Modelling the biological invasion of Prosopis juliflora using geostatistical-based bioclimatic variables under climate change in arid zones of southwestern Iran
Mohadeseh AMIRI, Mosfata TARKESH, Mohammad SHAFIEZADEH
1 Department of Natural Resources, Isfahan University of Technology, Isfahan 8415683111, Iran;
2 Natural Resources and Watershed Management Organization, Khuzestan 6134788439, Iran
Abstract: Invasive species have been the focus of ecologists due to their undesired impacts on the environment. The extent and rapid increase in invasive plant species is recognized as a natural cause of global-biodiversity loss and degrading ecosystem services. Biological invasions can affect ecosystems across a wide spectrum of bioclimatic conditions. Understanding the impact of climate change on species invasion is crucial for sustainable biodiversity conservation. In this study, the possibility of mapping the distribution of invasive Prosopis juliflora (Swartz) DC. was shown using present background data in Khuzestan Province, Iran. After removing the spatial bias of background data by creating weighted sampling bias grids for the occurrence dataset, we applied six modelling algorithms (generalized additive model (GAM), classification tree analysis (CTA), random forest (RF), multivariate adaptive regression splines (MARS), maximum entropy (MaxEnt) and ensemble model) to predict invasion distribution of the species under current and future climate conditions for both optimistic (RCP2.6) and pessimistic (RCP8.5)scenarios for the years 2050 and 2070, respectively. Predictor variables including weighted mean of CHELSA (climatologies at high resolution for the Earth's land surface areas)-bioclimatic variables and geostatistical-based bioclimatic variables (1979–2020), physiographic variables extracted from shuttle radar topography mission (SRTM) and some human factors were used in modelling process. To avoid causing a biased selection of predictors or model coefficients, we resolved the spatial autocorrelation of presence points and multi-collinearity of the predictors. As in a conventional receiver operating characteristic(ROC), the area under curve (AUC) is calculated using presence and absence observations to measure the probability and the two error components are weighted equally. All models were evaluated using partial ROC at different thresholds and other statistical indices derived from confusion matrix. Sensitivity analysis showed that mean diurnal range (Bio2) and annual precipitation (Bio12) explained more than 50%of the changes in the invasion distribution and played a pivotal role in mapping habitat suitability of P. juliflora.At all thresholds, the ensemble model showed a significant difference in comparison with single model.However, MaxEnt and RF outperformed the others models. Under climate change scenarios, it is predicted that suitable areas for this invasive species will increase in Khuzestan Province, and increasing climatically suitable areas for the species in future will facilitate its future distribution. These findings can support the conservation planning and management efforts in ecological engineering and be used in formulating preventive measures.
Keywords: invasive species; climate change scenarios; partial ROC; ensemble forecast; Kriging; spatial bias
1 Introduction
Invasive species are common focus of interest for ecologists, natural resource managers and biological conservationists due to their rapid spread, threat to biodiversity and damage to ecosystems (Kandwal et al., 2009). Bio-invasion has homogenized the world's flora and fauna,altered species composition and community structure and changed biogeochemical cycles. It is also recognized as one of the major drivers of global biodiversity loss and species extinction(Huang and Asner, 2009; Shrestha et al., 2018). Linders et al. (2019) proposed bio-invasion to be investigated as a substantial component in global change and one of the main reasons of species destruction. Bio-invasion has also been recognized as an important non-climatic driver of global change to affect the metabolism of ecosystems and change disturbance regimes (Huang and Asner,2009). Bio-invasion has become the top preference topic for more research and assessment(IPBES, 2015). Significant efforts from the local to global scale have been made in this context,including assessment and management of alien invasive species (Shrestha et al., 2018). Invasive species distribution models (iSDMs) and mapping techniques could significantly reduce control and eradication costs (Joshi et al., 2004). In the last decades, the development of advanced remote sensing technology and species distribution models (SDMs) has significantly contributed to vegetation mapping on different scales and offered remarkable possibilities (Oldeland et al.,2010).
In previous studies, SDMs were employed for mapping the invasive species habitat suitability at different scales by relating the field occurrence data with environmental factors, land cover and vegetation information (Joshi et al., 2004; Huang and Asner, 2009; Kandwal et al., 2009; Diao and Wang, 2014; Wakie et al., 2014; Choudhury et al., 2016; Peknicová and Berchová-Bímová,2016; Bazzichetto et al., 2018; Shrestha et al., 2018; Srivastava et al., 2018; Heshmati et al., 2019;Gong et al., 2020). According to previous studies, plant invasions occur across a wide range of bioclimatic conditions and have significant feedback with regional and global climates. Therefore,species climate models that are based on the statistical and geographical relationships between invasive species (presence/absence/abundance) and the environment (Srivastava et al., 2018), by emphasizing the preference susceptible locations and identifying the potential range of infestations, can greatly assist land managers in coordinating response for efficient eradication of invasive plants before they become extensively established and costly to contain (Diao and Wang,2014). The management of exotic species is becoming more challenging because of distracting effects of global warming on the distribution of those species (Shrestha et al., 2018). Species climatic niche evolves slowly because of the niche conservatism. This conservation has been a principle to expand species climatic niche based on the prevalent distribution of the species and project it to new areas to find suitable habitats for the presence of the species (Guisan et al., 2013).Invasive species increase the susceptibility of ecosystems to climate-related stressors. In many ways, bio-invasions have synergistic impacts with climate changes to create new suitable habitats for invasive plant establishment and intensify the invasion process (Bellard et al., 2016; Heshmati et al., 2019). According to Bronnimann et al. (2006), by 2050, perennial plants are more likely to be affected by climate change than annuals. Also in comparison with endemic species, invasive species have usually more abundance and are tolerant to a wide span of climatic conditions.Therefore, in management strategies for invasive species, there need to be a consideration of the climate change factors. Availability of global climate data offers an opportunity to predict the future distribution of species under different climate change scenarios (Srivastava et al., 2018).
The accuracy and performance of single model varied widely among different methods (Elith et al., 2006; Marmion et al., 2009). Basically, identifying any single modelling approach that is better than others has failed. Methods integrating multiple single model have the potential to provide more robust estimations of suitable habitat for a certain invasive species at a given time,leading to more reliable SDMs (Srivastava et al., 2018). The ensemble approach assumes that when averaging across multiple models that the true ''signal'' of interest will separate itself from the ''noise'' and bias associated with any single model (Araújoand and New, 2007). Lately, Naimi and Araújo (2016) created an R-based program to model species distribution, enabling ensemble of models to be fitted and evaluated.
