Master Quorum Sensing Regulator HapR Acts as A Repressor of the Mannitol Phosphotransferase System Operon in Vibrio cholerae*
ZHANG Yi Quan ,MA Li Zhi ,GAO Yue ,QIN Qin ,LI Jie ,LOU Jing ,ZHANG Miao Miao,5,XUE Xing Fan,5,KAN Biao,#,and GAO He,#
The phosphoenolpyruvate (PEP):carbohydrate phosphotransferase system (PTS) is widely used by bacteria to take up sugars or sugar derivatives[1].PTS is usually composed of a histidine protein (HPr),enzyme I (EI),and several enzyme II (EII) proteins,such as EIIA,EIIB,EIIC,and EIID[1].PEP,HPr,and one of several EIIA and EIIB pairs form a phosphorylation cascade where PEP acts as a phosphoryl donor[1].PEP transfers a phosphoryl group to HPr,which then phosphorylates EIIA. The phosphorylated EIIA transfer the phosphoryl group to EIIB. The phosphorylated EIIB finally phosphorylates the membrane-spanning protein EIIC or EIID,which then facilitates the transport of sugars into bacterial cells.
Vibrio cholerae,the causative agent of cholera,can utilize mannitol with the aid of PEP:PTS encoded by anmtlADRoperon[2].mtlAencodes the mannitolspecific EIICBA component (EIIMtl),mtlDencodes mannitol-1-phosphate-5-dehydrogenase,andmtlRencodes anmtlADRrepressor[2].MtlA is required forV.choleraebiofilm formation in the absence of mannitol[3].In addition,mtlADRtranscription is repressed by sRNA MtlS and the ferric uptake regulator (Fur),but it is activated by the cAMP receptor protein (CRP)[4,5].However,as a mannitol transport system,MtlADR expression should be strictly controlled by complex regulatory networks.
Quorum sensing (QS) is a cell-to-cell communication process widely used by bacteria to regulate gene expression in response to changes in bacterial cell density[6]. QS usually uses its downstream master regulators to regulate gene expression[6].LuxR orthologs,which are master QS regulators (MQSR) inVibriosp.,regulate multiple behaviors,including virulence factors production,biofilm formation,and motility[6].A 20 bp invertrepeat sequence TATTGATAAA-TTTATCAATA representing the DNA binding box of MQSR was constructed in a previous study[7].HapR is an LuxR ortholog inV.cholerae.An MQSR box-like sequence‘TATTGACAAAATAAAAAATA’ was detected within the regulatory DNA region ofmtlADR,suggesting thatmtlADRtranscription is probably under the direct control of HapR inV.cholerae[7].Therefore,theV.choleraestrain C7258 (wild type,WT),its nonpolarhapRmutant (designated ΔhapR)[8],and the complementary mutant (ΔhapR/pBAD24-hapR,designated C-ΔhapR)[8]were employed to investigate the regulatory actions of HapR onmtlADRtranscription.
Fresh colonies ofV.choleraestrains on LB agar[1% tryptone (Oxoid),0.5% yeast extract (Oxoid),1%NaCl (Merck Millipore),and 1.5% bacto agar (BD biosciences)] were inoculated into a 5-mL LB broth at 37 °C with shaking to grow overnight.The resultant cultures were diluted 50-fold into a 5-mL LB broth;these were statically grown at 37 °C to an OD600value of about 0.3.Then,the resultant cultures were diluted 30-fold into 5 mL of mannitol fermentation medium (0.5% NaCl,0.1% tryptone,and 0.2%mannitol;pH 8.0) containing 0.025% phenol red and 100 μg/mL ampicillin.This was statically cultured at 37 °C to investigate their abilities to ferment mannitol by observing the color changes of the medium[9].As shown in Figure 1,the color of the medium for ΔhapR/pBAD24 (ΔhapRharboring the empty pBAD24 plasmid) culture completely became yellow after 10 h,whereas those for WT/pBAD24 and C-ΔhapRculture manifested no significant color changes after 0 to 12 h relative to the control.The pH values of each culture medium were determined during the mannitol fermentation test[9].Results showed that the pH values of the mannitol fermentation medium of ΔhapR/pBAD24 dropped gradually over time,whereas those of WT/pBAD24 and C-ΔhapRhardly changed over time (Figure 2).These results indicated that ΔhapR/pBAD24 produced more organic acids than the other two strains.
To determine whether the phenotype ofhapRmutant that produced more organic acids was related to its growth,the growth curves of WT/pBAD24,ΔhapR/pBAD24,and C-ΔhapRin a mannitol fermentation medium were measured.Results showed that ΔhapR/pBAD24 had a lower growth rate than WT/pBAD24,whereas C-ΔhapRexhibited a restored growth rate (Supplementary Figure S1 available in www.besjournal.com).Thus,the higher level of organic acids produced by thehapRmutant was directly unrelated to its growth.

