained by genetic mapping. The information derived from genetic mapping is useful in understanding the allelism of genes conferring similar phenotypes. Linked markers have been used in marker-assisted selection (MAS) programs for developing improved rice cultivars, and new sources of resistance againstwere identified
Accessions of wild species of,such as,and,are recommended for utilizing in rice breeding programs (Akhtar et al, 2011). The differential virulence of isolates ofto diverse rice cultivars has also been reported (Thimmegowda et al, 2011). Several BB resistance genes have been associated with tightly linked DNA markers, and some of them have been cloned [,,,,,,,(t)and(t)]. Therefore, it is now possible to pyramid several genes into susceptible elite rice varieties.
Genetic resistance against bacterial leaf blight
Many studies have been carried out with respect to inheritance of BB resistance in different donor lines of rice. It is difficult to characterize and differentiate the Rgenes due to the diversity ofstrains in various countries (Chukwu et al, 2019). BB Rgenes were identified from wild relatives, landraces, mutants and cultivated species and they confer resistance against differentstrains. Out of the 46 Rgenes identified, 17 [,,,,,,,(t),(t),(t),(t),(t),(t),(t),,(t)and(t)] are recessive, while 29 are dominant, and 11 of these have been cloned and characterized [,/,,,,,,,(t),(t)and(t)] (Table 2). The cloned genes were identified to encode different types of proteins, suggesting multiple mechanisms of Rgene mediatedresistance. There are two major classes of R genes related to disease resistance. One is the nucleotide-binding site leucine-rich repeat (NBS-LRR) and the other is receptor kinase (RLK) class (Sekhwal et al, 2015). The first R gene of the RLK class with broad spectrum of resistance abundantly used in rice breeding programs is. The NBS-LRR class is the largest R gene class conferring resistance against several bacteria, fungi and viruses. The majority of these genes give complete race-specific resistance to. The Rgenes have been introgressed singly or pyramided in combination in breeding for BB resistance in rice (Sundaram et al, 2009; Pandey et al, 2014).
The dominant genewas discovered by Sakaguchi (1967) and was isolated by map-based cloning strategy by Yoshimura et al (1998) from donors Kogyoku and Java14. This gene shows high level of specific resistance to Japanese race 1. For, with the use of F2generation and RFLP (restriction fragment length polymorphism) markers, a high-resolution genetic map was constructed (Yoshimura et al, 2008). XNpb235, XNpb264 and C600 markers located on chromosome 4 are tightly linked to, while the marker U08750 maintains a 1.5 cM distance from. Similarly, another BB R genewas identified bySakaguchi (1967), in rice cultivar Tetep and Rantai Emas II. It is a dominant gene and conferred specific resistance to T7147 (Japaneserace 2). It has been mapped on chromosome 4, linked towith a recombination frequency of 2%–16% (Yoshimura et al, 1995). Fine mapping ofwas reported by He et al (2006). Similarly,gene was identified in avariety Wase Aikoku 3 (Ezuka et al, 1975). It is a dominant gene on the long arm of chromosome 11 and is tightly linked to another BB resistance gene(Yoshimura et al, 1992).confers resistance against the bacteria at both the seedling and maturity stages (Gao et al, 2013). Another geneon chromosome 11 is derived from cultivars TKM6, IR20, IR22 and IR72. The recessive genewas mapped on chromosome 5 (Petpisit et al, 1977), and Blair et al (2003) developed a high-resolution genetic map ofand mapped to a 0.5 cM interval between markers RS7 and RM611. The dominant genewas mapped on chromosome 11 in donor Malagkit Sungsong Zenith (Sidhu, 1978). Later on, by studying the genetics of resistance toin 74cultivarsusing PXO61 isolate from the Philippines, an additional dominant geneconferring resistance in DZ78 was identified (Sidhu et al, 1978). A single recessive geneshowingresistance in rice germplasm accession PI231129 was mapped between two SSR markers, RM21044 and RM21045, on chromosome 7 (Singh et al, 2002). Another dominant genewas discovered from donor Khaolay Nhay by Singh et al (1983) and was mapped on long arm of chromosome 11 (Ogawa et al, 1989).was identified from rice cultivar Cas 209 (Yoshimura et al, 1983),and roughly integrated to a large region between two RFLP markers RG103 (~83 cM) and RG1109 (~91.4 cM) on the long arm of rice chromosome 11.

Table 2. Cloned genes of bacterial leaf blight in rice and cognate Xanthomonas oryzae Avr genes.
NBS-LRR, Nucleotide binding site-leucine rich repeat; LRR-RLK, Leucine rich repeat-receptor-like protein kinase; WAK, Wall associated kinase; TM, Transmembrane; TAL, Transcription activator like.
Ogawa and Yamamoto (1986) identified another dominant BB R genefrom rice cultivar RP9-3. It confers specific resistance to strains T7147, T7133, T7156 and H75304 (Japaneseraces IB, II, IIIA and V, respectively), and was mapped on the long arm of chromosome 3 (Goto et al, 2009). Further studies reported that a single dominant gene,, controls the resistance in Kogyoku and Java14 to BB race V of Indonesia (Ogawa et al, 1978). The genethat specifically confers resistance to the Philippinerace 6was first discovered in the rice variety BJ1 (Ogawa et al, 1986).identified from Taichung Native 1 is a dominant gene and confers resistance to PXO112, BB race 5 of the Philippines (Taura et al, 1987). A series of genes (,,,,,,,and) with different resistance levels and resistance spectrums have been obtained from mutagenesis (Liu et al, 2010).
A dominant genewas identified from the donor(Khush et al, 1990). Genetic and physical analysis oflocalized it in an interval of 8.3 cM on chromosome 11 and the physical size of the region containinggene was estimated to be about 800 kb (Ronald et al, 1992). It is the first cloned R gene in rice (Song et al, 1995),and confers broad spectrum resistant to all the six Philippineraces (Ronald et al, 1992). Another dominant BB resistance gene(t)was identified from a landrace Zhachanglong from Southwest China, and shows high level of resistance to 16 of the 17 BB strains tested (Lin et al, 1996). Zhang et al (1998) identified the genefrom the donoron chromosome 11 between the two markers Lj138 and A83B4. Theresistance gene was identified from two donors DV86 and DV85. It confers resistance against the Philippineraces 4, 6 and 10 and Chinesestrains Zhe 173, JL691 and KS-1-21 (Mir and Khush, 1990).(t), a recessive resistance gene, was identified from the donor Minghui 63 (Lee et al, 2003). At both the seedling and maturity stages, this gene confers resistance to the Philippine race 9 (PXO330). Lee et al (2003) also reported another recessive gene(t), from donor Minghui 63. A resistance locus was identified from a progeny ofcv. IR31917-45-3-2 andaccession IRGC101141,and was designated as(t) (Amante-Bordeos et al, 1992). This gene was mapped between the markers M964 and M1197 on the long arm of chromosome 6 (Lee et al, 2003), and is resistant to the Philippineraces 1 and 2. The resistance gene identified in cultivar B5 was designated as(t) (Tan et al, 2004). Bulked segregant analysis of recombinant inbred lines from a cross between B5 and Minghui63 located the resistance gene within a 1.3 cM region flanked by RFLP markers C904 and R596 on chromosome 1. Jin et al (2007) identified a rice BB resistance gene from a wild rice species ofand designated it as(t) (Cheema et al, 2008).