The objectives of this study are: (1) to apply remote sensing and SDMs to map and model invasive species distribution; (2) to test whether ensemble modelling framework yields more accurate prediction of plant species distribution than single model; (3) to use partial ROC analysis to provide a consistent basis for assessment of predictions from SDMs; and (4) to survey climate change impacts on the distribution and potential invasion ofP. juliflora.
2 Materials and methods
2.1 Study area and species
Khuzestan Province is located in southwestern Iran, bordering the Persian Gulf and Iraq. It covers an area of 62,432 km2, which lies between the latitudes of 29°57′N and 33°00′N and the longitudes of 47°40′E and 50°33′E (Fig. 1). The elevation varies around 3740 m a.s.l. Climate varies extensively from arid to humid, but most parts of the province are arid. According to climate station data, mean annual precipitation is 266 mm, and the main period of precipitation occurs in winter. The maximum temperature in summer in the province is about 50°C (in July)and the minimum temperature in winter is 9°C (in March). Summer is from April to September,whereas winter is from October to March. The locations of climate stations within and around the study area are shown in Figure 1.
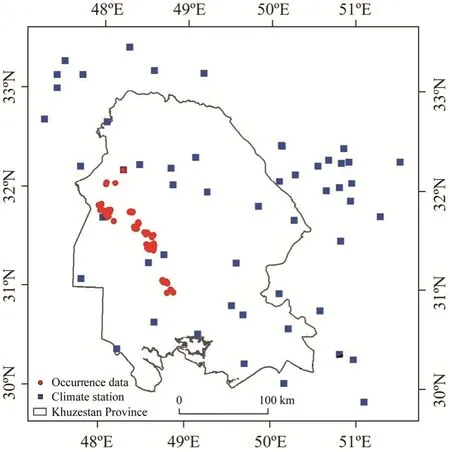
Fig. 1 Occurrence data of P. juliflora and climate stations in Khuzestan Province, Iran
Vegetation cover of the study area includes four types: wetland species, hygrophytes, terrestrial halophytes and psammophytes.Seidlitzia rosmarinusEhrenb. Ex Boiss.,Tamarix passerinoidesDel. ex Desv.,Atriplex leucocladaBoiss. andLycium depressumPojark. could be introduced as the species adapted to the climate and saline soil of the study area. Based on available information,1.565×106hm2of coastal plains in the study area are desert that are exposed to wind erosion. This soil condition complicates the forestation in the area, causing the burial and loss of newly planted seedlings and threatening quicksand stabilization projects. Tree and shrub species such asP.juliflora(Fabaceae) are used for biological restoration of arid and desert areas, combating desertification, preventing the movement of quicksand and creating windbreaks. However, this tree has been reported as invasive and has caused problems for farmers and ranchers by creating dense stands, regenerating in natural environments, invading rangelands and fields by transferring water and manure, as well as causing adverse effects on local livestock.
2.2 Data collection
A total of 115P. julifloraobservations with geographic coordinates were recorded in 2020 (Fig.1). We used a sampling approach based on experts and local knowledge. In addition to avoiding duplication of sampling points, this approach along with spatial filtering of presence points allowed us to reduce spatial autocorrelation and thus over-fitting of models. After avoiding biased selection of predictor variables and the spatial autocorrelation of presence data, we removed 13 auto-correlated occurrence data. About 75% of remaining data were randomly used to train the model (training dataset) and the remaining 25% data were used to evaluate the model (test dataset). We generated 3000 background points instead of absence points throughout the area. The number was set depending on the number of presence points and the level of sample prevalence,where sampling prevalence is defined as the number of presence relative to the entire sample(Santini et al., 2021). This is large enough to accurately represent the range of environmental conditions in the study area, i.e., more background samples don't improve model performance(Phillips and Dudík, 2008; Lamelas-López et al., 2020). Background samples are commonly chosen uniformly at random, whereas occurrence data are often spatially biased toward relatively accessible areas such as near roads, villages and rivers. Therefore, the sampled locations may not be proxy of the actual range of environmental conditions in which the species occurs. As the spatial bias usually causes environmental bias, the difference between background samples and occurrence data may lead to incorrect model. One approach to address this problem would be to manipulate the background data in order to remove the bias based on Phillips et al. (2009). We produced weighted sampling bias grids for the occurrence data and used the grids with environmental variables and geographical locations of species occurrence in modelling process.If true absence data are not available for modelling purposes, pseudo-absence data or randomly simulated background data are used by different SDMs. Absence data of species are of dubious utility in the process of species distribution modelling. This point is easily understandable when considering invasive species; if an invasive species distribution a few decades prior to its introduction in the novel area was modeled using absence data, its future distribution area would be counted as absence. This area was actually quite within the ecological niche dimensions of the species, as will be demonstrated by the later invasion. Generating background points in the vicinity of the occurrence data allows both the background and the occurrence points to carry similar types of bias (Peterson et al., 2008). In a conventional ROC, the AUC is calculated using the presence and absence observations to measure the probability and the two error components(omission and commission errors) are weighted equally. Also, as absence data have been eliminated, the 1-specificity axis was transformed to proportion of area predicted as present.There are two sources of problems in ROC analysis: The first limitation is that some algorithms cover broad spectra of feasible commission errors, while others are limited to smaller ranges and span only relatively small areas of the entire ROC curve, either by the inherent function of the algorithm or by design. The second limitation is related to the fact that ROC analyses don't recognize the meanings of ''absence'' in ecological niche modeling (ENM) versus SDM. Therefore,partial ROC analysis is used instead of AUC. This approach removes the emphasis on absence data and expresses ROC results as ratios of the area under the curve, i.e., the ratio of the model AUC to the null expectation (Elith et al., 2006; Peterson et al., 2008).
2.3 Environmental variables selection
Totally 19 bioclimatic variables (Table 1) were downloaded from the CHELSA dataset, available at 1-km spatial resolution. The data include the minimum, maximum and mean precipitation and temperature of climatology stations during the period 1979–2013, which have been interpolated for the whole world using accurate methods (https://chelsa-climate.org). The ''con'' function of raster calculator was used for ''no data'' values. During the period 2014–2020, a climatic set including 19 bioclimatic variables derived from mean annual precipitation, the maximum and minimum annual precipitation recorded in 70% data of the stations within and around the study area were analyzed using R software. Then, their spatial distribution maps were extracted in ArcMap v10.3 using altitude, longitude and latitude of stations as predictor variables under geostatistical methods (Kriging and inverse distance weight (IDW)) with a spatial resolution of 1-km. Before using these methods, we checked the normality of climatic data. If the data were not normal, logarithmic and square transformations were used to normalize the data. Inverse distance weighting method was used to map the spatial distribution of some abnormal bioclimatic variables. To evaluate the accuracy of Kriging interpolation methods and IDW, we used the data of other meteorological stations, mean bias error (MBE) and root mean of square error (RMSE):

wheremiis the measured data andsiis the values estimated by model. The lower RMSE and MBE represent the more statistically accurate model (Ruiz and Bandera, 2017). Then, the weighted average (1979–2020) of CHELSA bios and geostatistical-based bios were used as input of statistical models.
Physiographic factors within one elevation, aspect and slope as well as soil type are responsible for vegetation dynamics (Singh, 2018). Digital elevation model (DEM) extracted from SRTM with 90-m spatial resolution (http://srtm.csi.cgiar.org) was used to generate the elevation, slope and aspect layers. All physiographic factors were resampled using the nearest neighbor method with 1-km spatial resolution to match the resolution of the remote sensing and bioclimatic predictors. Using the DEM and the longitude and latitude coordinates, we also obtained distance to water lines and distance to villages as 1-km raster using Euclidean distance tool. The ENVI v5.1 and ArcMap v10.3 were used to create the spatial data layers.
The multi-collinearity test was done to examine the cross-correlation (Table S1) and establish a model that has better performance with fewer variables by using Pearson correlation coefficient(r). We selected one variable from highly paired correlated variables (|r|>0.8) based on biological importance and ease of interpretation for inclusion in the model. Qualitative evaluation of the distribution of values of each variable at all presence points and of the relation between the variable and species presence or pseudo-absence was used to remove the least important variables(Alvarez et al., 2017). After elimination of highly correlated and unessential variables, we reduced the number of potential predictors to seven variables including mean diurnal range in temperature (Bio2), the maximum temperature of the warmest month (Bio5), mean annual precipitation (Bio12) and precipitation seasonality (Bio15), slope as physiographic factor,distance to villages and distance to water lines as human factors.
2.4 SDM implementation and assessing variable importance
According to the results of recent studies about comparing models and their application in several ecological studies (Crimmins et al., 2013; Peknicová and Berchová-Bímová, 2016; Shrestha et al.,2018; Kaky et al., 2020), we ran 5 correlative models that have higher accuracy and precision to create ecological niche modelling using non-correlative variables derived from satellite data,occurrence data, background points and sampling bias grid. These models represent a broad range of analytical approaches and are included one classification method (CTA), two regression methods (GAM and MARS) and two machine learning methods (RF and MaxEnt). In order to increase the accuracy and efficiency, we considered 10 replications for each model (Barbet-Massin et al., 2012; Rueda-M et al., 2021). In every replicate, the data were indiscriminately divided and the mean prediction map was presented as the final map of each model using the weighted average method:

BIOMOD package was calibrated to assess variable importance. This importance was automatically calculated separately for all modelling techniques. The goal was to determine which environmental factor has the immense effect on species invasion at a local scale.
2.5 Model evaluation statistics
Since the complexities of the ecological niche are unknown and probably unknowable, we test the efficiency of several criteria from a set of models (Warren and Seifert, 2011). Therefore, after implementation of SDMs, we tested the model's performance with threshold-dependent measures(correct classification rate (CCR), sensitivity, specificity and true skill statistic (TSS)) and threshold-independent measures using features available with partial ROC analysis (Barve, 2008).This program is specifically developed for assessing the predictive performance of habitat models using background data and requires presence and area dependent suitability files. The AUC is an appropriate indicator for evaluation because it accepts the least impact from the sample size(Peterson et al., 2008). We begin by defining a user-selected parameter E, which refers to the amount of error admissible along the true-positives axis. This parameter refers to how much omission error is acceptable in applications in which the highest-quality occurrence data are used.It might be set at E=0%, or it might be higher when the occurrence data are known to include certain amounts of error. At each predictive threshold, sensitivity is calculated as 1-omission error,and the AUC comparisons are presented as the ratio of the model AUC to the null expansion(AUCratio).
Significant differences (P>0.05) in the performance of the models were investigated by performing Tukey's test on the partial ROC analysis. After rejecting the null hypothesis in the analysis of variance (ANOVA), this test scrutinizes the significant differences between the pairs of means. The critical value for comparing the mean difference is obtained from Equation 5:

whereais the number of treatments;fis the degree of freedom; and MSE is the mean square error obtained from ANOVA. The Tukey's test has an error of type I (α) for all pairwise comparisons.
The quality of a discrimination model is characterized by the CCR. The CCR is a measure of classification performance or diagnostic accuracy when the balanced data for two classes are involved:

It is generally agreed that the higher the CCR, the better the model. Additionally, one should consider sensitivity and selectivity of the model (Yin et al., 2021). The probability of occurrence data was transformed into binary presence-absence prediction by sensitivity-specificity equal approach that is popular in ecology to select prediction thresholds for models. Thus, four fractions were calculated: (1) true positive (TP): the species exists where predicted to exist; (2) false positive (FP): the species does not exist where predicted to exist; (3) false negative (FN): the species exists where not predicted to exist; and (4) true negative (TN): the species does not exist where not predicted to exist. To evaluate and predict the actual maps conformity, we also measured true skill statistic (TSS) using above mentioned fractions. For a 2×2 confusion matrix TSS is defined as follows:
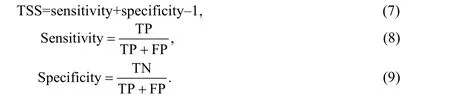
Specificity and sensitivity are applied to consider all elements of a confusion matrix (true and false presences, and absences). TSS considers both commission and omission errors ranging from–1 to +1, where +1 represents perfect agreement and scores range from 0.7 to 0.9, specifying fair to good model performance. The scores of zero or less demonstrate an efficiency no further than random (Allouche et al., 2006).
2.6 Ensemble model-fitting
Modelling algorithms can be easily integrated in the ensemble framework. Various ensemble methods are now available, but forecasts ofP. juliflorahabitat were combined using weighted average approach that weights each model outputs based on AUCratioas it offers a simple and straightforward way to unite various SDMs and its output is easy to interpret. The method based on average function algorithm is likely to increase the accuracy of species distribution forecasts.To fully comprehend the reasons for ensemble model behavior and results, we required a wider study that addresses which aspect of ensemble modelling has the most impact on species distribution forecasts. We examined the possibility of combining several independent but complete modelling methods to increase the accuracy of spatial predictions. We combined the results of the above five modelling algorithms by the weighted average approach of separate habitat maps based on partial ROC analysis to generate potential distribution.
The models produced by the weighted average approach (Eq. 4) were combined into a consensus niche model for the species as Equation 10:
wherec(x) is the weighted consensual niche model. But instead of a replicate model, a single model is considered (Masocha and Dube, 2018). Ensemble scores in the modelled output represent the grade of model agreement. In this model, a pixel with a value of zero indicates that none of the modelling techniques defines that area as a suitable habitat, while the number one indicates that all modelling techniques consider that area as a suitable habitat.