Wang et al (2009) identified the gene(t) for BB resistance from donor Zhachanglong, a regional rice variety from Southwest China, which confers high level of resistance to a broad spectrum ofisolates. Similarly,(t)from a wild rice () introgression line, C4064, is resistant tostrains PXO61, PXO71, PXO99, PXO145, PXO280, PXO339 and KX085. The gene is flanked by two SSR markers RM2064 and RM6293 on the long arm of chromosome 11 (Zheng et al, 2009).(t) on chromosome 6 was identified from rice cultivar Ba7 (Korinsak et al, 2009) andfrom wild accession of(Kumar et al, 2012). Ram et al (2010) identified the gene(t) fromon chromosome 1.(t) was another dominant gene identified from the wild rice species(Guo et al, 2010),with a high level of resistance to PXO61, PXO112 and PXO339. Miao et al (2010) identified a BB resistance gene(t) on chromosome 11 from rice germplasm C4059. Another dominant genewas identified from wild accession of(IRGC81825). It confers resistant to all the sevenpathotypes prevalent in North India (Kaur et al, 2005). A typical hypersensitive response was exhibited by a rice introgression line (IL), FF329 to all 21 representativestrains. The novel gene identified in FF329 was(Zhang et al, 2015). Kim et al (2015) identified a dominant gene(t) on chromosome 11 from the donor IR65482-7-216-1-2,and the gene is co-segregated with the markers RM27320 and ID55.
A major BB susceptibility genewas used by Hutin et al (2015), in screening the germplasm of 169 rice accessions for polymorphism.gene encodes a sugar transporter which is targeted by manystrains. Only three plant SWEET genes that encode putative sugar transporters are induced by transcription activator-like (TAL) effectors from ricepathogen (Streubel et al, 2013). Further, the gene is designated as(t). Thegene was identified in XM14, a mutant cultivar with resistance to all Japaneseraces (Busungu et al, 2016). Kim and Reinke (2019) identified another dominant BB resistancegene(t) from the donor P8 on chromosome 11. The single major recessive-gene designated as(t) was fine mapped to a region of 120 kb segment on chromosome 11 by Kim (2018). Another novel recessive BB resistance gene designated as(t) was identified fromaccession IRGC102600B, and was mapped using ddRAD sequencing approach by Neelam et al (2020). Recently, a BB resistance gene(t)was identified byChen et al (2020), in mutant line H120 derived fromrice Lijiangxintuanheigu. It is a dominant gene and confers resistance to all Chineseraces. It was mapped between the markers RM26981 and RM26984 within approximately 65.34 kb on chromosome 11.
MAS for pyramiding of BB resistance genes into rice cultivars
The improvement of rice varieties for resistance to the diseases that are prevalent and destructive is necessary for sustainable rice production. Due to high levels of variability in the disease populations in growing areas, past attempts in developing BB resistant varieties have been disappointing (Naveed et al, 2010). Pyramiding major or minor resistance genes into a single genetic background might be expected to give more durable disease resistance (Koide et al, 2010). The major BB resistance genes identified by different research groups have been extensively studied and utilized for developing BB resistant varieties.
Genes such as,,,,andhave been extensively used for pyramiding into many susceptible rice cultivars and landraces due to their broader spectrum of resistance (Huang et al, 1997; Chen et al,2000; Sundaram et al, 2008; Pandey et al, 2013; Dnyaneshwar et al, 2018) (Table 3). Huang et al (1997) have transferred four different R genes (,,and) to the recurrent IR24 background from four different resistant parents to enhance tolerance against BB. The introgression ofgene into an elite restorer line Minghui 63 (Chen et al,2000) is the first report of using MAS for improving the BB resistance of rice hybrids. Singh et al (2001) transferred the R genes (,and) to rice cultivar PR106. Toenniessen et al(2003) pyramided the R genes such asto the cultivar IR64. This results in the increased resistance to BB and enhancement of overall yield. Two genes, namely,and, were pyramided into well-known rice cultivar Pusa Basmati 1 from the resistant donor IRBB55 to enhance the resistance by using sequence-tagged site markers (Joseph et al, 2004). The same approach was also used to improve the varieties Pusa Basmati 1 (Gopalakrishnan et al, 2008), Samba Mahsuri and Triguna(Sundaram et al, 2008, 2009), Pusa 6B (Basavaraj et al, 2010), PRR78 (Singh et al, 2012), Tapaswini (Dokku et al, 2013), RD6 (Pinta et al, 2013), APMS6B (Yugander et al, 2018), Taraori Basmati and Basmati 386 (Pandey et al, 2013).
The discovery of multiple genes controlling the resistance against BB has become a boon to rice breeders for gene pyramiding by MAB, for example,(Win et al, 2013),(Guvvala et al, 2013),(Luo et al, 2014),(Arunakumari et al, 2016),(Ellur et al, 2016), and(Luo et al, 2017)Ni et al (2015) transferredto susceptible Chinese cultivars (Guangzhan63S and Liangyou6326). Yap et al(2016) transferred more than four genes (using Xa7F/7-1R/7-2R, RM604F/604R, Xa13F/13R, Xa21F/21R and Xa4F/4R markers) from the donor IRBB66 to eight Chinese rice cultivars. Dnyaneshwar et al (2018) pyramided three R genes (,and) to two elite rice cultivars Safri 17 and Dubraj from the donor parent RP-Bio-226. Gene combination ofwas found to impart good BB resistance when introgressed in the background of a deep-water rice variety Jalmagna (Pradhan et al, 2016) and a salt tolerant variety CSR-30 (Baliyan et al, 2018). Introgression of othergenes such as,(Balachiranjeevi et al, 2018) and(Mi et al, 2018) improves the BB resistance in different rice cultivars in various countries. Reinke et al (2018) transferred thegene (using ID55.WA3 and RM1233 markers) to the susceptible Korean variety Junam using marker-assisted backcrossing. Chukwu et al (2020) pyramided the genes,,andinto Putra 1 from the donor IRBB60 for enhancement of resistance against BB. Recently, Hsu et al (2020) pyramided five BB resistance genes,,,andfrom a donor parent IRBB66 into Tainung82, a popularvariety in Taiwan, China.

Table 3. Successful examples of marker-assisted breeding for bacterial blight disease resistance in rice.
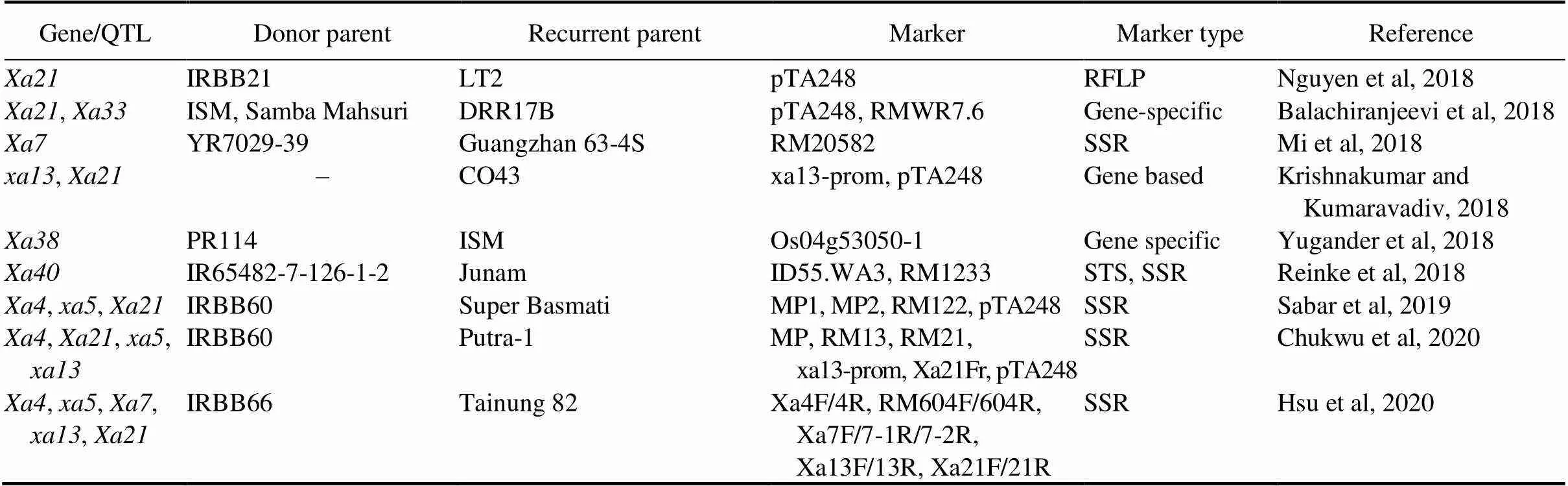
Table 3. Continued.
ISM, Improved Samba Mahsuri; CAPS, Cleaved amplified polymorphism sequence; EST,Expressed sequence tag; RAPD, Random amplified polymorphism DNA; RFLP,Restriction fragment length polymorphism; SSR, Simple sequence repeat; STS, Sequence tagged site.
Deployment of host plant resistance is considered to be the best option for managing multiple biotic stresses. Breeding rice varieties with multiple disease and insect resistance will broaden the resistance spectrum and increase the resistance durability for the varieties. By combining with other traits such as blast, brown planthopper, gall midge resistance, a total of 18 varieties resistant to multiple diseases and pests were developed (Table 4). For example, pyramiding of+(Narayanan et al, 2002),+(Suh et al, 2015),+++(Sagar et al, 2020) improve resistance against bacterial blight and blast.(Abhilash Kumar et al, 2017),++(He et al, 2019), and++++++(Dixit et al, 2020) improve resistance against bacterial blight, blast, gall midge and brown planthopper. Similarly, marker- assisted pyramiding was carried out to introgress the genes for bacterial blight, aroma and submergence (Salgotra et al, 2012; Pradhan et al, 2019; Raina et al, 2019; Hairmansis et al, 2020).
The BB resistant popular rice varieties with many gene combinations developed by marker-assisted backcrossing would provide an economical and sustainable assurance for the farmers against losses due to various pests and diseases. This scheme requires less than three years, which is much more efficient than the conventional breeding method. The improved lines show recovery of more than 99% genome background of the recurrent parent genome and record a broad-spectrum resistance towithout any significant difference in agronomic traits (Chukwu et al, 2020; Zheng et al, 2021). Further, development of near-isogenic lines containing single and multiple R genes and the rotation of the R genes can resist the pathogen much better and can also maximize the life span of the individual R genes. Multi-lines containing differentgenes can also provide broad-spectrum and durable resistance against BB.
Perspective
In the present situation, it is very difficult to meet the growing challenges in sustaining rice production and productivity only with the application of conventional plant breeding techniques and tools due to the limitations of conventional, phenotype-based selections. In addition, there is a high level of variability in the BB pathogen and the constant threat of evolution of virulentisolates, hence there is a continuing need for identification of new BB resistance genes and their incorporation into popular varieties. Marker-assisted backcross breeding has been successfully used for incorporation of major genes conferring resistance to BB and other biotic stress into popular varieties, thus meeting the enhanced production with resistance against multiple pests and diseases. Identification of resistance genes, their characterization, mapping, tagging and introgression of many BB resistance genes to elite rice varieties have been instrumental in achieving durable resistance against BB in many countries. The deployment of molecular approaches to combat BB disease in rice has been encouraging and these efforts can significantly enhance the rice productivity, quality and overall rice production in the coming decades. This review will be helpful in exploiting the full range of variability existing in nature with respect to the major BB resistance genes across the world for incorporation into popular varieties to develop durable, broad-spectrum BB resistant rice varieties through innovative breeding technologies.

Table 4. Successful examples of marker-assisted breeding for blight disease along with other traits in rice.
ISM, Improved Samba Mahsuri;SCAR, Sequence characterized amplified region; SNP, Single nucleotide polymorphism; SSR, Simple sequence repeat; STS, Sequence tagged site.
Abhilash Kumar V, Balachiranjeevi C H, Bhaskar Naik S, Rambabu R, Rekha G, Harika G, Hajira S K, Pranathi K, Vijay S, Anila M, Mahadevaswamy H K, Kousik M, Yugander A, Aruna J, Hari Prasad A S, Madhav M S, Laha G S, Balachandran S M, Prasad M S, Ravindra Babu V, Sundaram R M. 2016. Marker-assisted improvement of the elite restorer line of rice, RPHR-1005 for resistance against bacterial blight and blast diseases., 95: 895‒903.
Abhilash Kumar V, Balachiranjeevi C H, Bhaskar Naik S, Rekha G, Rambabu R, Harika G, Pranathi K, Hajira S K, Anila M, Kousik M, Kale R, Dilip Kumar T, Prasad M S, Hari Prasad A S, Padmakumari A P, Laha G S, Balachandran S M, Madhav M S, Senguttuvel P, Kemparajau K B, Fiyaz A R, Bentur J S, Viraktamath B C, Ravindra Babu V, Sundaram R M. 2017. Marker-assisted pyramiding of bacterial blight and gall midge resistance genes into RPHR-1005, the restorer line of the popular rice hybrid DRRH-3.,37:86.
Akhtar M A, Abbasi F M, Ahmad H, Shahzad M, Shah M A, Shah A H. 2011. Evaluation of rice germplasm againstcausing bacterial leaf blight., 43(6):3021‒3023.
Amante-Bordeos A, Sitch L A, Nelson R, Dalmacio R D, Oliva N P, Aswidinnoor H, Leung H. 1992. Transfer of bacterial blight and blast resistance from the tetraploid wild riceto cultivated rice,., 84:345‒354.
Arunakumari K, Durgarani C V, Satturu V, Sarikonda K R, Chittoor P D R, Vutukuri B, Laha G S, Nelli A P K, Gattu S, Jamal M, Prasadbabu A, Hajira S, Sundaram R M. 2016. Marker-assisted pyramiding of genes conferring resistance against bacterial blight and blast diseases into Indian rice variety MTU1010., 23(6):306‒316.