2.7 Mapping invasion risk under climate change
At present, global climate models (GCMs) are the most reliable tools for prediction the future climate of the world, despite some deficiencies. The future climatic data are climate projections for the years 2050 (2040–2060) and 2070 (2060–2080) from GCMs that are obtained from the following website https://chelsa-climate.org. In the fifth IPCC report, four representative concentration pathways (RCPs) were considered, representing scenarios in which the total radiative forcing in 2100 had reached 2.6, 4.5, 6.0 and 8.5 W/m2(IPCC, 2013). Here, to observe the response of species to future climate, we used MPI-ESM-P, a global circulation model. This model is one of the most suitable and practical models of GCMs in the Middle East (Pal and Eltahir, 2015). MPI-ESM-P data for the RCP2.6 (reduction of atmospheric CO2as a result of green employment) and RCP8.5 (industrial development) scenarios for two different periods were downloaded to derive an accurate estimation of changes in the invasion pattern ofP. juliflora. The purpose of selecting these scenarios was to predict the maximum and minimum impacts of future climate change. After species distribution mapping under these scenarios and climatic model for the present and future time periods, the discrepancy between the two ensemble maps was considered as the basis to determine the amount and direction of species habitat relocation.
The habitat suitability maps of the current condition were subtracted from the projected maps of the future climate in order to assess the potential changes in the range size under climate change scenarios. In other words, by preparing the model outputs for the present and comparing it with the output of models for the years 2050 and 2070, we investigated the impact of global warming on the studied species and changes in its habitat extent.
3 Results
3.1 Validation of geostatistical-based bioclimatic variables
The validation results of bioclimatic variables based on the best interpolation method are given in Table 1. RMSE values ranged from 0.72 to 17.74, MBE values from –1.27 to 2.12. Each of the interpolation methods that had the minimum value of error criteria was selected as the best method to calculate given climatic variables. CoKriging result showed that a better efficiency compared with other methods for temperature-based bioclimatic variables, such as mean temperature of the driest quarter, mean temperature of the warmest quarter and mean temperature of the coldest quarter.
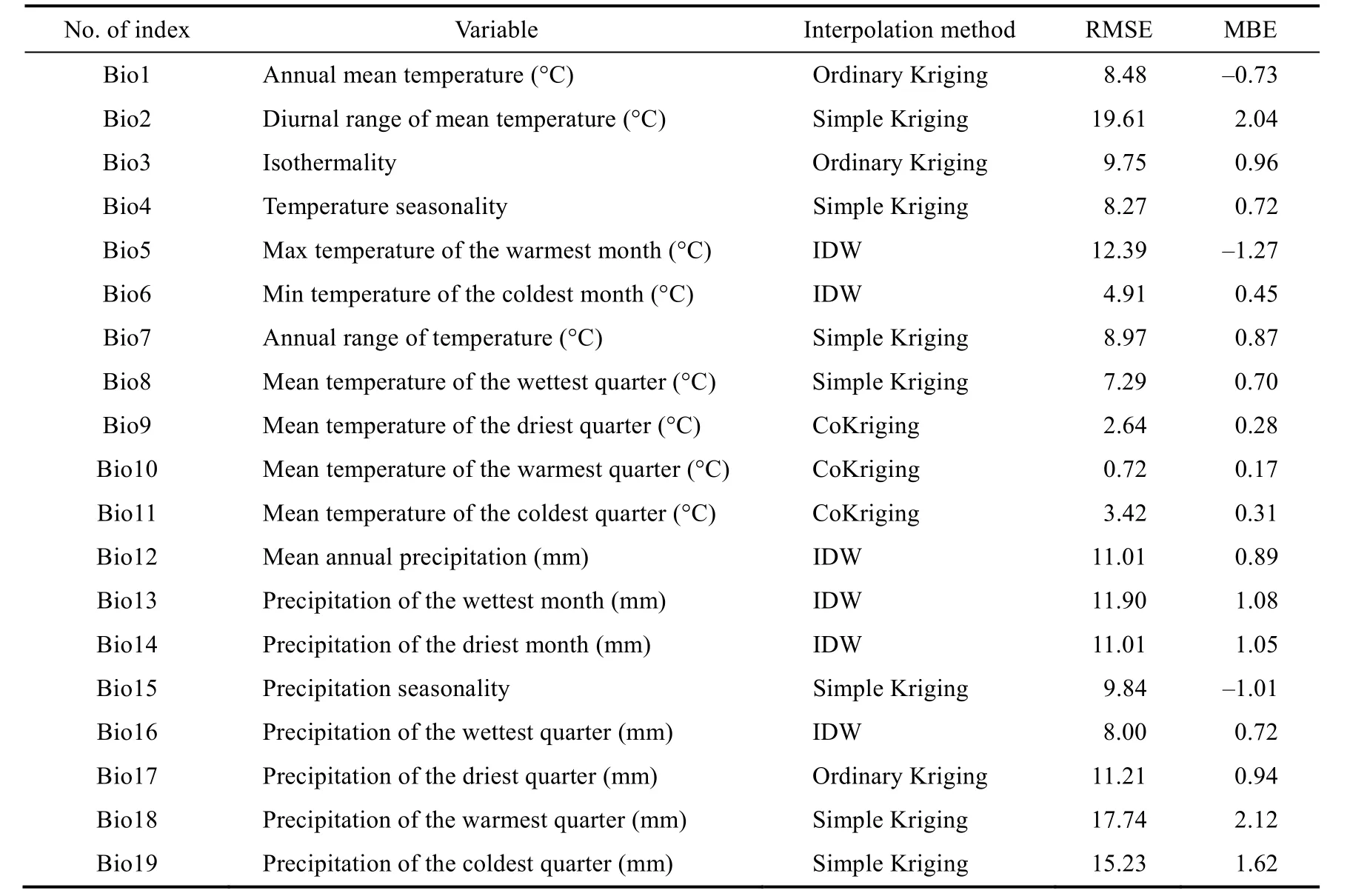
Table 1 Validation of the bioclimatic variables produced by geostatistical method (2014–2020)
3.2 Model evaluation and partial ROC analysis
The models represented the variations in the discriminatory abilities. In general, all the models performed well for detectingP. julifloraand gave us confidence in the overall accuracies of the potential distribution maps. Based on accuracy statistics, among the four group discrimination models and one profile model used, MaxEnt and RF predicted the climatic habitat of this invasive species with a higher discriminatory ability and accuracy compared with other models, especially GAM. Indeed, these models showed less errors in determining suitable and unsuitable climatic areas for the presence of the species. We developed partial ROC analysis for each of all model outputs at three levels of E, including 100% (in which the models are accepted across the entire spectrum of areas predicted as present, equivalent to the traditional ROC analysis), 10% and 5%.In Table 2, the results of partial ROC analysis are represented as ratios of the AUC.