Balachiranjeevi C H, Bhaskar Naik S, Abhilash Kumar V, Harika G, Mahadev Swamy H K, Hajira Sk, Dilip Kumar T, Anila M, Kale R R, Yugender A, Pranathi K, Koushik M B V N, Suneetha K, Bhadana V P, Hariprasad A S, Laha G S, Rekha G, Balachandran S M, Madhav M S, Senguttuvel P, Fiyaz A R, Viraktamath B C, Giri A, Swamy B P M, Jauhar Ali, Sundaram R M. 2018.Marker-assisted pyramiding of two major, broad-spectrum bacterial blight resistance genes,andinto an elite maintainer line of rice, DRR17B., 13(10): e0201271.
Baliyan N, Malik R, Rani R, Mehta K, Vashisth U, Dhillon S, Boora K S. 2018. Integrating marker-assisted background analysis with foreground selection for pyramiding bacterial blight resistance genes into Basmati rice., 341(1):1‒8.
Basavaraj S H, Singh V K, Singh A, Singh A, Singh A, Anand D, Yadav S, Ellur R K, Singh D, Gopala Krishnan S, Nagarajan M, Mohapatra T, Prabhu K V, Singh A K. 2010. Marker-assisted improvement of bacterial blight resistance in parental lines of Pusa RH10, a superfine grain aromatic rice hybrid.,26: 293‒305.
Bharani M, Nagarajan P, Rabindran R, Saraswathi R, Balasubramanian P, Ramalingam J. 2010. Bacterial leaf blight resistance genes (,and) pyramiding through molecular marker assisted selection into rice cultivars., 43(10): 1032‒1043.
Bhatia D, Sharma R, Vikal Y, Mangat G S, Mahajan R, Sharma N, Lore J S, Singh N, Bharaj T S, Singh K. 2011. Marker-assisted development of bacterial blight resistant, dwarf, and high yielding versions of two traditional Basmati rice cultivars., 51(2): 759‒770.
Bharathkumar S, David Paulraj R S, Brindha P V, Kavitha S, Gnanamanickam S S. 2008. Improvement of bacterial blight resistance in rice cultivars Jyothi and IR50 via marker-assisted backcross breeding.,21(1):101‒116.
Blair M W, Garris A J, Iyer A S, Chapman B, Kresovich S, McCouch S R. 2003. High resolution genetic mapping and candidate gene identification at thelocus for bacterial blight resistance in rice (L.).,107:62‒73.
Busungu C, Taura S, Sakagami J I, Ichitani K. 2016. Identification and linkage analysis of a new rice bacterial blight resistance gene from XM14, a mutant line from IR24.,66(4):636‒645.
Cheema K K, Grewal N K, Vikal Y, Sharma R, Lore J S, Das A, Bhatia D, Mahajan R, Gupta V, Bharaj T S, Singh K. 2008. A novel bacterial blight resistance gene frommapped to 38 kb region on chromosome 4L and transferred toL., 90(5): 397‒407.
Chen S, Lin X H, Xu C G, Zhang Q F. 2000. Improvement of bacterial blight resistance of ‘Minghui 63’, an elite restorer line of hybrid rice, by molecular marker-assisted selection.,40(1):239‒244.
Chen S, Xu C G, Lin X H, Zhang Q F. 2001. Improving bacterial blight resistance of ‘6078’, an elite restorer line of hybrid rice, by molecular marker-assisted selection., 120: 133‒137.
Chen S, Wang C Y, Yang J Y, Chen B, Wang W J, Su J, Feng A Q, Zeng L X, Zhu X Y. 2020. Identification of the novel bacterial blight resistance gene(t) by mapping and expression analysis of the rice mutant H120., 10(1):12642.
Chu Z H, Yuan M, Yao J L, Ge X J, Yuan B, Xu C G, Li X H, Fu B Y, Li Z K, Bennetzen J L, Zhang Q F, Wang S P. 2006. Promoter mutations of an essential gene for pollen development result in disease resistance in rice., 20(10): 1250‒1255.
Chukwu S C, Rafii M Y, Ramlee S I, Ismail S I, Oladosu Y, Okporie E, Onyishi G, Utobo E, Ekwu L, Swaray S, Jalloh M. 2019. Marker-assisted selection and gene pyramiding for resistance to bacterial leaf blight disease of rice (L.)., 33: 440‒455.
Chukwu S C, Rafii M Y, Ramlee S I, Ismail S I, Oladosu Y, Kolapo K, Musa I, Halidu J, Muhammad I, Ahmed M. 2020. Marker-assisted introgression of multiple resistance genes confers broad spectrum resistance against bacterial leaf blight and blast diseases in Putra-1 rice variety.,10(1):42.
Davierwala A P, Reddy A P, Lagu M D, Ranjekar P K, Gupta V S. 2001. Marker assisted selection of bacterial blight resistance genes in rice.,39:261‒278.
Dixit S, Singh U M, Singh A K, Alam S, Venkateshwarlu C, Nachimuthu V V, Yadav S, Abbai R, Selvaraj R, Devi M N, Ramayya P J, Badri J, Ram T, Lakshmi J, Lakshmidevi G, Lrk J V, Padmakumari A P, Laha G S, Prasad M S, Seetalam M, Singh V K, Kumar A. 2020. Marker assisted forward breeding to combine multiple biotic-abiotic stress resistance/tolerance in rice., 13(1): 29.
Dnyaneshwar S U, Agrawa T, Kadu T, Pradhan A, Verulkar A. 2018. Improvement of Dubraj and Safri-17 varieties for conferring resistance against bacterial leaf blight through marker assisted selection approach., 6:1785‒1790.
Dokku P, Das K M, Rao G J N. 2013. Pyramiding of four resistance genes of bacterial blight in Tapaswini, an elite rice cultivar, through marker-assisted selection., 192:87–96.
Ellur R K, Khanna A, GopalaKrishnan S, Bhowmick P K, Vinod K K, Nagarajan M, Mondal K K, Singh N K, Singh K, Prabhu K V, Singh A K. 2016. Marker-aided incorporation of, a novel bacterial blight resistance gene, in PB1121 and comparison of its resistance spectrum with+.,6: 29188.
Ezuka A, Horino O, Toriyama K, Shinoda H, Morinaka T. 1975. Inheritance of resistance of rice variety Wase Aikoku 3 to.,28:124‒130.
Fu C Y, Wu T, Liu W G, Wang F, Li J H, Zhu X Y, Huang H J, Liu Z R, Liao Y L, Zhu M S, Chen J W, Huang Y J. 2012. Genetic improvement of resistance to blast and bacterial blight of the elite maintainer line Rongfeng B in hybrid rice (L.) by using marker-assisted selection., 11(67): 13104‒13114.
Gao L F, Cao Y H, Xia Z H, Jiang G H, Liu G Z, Zhang W X, Zhai W X. 2013. Do transgenesis and marker-assisted backcross breeding produce substantially equivalent plants? A comparative study of transgenic and backcross rice carrying bacterial blight resistant gene.,14:738.
Gopalakrishnan S, Sharma R K, Anand Rajkumar K, Joseph M, Singh V P, Singh A K, Bhat K V, Singh N K, Mohapatra T. 2008. Integrating marker assisted background analysis with foreground selection for identification of superior bacterial blight resistant recombinants in Basmati rice., 127(2):131‒139.
Goto T, Matsumoto T, Furuya N, Tsuchiya K, Yoshimura A. 2009. Mapping of bacterial blight resistance geneon rice chromosome 3., 43(3): 221‒225.