Table 2 Mean values of used accuracy statistics to evaluate the performance of ensemble map and SDMs to predict the distribution of P. juliflora
The box plots of the models using Tukey's test (P<0.05) showed that atE=100% threshold, all models except RF and CTA had significant differences (Fig. 2). At E=5% threshold, RF had no statistically significant difference with CTA and MaxEnt; and at E=2% threshold, MaxEnt with CTA and RF. At these thresholds, no significant difference was observed between the models with a higher performance, i.e., MaxEnt and RF.
All algorithms predicted responses from 0% to 100% of false positive, but GAM predicted only in the range of 7%–84%, and MARS predicted at the range above 18%. Since these two models only predicted a part of geographical areas, their ROC curves were defined only within a subset of possible areas. In fact, this procedure in effect penalized algorithms with ROC curves that didn't begin at or near the origin.
In Figure 3, we considered the portion of ROC curves that lied within the predictive range of the modelling algorithm and within the range of acceptable models in terms of omission error(0.1–1.0). In case, models' AUC was compared with the AUC for a line of null expectations(=0.5), elevation of these curves above the straight line of random expectation is a measure of the discriminatory capacity of the algorithms (i.e., their capacity to classify correctly true presences and true absences). Here, for the efficiency of ensemble model, MaxEnt, RF and CTA were clearly better than MARS and GAM, because they were further away from the line of null expectations. At E=5% and E=2% thresholds, the relative positions of the curves shifted and theXandYaxes differed from those of a conventional ROC curve. GAM provided no predictions at E=2% threshold, and so was excluded from calculation. Also, the validity criteria in the ensemble prediction had allocated a higher average than all single modelling algorithm. Of course, the efficiency of consensus method is the maximum when all the models used in it have the high accuracy and precision, because the weakness of one model will reduce the efficiency of other models (Araújo and New, 2007).
3.3 Contribution of environmental variables and model application

Fig. 2 Box plot of partial receiver operating characteristic (ROC) analysis in invasion distribution models under different E thresholds. (a), E=100%; (b), E=5%; (c), E=2%. Different lowercase letters indicate significant difference among different models.

Fig. 3 ROC curves under different E thresholds. (a), E=100%; (b), E=5%; (c), E=2%.
The relative importance of each environmental variable used in the modelling process or their extent of impact on the output of the model based on the results of sensitivity analysis has been reported for each model (Table 3). Due to the type of algorithm used in each model, the degree of ecological significance of the variables varies from model to model, because as mentioned earlier,the nature of the models used in this study is of the correlated type, i.e., they are used to obtain the best prediction of species presence area. Factors that are introduced as important factors in the study of species distribution do not necessarily have a cause-and-effect relationship with the presence of the species and are merely factors that increase the predictive accuracy of the model.
The most significant predictors were found to be Bio2 (diurnal range in mean temperature),Bio12 (mean annual precipitation) and Bio5 (maximum temperature of the warmest month),respectively, then slope and Bio15 were placed with almost the same degree of importance, while distance to villages and water lines did not contribute much in the prediction of potential habitat.Bio2 and Bio12 explained about 50% of the changes in the ensemble model (Table 3).
Maps of species occurrence probability were prepared after generalizing the models from the mathematical space to the geographical space to show the distribution ofP. julifloraunder current climate conditions (Fig. 4). In determining the potential range of species habitat by different models, spatial uncertainties could not be completely eliminated. In other words, the overall areas of suitable habitats using different models are different. Although habitat suitability maps or spatial patterns differ in prediction occurrence between model algorithms, they overlap a lot. The models express a probability distribution where each grid cell has a predicted suitability of conditions for the species. Areas with higher habitat suitability scores indicate a higher risk. To better understand the impacts of climate change, we classified physiography and human factors and the suitability values into four categories of potential habitats including high potential(0.75–1.00), good potential (0.50–0.75), moderate potential (0.25–0.50) and least potential(0.00–0.25).
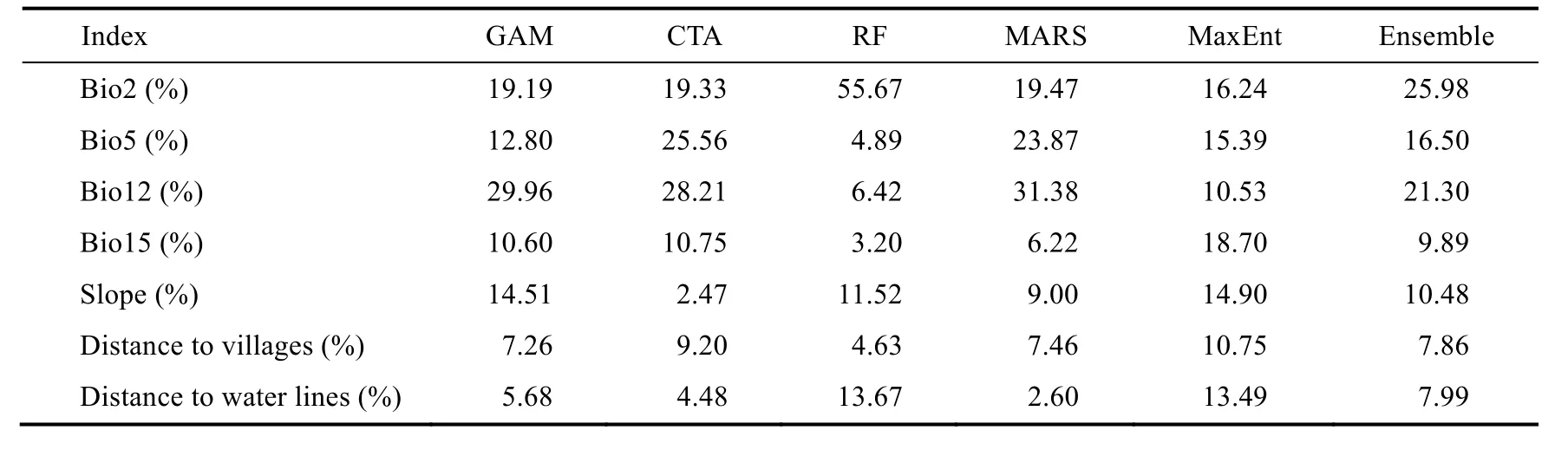
Table 3 Relative importance of environmental variables included in modelling
Although the predictive capacity of single model differed (Table 2), area predicted as suitable habitats and unoccupied habitats accorded with species' ecology and field study area in all models.Using the consensus technique and summarizing the results of all fitted models, we modelled a map of suitable areas for the distribution ofP. julifloraunder current climate conditions (Fig. 4).According to the current ensemble model, the current presence and absence areas of the species are 4863 and 57,569 km2, respectively. The habitats of the species occupy 7.79% of the study area and are relatively integrated in the western area, where the altitude and precipitation are less than the average values in the whole area. In these areas, the presence of this species can be observed in the form of pure communities and sometimes mixed with other species.