Gu K, Tian D, Yang F, Wu L, Sreekala C, Wang D, Wang G L, Yin Z. 2004. High-resolution genetic mapping of(t), a new bacterial blight resistance gene in rice,L., 108(5): 800‒807.
Guo S B, Zhang D P, Lin X H. 2010. Identification and mapping of a novel bacterial blight resistance gene(t) originated from.,43(13):2611‒2618. (in Chinese with English abstract)
Guvvala L D, Koradi P, Shenoy V, Marella L S. 2013. Improvement of resistance to bacterial blight through marker assisted backcross breeding and field validation in rice ().,1:52‒66.
Hairmansis A, Warsono, Supartopo, Yullianida, Nasution A, Utami D W, Suwarno. 2020. Introgression of bacterial blight resistance geneinto popular Indonesian rice varieties through backcross and molecular breeding.,457:012050.
Hari Y, Srinivasarao K, Viraktamath B C, Hariprasad A S, Laha G S, Ahmed M I, Natarajkumar P, Ramesha M S, Neeraja C N, Balachandran S M, Rani N S, Balaji Suresh P, Sujatha K, Pandey M, Ashok Reddy G, Madhav M S, Sundaram R M. 2011. Marker- assisted improvement of a stable restorer line, KMR-3R and its derived hybrid KRH2 for bacterial blight resistance and grain quality., 130(6): 608‒616.
Hari Y, Srinivasarao K, Viraktamath B C, Hari Prasad A S, Laha G S, Ahmed M I, Natarajkumar P, Sujatha K, Srinivas Prasad M, Pandey M, Ramesha M S, Neeraja C N, Balachandran S M, Rani N S, Kemparaju B, Madhan Mohan K, Sama V S A K, Shaik H, Balachiranjeevi C, Pranathi K, Ashok Reddy G, Madhav M S, Sundaram R M. 2013. Marker-assisted introgression of bacterial blight and blast resistance into IR 58025B, an elite maintainer line of rice., 132: 586‒594.
He C, Xiao Y L, Yu J H, Li J J, Meng Q C, Qing X G, Xiao G Y. 2019. Pyramiding,, andgenes into the elite restorer line Yuehui9113 increases resistance to bacterial blight and the brown planthopper in rice.,115:31‒39.
He Q, Li D B, Zhu Y S, Tan M P, Zhang D P, Lin X H. 2006. Fine mapping of, a bacterial blight resistance gene in rice.,17:1‒6.
Hsu Y C, Chiu C H, Yap R, Tseng Y C, Wu Y P. 2020. Pyramiding bacterial blight resistance genes in Tainung82 for broad- spectrum resistance using marker-assisted selection.,21(4):1281.
Hu K M, Cao J B, Zhang J, Xia F, Ke Y G, Zhang H T, Xie W Y, Liu H B, Cui Y, Cao Y L, Sun X L, Xiao J H, Li X H, Zhang Q L, Wang S P. 2017. Improvement of multiple agronomic traits by a disease resistance gene via cell wall reinforcement., 3(3): 17009.
Huang C J, Tsay J F, Chang S Y, Yang H P, Wu W S, Chen C Y. 2012. Dimethyl disulfide is an induced systemic resistance elicitor produced byC1L., 68(9): 1306‒1310.
Huang N, Angeles E R, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadivel N, Bennett J, Khush G S. 1997. Pyramiding of bacterial blight resistance genes in rice: Marker-assisted selection using RFLP and PCR., 95:313‒320.
Hutin M, Sabot F, Ghesquière A, Koebnik R, Szurek B. 2015. A knowledge-based molecular screen uncovers a broad-spectrumresistance allele to bacterial blight from wild rice.,84(4):694‒703.
Ishiyama S. 1922. Studies on the white leaf disease of rice plants.,47:233‒251.
Jiang J F, Mou T M, Yu H H, Zhou F S. 2015. Molecular breeding of thermo-sensitive genic male sterile (TGMS) lines of rice for blast resistance usinggene.,8: 11.
Jin X W, Wang C L, Yang Q, Jiang Q X, Fan Y L, Liu G C, Zhao K J. 2007. Breeding of near-isogenic line CBB30 and molecular mapping of(t), a new resistance gene to bacterial blight in rice., 40(6):1094–1100. (in Chinese with English abstract)
Joseph M, Gopalakrishnan S, Sharma R K, Singh V P, Singh A K, Singh N K, Mohapatra T. 2004. Combining bacterial blight resistance and Basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice.,13:377‒387.
Kaur R, Grewal N, Das A, Vikal Y, Singh J, Bharaj T S, Sidhu J S, Singh K. 2005. Inheritance of bacterial blight resistance in two accessions of wild rice,., 22:78–82.
Khush G S. 2005. What it will take to feed 5.0 billion rice consumers in 2030., 59:1–6.
Khush G S, Bacalangco E, Ogawa T. 1990. A new gene for resistance to bacterial blight from., 7:121–122.
Kim S M. 2018. Identification of novel recessive gene(t) conferring resistance to bacterial blight races in rice by QTL linkage analysis using an SNP chip., 131(12):2733‒2743.
Kim S M, Reinke R F. 2019. A novel resistance gene for bacterial blight in rice,(t) identified by GWAS, confirmed by QTL mapping using a bi-parental population.,14(2): e0211775.
Kim S M, Suh J P, Qin Y, Noh T H, Reinke R F, Jena K K. 2015. Identification and fine-mapping of a new resistance gene,, conferring resistance to bacterial blight races in rice (L.)., 128:1933‒1943.
Koide Y, Kawasaki A, Telebanco-Yanoria M J, Hairmansis A, Nguyet N T M, Bigirimana J, Fujita D, Kobayashi N, Fukuta Y. 2010. Development of pyramided lines with two resistance genes,and, for blast disease (B. Couch) in rice (L.)., 129(6):670‒675.
Korinsak S, Sriprakhon S, Sirithanya P, Jairin J, Korinsak S, Vanavichit A, Toojinda T. 2009. Identification of microsatellite markers (SSR) linked to a new bacterial blight resistance gene(t) in rice cultivar ‘Ba7’.,3(2):235‒247.
Krishnakumar R, Kumaravadivel N. 2018. Marker-assisted selection for biotic stress (bacterial leaf blight and gall midge) tolerance in BC4F4generation of rice (L.).,9(1):275‒282.
Kumar A, Kumar R, Sengupta D, Das S N, Pandey M K, Bohra A, Sharma N K, Sinha P, Sk H, Ghazi I A, Laha G S, Sundaram R M. 2020. Deployment of genetic and genomic tools toward gaining a better understanding of rice-pv.interactions for development of durable bacterial blight resistant rice.,11:1152.
Kumar P N, Sujatha K, Laha G S, Rao K S, Mishra B, Viraktamath B C, Hari Y, Reddy C S, Balachandran S M, Ram T, Madhav M S, Rani N S, Neeraja C N, Reddy G A, Shaik H, Sundaram R M. 2012. Identification and fine-mapping of, a novel gene for resistance topv.., 102(2): 222‒228.
Lee K S, Rasabandith S, Angeles E R, Khush G S.2003. Inheritance of resistance to bacterial blight in 21 cultivars of rice., 93(2):147–152.