3.4 Invasion distribution under global warming scenarios
Using fitted equations for each model, we modelled the potential distribution ofP. juliflorafor the years 2050 and 2070 under RCP2.6 and RCP8.5 climate change scenarios (Fig. 5). The geographic distribution of the species would increase under predicted levels of climate warming.Some parts of unsuitable habitat under future climate scenarios would become suitable, resulting in its local expansion. The level of species distribution did not differ much in different scenarios.The displacement will generally be around the current nest of the species and the species will not occupy a new place in the future. Of course, the response to climate change may not be the same at local and regional scales for different species. Few studies have shown that on a regional scale there will be a decrease in suitable habitats, but on a small or local scale, the situation is quite different. Habitat suitability classes vary in size depending on their values, and the increase in species range size can be better understood under climate change.

Fig. 4 Maps of species habitat suitability of P. juliflora under different models. (a), GAM; (b), CTA; (c), RF; (d),MARS; (e), MaxEnt; (f), Ensemble. 0.75–1.00, high potential; 0.50–0.75, good potential; 0.25–0.50, moderate potential; 0.00–0.25, least potential.
The spatial distribution ofP. julifloraobtained from different models under optimistic and pessimistic scenarios of climate change models was investigated (Fig. S1) and the values of change in the species distribution range in the two time periods were calculated under the above-mentioned scenarios (Table 4). In Table 4, residual appropriate area (stable presence) is an area of the current presence that is also appropriate for the species in the future. Residual inappropriate area (stable absence) is an area of current absence that is also inappropriate for the species in the future. Suitable and unsuitable areas are the areas of habitats that have become adapted to the species and the area of habitats that have lost their suitability due to climate change,respectively. In order to determine the suitable and unsuitable areas, we classified the species distribution map in the present and future conditions into two suitable and unsuitable classes using a critical level that maximizes the accuracy of the model according to the AUCratiocriterion,and then compared. The areas of presence in the future (sum of stable presence and adapted) and absence in the future (sum of stable absence and inappropriate) are also listed in Table 4. In all cases, the changes will be positive or incremental, i.e., the size of low suitability classes or inappropriate habitats will be reduced, and the size of high suitability classes or adapted habitats will be increased. The lowest and highest displacements will occur under RCP2.6 and RCP8.5 scenarios in 2070 with a ratio of 1:2, respectively.

Fig. 5 Maps of spatial distribution of P. juliflora under climatic scenarios for the years 2050 and 2070. (a),RCP2.6-2050; (b), RCP8.5-2050; (c), RCP2.6-2070; (d), RCP8.5-2070. 0.75–1.00, high potential; 0.50–0.75,good potential; 0.25–0.50, moderate potential; 0.00–0.25, least potential.

Table 4 Area changes of P. juliflora distribution under climate change scenarios for the years 2050 and 2070
The trend of changes in annual mean temperature and mean annual precipitation for the years 2050 and 2070 by MPI-ESM-P climate model in the study area is shown in Figure 6. Under both scenarios and in both time periods, the temperature will increase compared with normal state(1979–2020) and the precipitation will decrease. In the studied climate model, the highest amount of precipitation will be observed under the pessimistic emission scenario in 2070, when the amount of precipitation will decrease by more than 175 mm. The least amount of precipitation changes will occur under RCP2.6 in 2070, which is approximately 97 mm. Similar to precipitation, the highest and lowest rates of increasing in temperature are related to RCP8.5 in 2070 and RCP2.6 in 2070, respectively. In 2070, the minimum temperature will increase by 3.88°C and the maximum temperature will increase by 4.60°C.

Fig. 6 Trends of temperature (a) and precipitation (b) variables under climate change scenarios
4 Discussion
Better efficiency of coKriging for temperature-based bioclimatic variables interpolation indicated the appropriateness of elevation as an auxiliary variable to interpolate these bioclimatic variables in Khuzestan Province, Iran. This finding was also found in the study of Aznar et al. (2012).Khosravi and Balyani (2019) indicated that coKriging has provided the best performance for areas in Iran where the correlation coefficient between temperature and altitude or latitude is high,which is consistent with the results of our study. IDW has been extensively used as an effective interpolation method for precipitation data (Perry and Hollis, 2005). Yang et al. (2015) offered IDW for interpolation point-based precipitation data over Greater Sydney region, too.
All models indicated differences in spatial habitat predictions, referring the intrinsic inter-model uncertainty, as shown by Elith et al. (2006). Evidence comparing different modelling approaches also suggested that the accuracy of predictions between different modelling algorithms can be fundamentally different (Kumar et al., 2009; Dobrowski et al., 2011). The most comprehensive model comparison set was done by Elith et al. (2006). The researchers compared 16 modelling methods using 226 species in 6 regions of the world, and by comparison of modelling techniques concluded that Boosted Regression Trees (BRT) and the maximum entropy were among the most efficient methods. Heshmati et al. (2019) also focused on climatic variables and emphasized the effectiveness of the MaxEnt model in invasion distribution ofP. juliflora.MaxEnt based on presence-background data instead of presence-absence data, and most importantly, does not undertake that background data preclude the probability of occurrence(Evangelista et al., 2008). This particularly makes MaxEnt highly appropriate method for modelling the invasion distribution as invasive species tend to establish and expand their range to new areas beyond their native habitats (Heshmati et al., 2019). MaxEnt and RF models showed the highest efficiency after ensemble model in predictingP. julifloradistribution. MaxEnt has been extensively used to investigate vulnerability of natural ecosystems to be invaded by alien invasive species under current and future climate change scenarios (Kumar et al., 2015). Its capability to control over-fitting through regularization and retaining the similar bias between the background and presence points increases its discriminatory capability (Phillips et al., 2006). One basic limitation of MaxEnt is that sample selection bias has much more effect on profile models than on group discrimination models (Phillips et al., 2009). Another limitation is that prevalence cannot be accurately specified from presence-only data in isolation, irrespective of sample size. In other words, prevalence is not identifiable (Elith et al., 2010). Marmion et al. (2009) also found that RF model presented correct predictions of the withheld data. But, regression models are weak in explaining the complexity of associations between species and environment and predicted with a low discriminatory ability (Table 2).
Limitation by environmental conditions, dispersal limitation and biotic interactions are substantial ecological processes that specify whether a species may occupy a site. But, SDMs can't distinguish environmental effects from biotic interactions. Therefore, the effects of environmental and climatic factors are not separated from the effects of vegetation interactions and the fundamental niche remains unknown (Poggiato et al., 2021). In addition to this limitation,model performance has apparently divergent patterns that are probably to be relevant to variations in the methods abilities to recover helpful relationships between species and environmental variables with distinct strengths and lengths of gradients.
Environmental conditions will change with respect to climate and land use change and many other environmental factors. Thus, modeling and mapping of invasive plants must be considered an iterative process (Stohlgren et al., 2010). At a regional scale, the spread of alien invasive species is primarily shaped by spatiotemporal interactions with the invaded habitats (Peknicová and Berchová-Bímová, 2016). The distribution ofP. juliflorais unlikely in low invasive sites due to the overall unfavorable contribution of environmental factors to the occurrence of invasive plants. Because of the limited range of bioclimatic variables affecting the habitat suitability of the species resulting narrow niches ofP. juliflora, high fluctuations of efficient bioclimatic variables can lead to loss of potential habitat suitability. For example, the study by Behnamfar et al. (2019)indicated that a decrease in the temperature by –1°C for 3 h reduces plant yield by 52% and the continuation of this condition can lead to plant death. Despite the loss of distribution area for the species, the changes will be positive or incremental in case of global warming.
In this study, by performing iterations and changing coefficients of each variable, we determined percent contribution of any single variable in the models. The relationship between the predictors and invasion distribution was based solely on empirical observations and could not be interpreted as a cause. We found that the combined factors of temperature and precipitation among bioclimatic factors were responsible for the spread of this invasive species, which is consistent with Wakie et al. (2014). Adhikari et al. (2012), through studying the potential spatial distribution ofIlex khasianaPork., has also chosen precipitation and temperature along with the topographic variables ''as these have direct relevance to invasive species''. The temperature variables (such as Bio5) explained the thermal tolerance of the species, and mean annual precipitation (Bio12) and precipitation seasonality (Bio15) described water availability(Choudhury et al., 2016). Diurnal range of mean temperature (Bio2) showed a significant effect on the distribution ofP. juliflora. This probably explains the higher energy demand of the species for physiological activities. Although slope didn't contribute much in the prediction of biological invasion, it was the fourth contributed variable based on ensemble model, indicating incidence of topographic preferences in the spread ofP. juliflora. The suitable habitats are also distributed along the villages and water lines, with suitability increasing as the distance to the villages and water lines decreases. Although the distance to villages plays a lesser role in modelling than other variables, but as a socio-economic variable in species distribution is very important. The high settlement success ofP. juliflorain the area compared with native plant species has led to extreme planting of species around villages with the aim of combating desertification and mitigation the negative impacts of dust storms on residential areas. This is the reason why the model fits around rural areas.
The advantage of modelling approach is that the probability of invasions can be evaluated before the actual entry of the species (Choudhury et al., 2016). Since, all statistical models have uncertainties, weak base models in an ensemble model can attenuate overall results (Stohlgren et al., 2010). Of course, we considered the most commonly used models for our dataset. Ensemble models may also be exclusively useful in modeling recently arrived invasive species because species may not yet have spread to all suitable habitats, leaving species-environment relationships difficult to specify. Srivastava et al. (2018), Gong et al. (2020) and Kaky et al. (2020) also acknowledged that ensemble models are more powerful to analyze invasion risk than single adaptive species-environment model. Ensemble model has two fundamental flaws in the context of predicting species distribution through time. The first is that this approach is mostly based on evaluations of prediction precision among modelling approaches rather than actual prediction accuracy. The latter is that, when actual model accuracy is evaluated, such studies have not used temporary autonomous data to validate their predictions, and thus conclusions of improved accuracy are based only on data partitioning approaches that may not ever exactly reflect model performance in new temporal domains (Crimmins et al., 2013). For this reason, the method was criticized for the inclusion of unnecessary parameters leading to omission errors, i.e., areas being classified as climatically unsuitable, while the invasive species was in fact able to get established there. We didn't calculate confidence intervals for predictions. So similar advance studies in the future would validate our choice of modelling.
As mentioned earlier, while some modelling algorithms predicted across the whole spectrum of proportional areas, others predicted only over a range of spectrum. Therefore, partial ROC analysis underscores the key role of omission error in evaluating iSDMs prediction performance.
The main purpose of this study was to predict the impact of climate change on the invasion ofP.julifloravia relating current species distribution to climate, and then to predict future distribution under climate change scenarios. Bakkenes et al. (2002) also stated that predicting species distribution in the future using SDMs based on current environmental variables can provide the best description of species distribution under climate change scenarios. In literature, it was found that climate changes have affected the spread of plant invasion causing expansions in climatically appropriate areas (Bellard et al., 2013), our study also showed that future climate change would cause increase in the suitable habitats for this species in Khuzestan Province, Iran. One of challenges in predicting future distributions of invasive plant species is that model accuracy is usually specified by current species distribution, but since exotic species are not at equilibrium with the environment, high current accuracy may not represent high future accuracy (Jones, 2012).Therefore, the results must be used cautiously. The implicit assumption in many studies seems to be that models that best predict the current distribution will also best predict the future distribution. Future prediction ofP. juliflorainvasion in the study area was conducted quantitatively, since its dispersal mechanisms are zoochory and hydrochory, i.e., its seeds are easily dispersed by wild and domestic animals, streams and runoffs. Climate change due to global warming provides favorable conditions to species that preferring warm and humid regions by offering them wider geographical extents in the future. As a result,P. julifloramore likely invade additional areas in the future because of climate change. In global circulation models, it seems that Khuzestan Province will likely to become warmer in the future (Fig. 6). Therefore, additional areas at higher altitude and latitude will be suitable for the invasive species (Fig. 7), which is consistent with the results of Parmesan (2006) and Harsch et al. (2017). AsP. juliflorais sensitive to long-term frostbite (Pasiecznik et al., 2001), this is probably the only environmental factor limiting its spread at higher latitudes.

Fig. 7 Habitat suitability shifts of P. juliflora in relation to altitude under current and future climate change scenarios. MPI, Max Planck Institute.