Li L, Mo X Y, Li T T, Zhang L Y, Dong H S. 2020. Effector XopN ofpv.plays a virulent role in rice varieties harboringhomologs., 34(4): 368–382. (in Chinese with English abstract)
Lin X H, Zhang D P, Xie Y F, Gao H P, Zhang Q F. 1996. Identifying and mapping a new gene for bacterial blight resistance in rice based on RFLP markers., 86:1156–1159.
Liu J L, Wang X J, Mitchell T, Hu Y J, Liu X L, Dai L Y, Wang G L. 2010. Recent progress and understanding of the molecular mechanisms of the rice-interaction.,11(3):419‒427.
Luo S, Zhang Y, Hu Q, Chen J J, Li K P, Lu C, Liu H, Wang W, Kuang H H. 2012. Dynamic nucleotide-binding site and leucine-rich repeat-encoding genes in the grass family.,159(1):197‒210.
Luo W L, Huang M, Guo T, Xiao W M, Wang J F, Yang G L, Liu Y Z, Wang H, Chen Z Q, Zhuang C X. 2017. Marker-assisted selection for rice blast resistance genesandthrough high-resolution melting of a gene-targeted amplicon.,136(1):67‒73.
Luo Y C, Yin Z C. 2013. Marker-assisted breeding of Thai fragrance rice for semi-dwarf phonetype, submergence tolerance and disease resistance to rice blast and bacterial blight., 32: 709–721.
Luo Y C, Zakaria S, Basyah B, Ma T C, Li Z F, Yang J B, Yin Z C. 2014. Marker-assisted breeding of Indonesia local rice variety Siputeh for semi-dwarf phenotype, good grain quality and disease resistance to bacterial blight.,7:33.
Luo Y C, Ma T C, Teo J, Luo Z X, Li Z F, Yang J B, Yin Z C. 2021. Marker-assisted breeding of thermo-sensitive genic male sterile line 1892S for disease resistance and submergence tolerance., 28(1): 89‒98.
Mi J M, Yang D B, Chen Y, Jiang J F, Mou H P, Huang J B, Ouyang Y D, Mou T M. 2018. Accelerated molecular breeding of a novel P/TGMS line with broad-spectrum resistance to rice blast and bacterial blight in two-line hybrid rice., 11(1):11.
Miao L L, Wang C L, Zheng C K, Che J Y, Gao Y, Wen Y C, Li G Q, Zhao K J. 2010. Molecular mapping of a new gene for resistance to rice bacterial blight.,43(15):3051‒3058.(in Chinese with English abstract)
Mir G N, Khush G S. 1990. Genetics of resistance to bacterial blight in rice cultivar DV86., 3(2):194‒198.
Mizukami T, Wakimoto S. 1969. Epidemiology and control of bacterial leaf blight of rice., 7(1): 51‒72.
Nakai H, Nakamura K, Kuwahara S, Saito M. 1998. Genetic studies of an induced rice mutant resistant to multiple races of bacterial leaf blight., 5: 101–103.
Narayanan N N, Baisakh N, Vera Cruz C M, Gnanamanickam S S, Datta K, Datta S K. 2002. Molecular breeding for the development of blast and bacterial blight resistance in rice cv. IR50.,42(6):2072‒2079.
Naveed SA, Babar M, Arif A, Zafar Y, Sabar M, Ali I, Chragh M, Arif M. 2010. Detection of bacterial blight resistant geneusing linked marker approaches., 9:3549‒3554.
Neelam K, Mahajan R, Gupta V, Bhatia D, Gill B K, Komal R, Lore JS, Mangat G S, Singh K.2020. High-resolution genetic mapping of a novel bacterial blight resistance gene(t) identified fromand transferred to., 133(3): 689‒705.
Nguyen H T, Vu Q H, van Mai T, Nguyen T T, Vu L D, Nguyen TT, Nguyen L V, Vu H T T, Nong H T, Dinh T N, Toshitsugu N, van Vu L. 2018. Marker-assisted selection ofconferring resistance to bacterial leaf blight inrice cultivar LT2, 25(1):52–56.
Ni D H, Song F S, Ni J L, Zhang A F, Wang C L, Zhao K J, Yang Y C, Wei P C, Yang J B, Li L. 2015. Marker-assisted selection of two-line hybrid rice for disease resistance to rice blast and bacterial blight., 184:1–8.
Ogawa T, Yamamoto T.1986. Inheritance of resistance to bacterial blight in rice.: Banta S J. Rice Genetics I.World Scientific Publishing Co. Ltd: 471‒479.
Ogawa T, Morinaka T, Fujii K, Kimura T. 1978. Inheritance of resistance of rice varieties Kogyoku and Java 14 to bacterial group V of.,44(2): 137‒141.
Ogawa T, Tabien RE, Busto GA, Khush GS, Mew TW. 1986. The relationships between,, andXa-4for resistances to rice bacterial blight.,3:83‒84.
Ogawa T, Kaku H, Yamamoto T. 1989. A resistance gene of rice cultivar Asaminori to bacterial blight of rice.,39:196‒197.
Pandey M K, Shobha Rani N, Sundaram R M, Laha G S, Madhav M S, Srinivasa Rao K, Sudharshan I, Hari Y, Varaprasad G S, Subba Rao L V, Suneetha K, Sivaranjani A K P, Viraktamath B C. 2013. Improvement of two traditional Basmati rice varieties for bacterial blight resistance and plant stature through morphological and marker-assisted selection., 31: 239‒246.
Pandey S, Singh B, Kumar J. 2014. DNA typing and virulence determination ofpv.population for the management of bacterial leaf blight of rice in Udham Singh Nagar, India.,138:847‒862.
Petpisit V, Khush G S, Kauffman H E. 1977. Inheritance of resistance to bacterial blight in rice., 17(4):551–554.
Pinta W, Toojinda T, Thummabenjapone P, Sanitchon J.2013. Pyramiding of blast and bacterial leaf blight resistance genes into rice cultivar RD6 using marker assisted selection.,12: 4432–4438.
Pradhan S K, Barik S R, Sahoo A, Mohapatra S, Nayak D K, Mahender A, Meher J, Anandan A, Pandit E.2016. Population structure, genetic diversity and molecular marker-trait association analysis for high temperature stress tolerance in rice., 11(8): e0160027.
Pradhan S K, Pandit E, Pawar S, Baksh S Y, Mukherjee A K, Mohanty S P.2019. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’through marker-assisted backcross breeding.,9(1):12810.
Raina M, Salgotra R K, Pandotra P, Rathour R, Singh K. 2019. Genetic enhancement for semi-dwarf and bacterial blight resistance with enhanced grain quality characteristics in traditional Basmati rice through marker-assisted selection.,342: 142‒153.
Rajpurohit D, Kumar R, Kumar M, Paul P, Awasthi A, Osman Basha P, Puri A, Jhang T, Singh K, Dhaliwal H S.2011. Pyramiding of two bacterial blight resistance and a semidwarfing gene in Type 3 Basmati using marker-assisted selection.,178:111‒126.
Ram T, Laha G S, Gautam S K, Deen R, Madhav M S, Brar D S, Viraktamath B C.2010. Identification of a new gene fromwith broad-spectrum resistance to bacterial blight of rice in India.,25:57‒58.