5 Conclusions
Our results reinforce the use of iSDM as an evaluation tool for the risk caused by invasive alien species in Khuzestan Province, Iran. Considering the differences between all algorithms, the results generally confirm the use of ensemble model over single SDM. The accuracy of ensemble predictions was similar to those of MaxEnt and RF models at all thresholds of partial ROC analysis and across threshold independent evaluation metrics. Also, suitable ranges forP. julifloraincrease under both pessimistic and optimistic climate change scenarios and in higher altitudes.But since climate warming is increasing in global, there is no guarantee that these areas will not be affected by species invasion in the future. In addition to climate change, migration of species to mountain areas is also facilitated by infrastructure development such as road, villages, tourism and increasing anthropogenic disturbances. However, our study was based on the concept of ecological amplitude, in other words, it was assumed that the species is in balance with current environmental conditions (climate) and factors such as migration, biological interactions, human factors and so on are not considered. Currently, invasive species management aims at control of invaders and mitigation of their impacts rather than eradication. OnceP. julifloraestablishes its dominance, controlling and restoration efforts can be extremely labor intensive and costly.Therefore, detecting the species in the early steps of infestation and mapping their distribution are essential steps in cost-effective resource management and stewardship. Our findings can be used as a baseline for future monitoring activities in ecological engineering and assist conservative policy and decision makers to ensure that their efforts are effective in eradication or preventing further spread of alien species. In other words, these findings can be used to direct management and prevention practices toward areas with the greatest risk of invasion. Assessing the impacts of climate change, the result may efficiently help for early detection, minimize their threat in the future and priority-setting monitoring programs in natural ecosystems. However, detailed development of the input data is needed, which include soil and hydrological predictors in the analyses. High resolution remote sensing products and other SDMs may also provide new insights on the distribution ofP. juliflorain the study area. Broad climatic tolerance and extensive geographic distribution as inherent characteristics of many invasive plants may influence their responses to climate change. However, adding other factors such as propagule pressure, dispersal capacity, biotic interactions of facilitation and competition can also strengthen the findings.
Appendix

Table S1 Multi-collinearity test among bioclimatic variables by using Pearson correlation coefficient
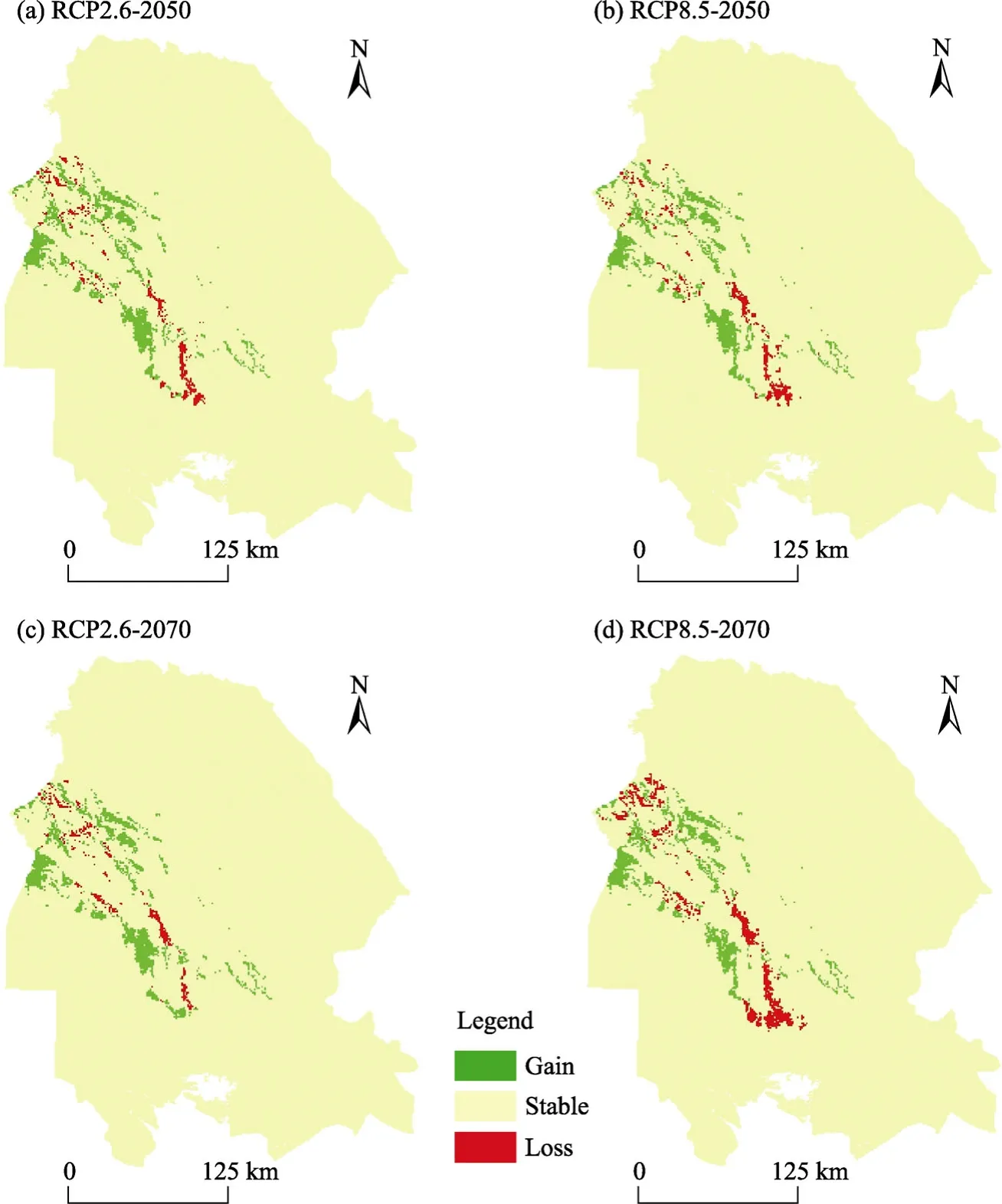
Fig. S1 Maps of spatial distribution of P. juliflora under optimistic and pessimistic scenarios for the years 2050 and 2070. (a), RCP2.6-2050; (b), RCP8.5-2050; (c), RCP2.6-2070; (d), RCP8.5-2070.
- Journal of Arid Land的其它文章
- Modeling and analyzing supply-demand relationships of water resources in Xinjiang from a perspective of ecosystem services
- Economic losses from reduced freshwater under future climate scenarios: An example from the Urumqi River,Tianshan Mountains
- Assessment of water resources in Yarmouk River Basin using geospatial technique during the period 1980–2020
- Assessment of river basin habitat quality and its relationship with disturbance factors: A case study of the Tarim River Basin in Northwest China
- Integrating multiple electromagnetic data to map spatiotemporal variability of soil salinity in Kairouan region, Central Tunisia
- Dieback intensity but not functional and taxonomic diversity indices predict forest productivity in different management conditions: Evidence from a semi-arid oak forest ecosystem