Ramalingam J, Basharat H S, Zhang G. 2002. STS and microsatellite marker-assisted selection for bacterial blight resistance and waxy genes in rice,L., 127(2): 255‒260.
Ratna Madhavi K, Rambabu R, Abhilash Kumar V, Vijay Kumar S, Aruna J, Ramesh S, Sundaram R M, Laha G S, Sheshu Madhav M, Ravindra babu V, Srinivas Prasad M S. 2016. Marker assisted introgression of blast (and) genes in to the genetic background of elite, bacterial blight resistantrice variety, Improved Samba Mahsuri., 212:331‒342.
Reinke R, Kim SM, Kim BK. 2018. Developingrice introgression lines with multiple resistance genes for brown planthopper, bacterial blight, rice blast, and rice stripe virus using molecular breeding., 293:1565‒ 1575.
Rekha G, Abhilash Kumar V, Viraktamath B C, Pranathi K, Kousik M B V N, Laxmi Prasanna B, Backiyalakshmi C, Sinha P, Ravindra R K, Bhaskar S, Hajira S K, Balachiranjeevi C H, Swapnil K, Rambabu R, Harika G, Punniakotti E, Anila M, Mahadev H K, Dilip Kumar T, Yugander A, Chaitra K, Praveen M, Madhavi K R, Prasad M S, Laha G S, Neeraja C N, Balachandran S M, Senguttuvel P, Fiyaz R A, Badri J, Giri A, Subba Rao L V, Ravindra Babu V, Sundaram R M. 2018. Improvement of blast resistance of the popular high-yielding, medium slender-grain type, bacterial blight resistant rice variety, improved Samba Mahsuri by marker-assisted breeding.,27(4):463‒472.
Ronald P C, Albano B, Tabien R, Abenes L, Wu K S, McCouch S, Tanksley S D.1992. Genetic and physical analysis of the rice bacterial blight disease resistance locus,.,236:113‒120.
Ryan E P, Heuberger A L, Weir T L, Barnett B, Broeckling C D, Prenni J E. 2011. Rice bran fermented withgenerates novel metabolite profiles with bioactivity., 59(5):1862‒1870.
Sabar M, Akhter M, Bibi T, Riaz A, Haider Z, Khan A R, Bibi A. 2019. Basmati rice lines development carrying multiple bacterial blight resistance genes pyramided using the marker-assisted backcross breeding approach.,39:155.
Sagar V, Dhawan G, Gopala Krishnan S, Vinod K K, Ellur R K, Mondal K K, Rathour R, Prakash G, Nagarajan M, Bhowmick P K, Bollinedi H, Singh A K. 2020. Marker assisted introgression of genes governing resistance to bacterial blight and blast diseases into an elite Basmati rice variety,‘Pusa Basmati 1509’., 216:16.
Sakaguchi S.1967. Linkage studies on the resistance to bacterial leaf blight,(Uyeda et Ishiyama) Dowson, in rice.,16:1‒18. (in Japanese with English abstract)
Salgotra R K, Gupta B B, Millwood R J, Balasubramaniam M, Stewart Jr C N.2012. Introgression of bacterial leaf blight resistance and aroma genes using functional marker-assisted selection in rice (L.),187:313‒323.
Sanchez A C, Ilag L L, Yang D, Brar D S, Ausubel F, Khush G S, Yano M, Sasaki T, Li Z, Huang N. 1999. Genetic and physical mapping of, a recessive bacterial blight resistance gene in rice., 98:1022‒1028.
Sanchez AC, Brar DS, Huang N, Li Z, Khush GS. 2000. Sequence tagged site marker-assisted selection for three bacterial blight resistance genes in rice., 40(3):792‒797.
Sekhwal MK, Li P C, Lam I, Wang X E, Cloutier S, You FM. 2015. Disease resistance gene analogs (RGAs) in plants.,16:19248‒19290.
Sidhu G S. 1978. Dominance reversal of a bacterial blight resistance gene in some rice cultivars.,63:461‒463.
Sidhu G S, Khush G S, Mew T W. 1978. Genetic analysis of bacterial blight resistance in seventy-four cultivars of rice,L., 53:105‒111.
Singh K, Vikal Y, Singh S, Leung H, Dhaliwal H S, Khush G S.2002. Mapping of bacterial blight resistance geneusing microsatellite markers., 19:94‒97.
Singh R J, Khush G S, Mew T W. 1983. A new gene for resistance to bacterial blight in rice., 23(3):558‒560.
Singh S, Sidhu J S, Huang N, Vikal Y, Li Z, Brar D S, Dhaliwal H S, Khush G S. 2001. Pyramiding three bacterial blight resistance genes (,and) using marker-assisted selection intorice cultivar PR106., 102(6): 1011‒1015.
Singh U D, Gogoi R, Mondal K K. 2011. Molecular approach for augmenting disease resistance in cereals: Rice and maize., 20:21‒30.
Singh V K, Singh A, Singh S P, Ellur R K, Choudhary V, Sarkel S, Singh D, Krishnan S G, Nagarajan M, Vinod K K, Singh U D, Rathore R, Prashanthi S K, Agrawal P K, Bhatt J C, Mohapatra T, Prabhu K V, Singh A K. 2012. Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding., 128: 8‒16.
Song W Y, Wang G L, Chen L L, Kim H S, Pi L Y, Holsten T, Gardner J, Wang B, Zhai W X, Zhu L H, Fauquet C, Ronald P.1995. A receptor kinase-like protein encoded by the rice disease resistance gene,.,270:1804‒1806.
Streubel J, Pesce C, Hutin M, Koebnik R, Boch J, Szurek B. 2013. Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility topv.., 200(3): 808‒819.
Suh J P, Cho Y C, Won Y J, Ahn E K, Baek M K, Kim M K, Kim B K, Jena K K.2015. Development of resistant gene-pyramidedrice for multiple biotic stresses using molecular marker-assisted selection.,3:333‒345.
Sundaram R M, Vishnupriya M R, Biradar S K, Laha G S, Reddy G A, Rani N S, Sarma N P, Sonti RV. 2008. Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an eliterice variety.,160:411‒422.
Sundaram R M, Vishnupriya M R, Laha G S, Rani N S, Rao P S, Balachandran S M, Reddy G A, Sarma N P, Sonti R V. 2009. Introduction of bacterial blight resistance into Triguna, a high yielding, mid-early duration rice variety.,4(3):400‒407.
Sundaram R M, Chatterjee S, Oliva R, Laha G S, Cruz C V, Leach J E, Sonti R V. 2014. Update on bacterial blight of rice: Fourth international conference on bacterial blight., 7(1): 12.
Swathi G, Durga Rani C V, Md J, Madhav M S, Vanisree S, Anuradha C, Kumar N R, Kumar N A P, Kumari K A, Bhogadhi S C, Ramprasad E, Sravanthi P, Raju S K, Bhuvaneswari V, Rajan C P D, Jagadeeswar R. 2019. Marker-assisted introgression of the major bacterial blight resistance genes,and, and blast resistance gene,, into the popular rice variety, JGL1798., 39: 58.
Tagami Y, Mizukami T. 1962. Historical review of the researches on bacterial blight of rice caused by(Uyede and Ishiyama) Dowson. Spec. Rep. Plant Dis. Ins. Pests Forecasting Serv. 10. Kyushu Agric. Station, Japan: 112.
Tan G X, Ren X, Weng Q M, Shi Z Y, Zhu L L, He G C. 2004. Mapping of a new resistance gene to bacterial blight in rice line introgressed from,31(7):724‒729.(in Chinese with English abstract)
Taura S, Ogawa T, Tabien R, Khush G, Yoshimura A, Omura T. 1987. The specific reaction of Taichung Native 1 to Philippine races of bacterial blight and inheritance of resistance resistance to race 5 (PX0112).,4:101‒102.
Taura S, Ogawa T, Yoshimura A, Ikeda R, Iwata N. 1992. Identification of a recessive resistance gene to rice bacterial blight of mutant line XM6,L., 42:7‒13.
Thimmegowda P R, Ambika D S, Manjunatha L, Arun R S, Prasad P S, Chandrashekar M. 2011. Screening germplasm for resistance to bacterial blight of rice caused bypv..,2(3):659‒661.
Tian D S, Wang J X, Zeng X, Gu K Y, Qiu C X, Yang X B, Zhou Z Y, Goh M, Luo Y C, Murata-Hori M, White F F, Yin Z C. 2014. The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum., 26(1): 497‒515.
Toenniessen G H, O’Toole J C, DeVries J. 2003. Advances in plant biotechnology and its adoption in developing countries., 6(2):191‒198.
Wang C L, Zhang X P, Fan Y L, Gao Y, Zhu Q L, Zheng C K, Qin T F, Li Y Q, Che J Y, Zhang M W, Yang B, Liu Y G, Zhao K J. 2015. XA23 is an executor R protein and confers broad- spectrum disease resistance in rice., 8(2): 290‒302.
Wang C T, Wen G S, Lin X H, Liu X Q, Zhang D P. 2009. Identification and fine mapping of the new bacterial blight resistance gene,(t), in rice., 123:235‒240.
White F F, Yang B. 2009. Host and pathogen factors controlling the rice-interaction., 150(4): 1677‒1686.
Win K M, Korinsak S, Sirithunya P, Lanceras-Siangliw J, Jamboonsri W, Da T, Patarapuwadol S, Toojinda T. 2013. Marker assisted introgression of multiple genes for bacterial blight resistance into aromatic Myanmar rice MK-75.,154:164‒171.
Yap R, Hsu Y C, Wu Y P, Lin Y R, Kuo C W. 2016. Multiplex PCR genotyping for five bacterial blight resistance genes applied to marker-assisted selection in rice ().,135(3):309‒317.
Yoshimura A, Mew T W, Khush GS, Moura T. 1983. Inheritance of resistance to bacterial blight in rice cultivar Cas 209.,73(10): 1409–1412.
Yoshimura A, Lei J X, Matsumoto T, Tsunematsu H, Yoshimura S, Iwata N, Baraoidan M R, Mew T W, Nelson R J. 2008. Analysis and pyramiding of bacterial blight resistance genes in rice by using DNA markers.: Khush G S, Hettel G, Rola T. Rice Genetics III.World Scientific Publishing Co. Ltd: 577‒581.
Yoshimura S, Yoshimura A, Saito A, Kishimoto N, Kawase M, Yano M, Nakagahra M, Ogawa T, Iwata N. 1992. RFLP analysis of introgressed chromosomal segments in three near-isogenic lines of rice for bacterial blight resistance genes,,and., 67(1):29‒37.
Yoshimura S, Yoshimura A, Iwata N, McCouch S R, Abenes M L, Baraoidan M R, Mew T W, Nelson R J. 1995. Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers., 1:375‒387.
Yoshimura S, Yamanouchi U, Katayose Y, Toki S, Wang Z X, Kono I, Kurata N, Yano M, Iwata N, Sasaki T. 1998. Expression of, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation.,95:1663‒1668.
Yu X Y, Liang X Y, Liu K X, Dong W X, Wang J X, Zhou M G. 2015. Thegene is required for full virulence ofpv.by preventing cell aggregation.,10(7): e0134237.
Yugander A, Sundaram R M, Singh K, Senguttuvel P, Ladhalakshmi D, Kemparaju K B, Madhav M S, Prasad M S, Hariprasad A S, Laha G S. 2018. Improved versions of rice maintainer line, APMS 6B, possessing two resistance genes,and, exhibit high level of resistance to bacterial blight disease.,38: 100.
Zhang F, Zhuo D L, Zhang F, Huang L Y, Wang W S, Xu J L, Vera Cruz C, Li Z K, Zhou Y L. 2015., a novel dominant gene conferring broad-spectrum resistance topv.in rice.,64(3):568‒575.
Zhang G, Angeles E R, Abenes M L P, Khush G S, Huang N. 1996. RAPD and RFLP mapping of the bacterial blight resistance genein rice., 93:65‒70.
Zhang J, Li X, Jiang G, Xu Y, He Y. 2006. Pyramiding ofandfor the improvement of disease resistance to bacterial blight in hybrid rice.,125(6): 600‒605.
Zhang Q, Lin S C, Zhao BY, Wang C L, Yang W C, Zhou Y L, Li D Y, Chen C B, Zhu L H. 1998. Identification and tagging a new gene for resistance to bacterial blight (pv.) from., 15:138‒142.
Zheng C K, Wang C L, Yu Y J, Liang Y T, Zhao K J. 2009. Identification and molecular mapping of(t), a novel resistance gene for bacterial blight (pv.) in rice.,35(7):1173‒1180. (in Chinese with English abstract)
Zheng E S, Wang X M, Xu R M, Yu F B, Zheng C, Yang Y, Chen Y, Chen J P, Yan C Q, Zhou J. 2021. Regulation ofpromoter activity by phytohormone and pathogen stimulation in rice., 28(5): 442‒456.
Zhou Y L, Xu J L, Zhou S C, Yu J, Xie X W, Xu M R, Sun Y, Zhu L H, Fu B Y, Gao Y M, Li Z K. 2009. Pyramidingandfor resistance to two bacterial diseases into an eliterice variety using molecular approaches., 23(2): 279‒287.
Zou H S, Yuan L, Guo W, Li YR, Che Y Z, Zou L F, Chen G Y. 2011. Construction of a Tn5-tagged mutant library ofpv.as an invaluable resource for functional genomics., 62:908‒916.
16 January 2021;
24 August 2021
R. Abdul Fiyaz (genefiyaz@gmail.com; fiyaz.ra@icar.gov.in)
Copyright © 2022, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2021.08.002
(Managing Editor: Li Guan)
- Rice Science的其它文章
- Functions of Nitrogen, Phosphorus and Potassium in Energy Status and Their Influences on Rice Growth and Development
- Epoxiconazole Improved Photosynthesis, Yield Formation, Grain Quality and 2-Acetyl-1-Pyrroline Biosynthesis of Fragrant Rice
- Comparisons of Metabolic Profiles for Carbohydrates, Amino Acids, Lipids, Fragrance and Flavones During Grain Development in india Rice Cultivars
- Cloning and Functional Analysis of Calcineurin Subunits A and B in Development and Fecundity of Nilaparvata lugens (Stål)
- Improving Rice Blast Resistance by Mining Broad-Spectrum Resistance Genes at Pik Locus
- Pectin Methylesterases Enhance Root Cell-Wall Phosphorus Remobilization in Rice