Figure 1.Mannitol fermentation phenotypes.V.cholerae strains were cultured at 37 °C in a mannitol fermentation medium containing phenol red as an indicator.The color of the medium shows purple-red if the pH value is >7.0,and it turns yellow if the pH value is< 6.5[9].A blank medium was used as a negative control.The pictures illustrated here represent three independent experiments with three replicates each.
ThemtlADRoperon has been shown to be required for mannitol utilization inV.cholerae[2].To detect whether HapR had regulatory actions onmtlADRexpression,the 558 bp upstream DNA region ofmtlADRwas cloned into the pBBRlux vector harboring a promoterlessluxCDABEreporter gene.The primers used in this study are listed in Supplementary Table S1 available in www.besjournal.com.Then,the recombinant plasmid was transferred into WT and ΔhapRto determine the luminescence activities in each strain.As shown in Figure 3A,the promoter activities ofmtlAin both WT and ΔhapRconsiderably increased with the increase in OD600from 0.05 to 0.25.Then,these significantly declined with a further increase in cell density.In addition,the promoter activities ofmtlAsignificantly decreased in WT relative to those in ΔhapRat any cell density analyzed,suggesting that HapR acted as a negative regulator ofmtlADRexpression.
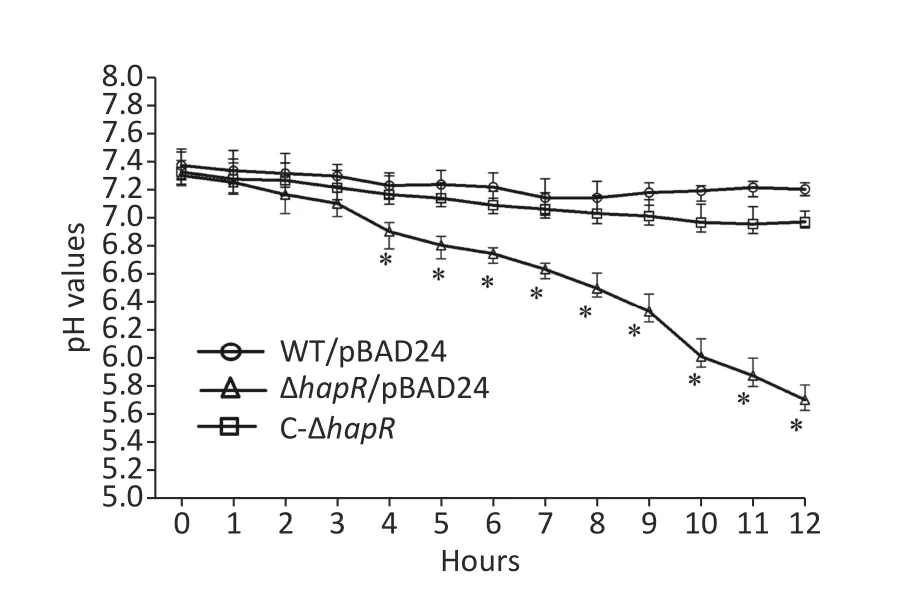
Figure 2.pH-time curves of WT/pBAD24,ΔhapR/pBAD24,and C-ΔhapR in mannitol fermentation media.The pH values of each culture medium were measured during the mannitol fermentation test with a 1-h interval.The assay was performed in at least three independent bacterial cultures,with values expressed as the mean ± standard deviation.Paired Student’s t-test was used to calculate the statistical significance,with P < 0.01 considered as significant.The asterisks indicate the statistical significance between ΔhapR/pBAD24 and WT/pBAD24.
To determine whether HapR directly regulated themtlADRtranscription,a DNase I footprinting assay was performed to detect the binding activity of His-HapR on the promoter-proximal DNA fragment ofmtlADRin vitro[8,9].Results showed that His-HapR protected a single region within the DNA fragment,which was located from 244 to 206 bp upstream of the start codon ofmtlADR(Figure 3B).Notably,the HapR site was located far upstream of the transcription start site and -35 element,and it partly overlaps with the CRP site 2 and almost completely overlaps with the Fur site (Supplementary Figure S2 available in www.besjournal.com).It is unclear whether the binding of HapR to themtlADRpromoter can interfere with the binding of CRP and Fur.However,it has been shown that CRR was required for HapR expression[10],whereas HapR acted as afurrepressor[8].Thus,mtlADRexpression should be under the strict regulation of the regulatory circuit composed of CRP,Fur,and HapR,which may lead to the expression ofmtlADRalways at a reasonable level.
In summary,our data showed that the capacity for mannitol fermentation was significantly enhanced in ΔhapRrelative to that in WT and its complementary mutant.The level of organic acids produced by ΔhapRwas also enhanced in comparison to that produced by WT or its complementary mutant.The data also showed that HapR can bind to the promoter-proximal DNA region ofmtlADRto repress its transcription.Thus,the HapR-dependent regulation of mannitol utilization inV. choleraewas through direct regulation ofmatlADRtranscription.The data presented here provided a deeper understanding of the regulatory network ofmtlADRinV.cholerae.
#Correspondence should be addressed to KAN Biao,Email:kanbiao@icdc.cn;GAO He,E-mail:gaohe@icdc.cn
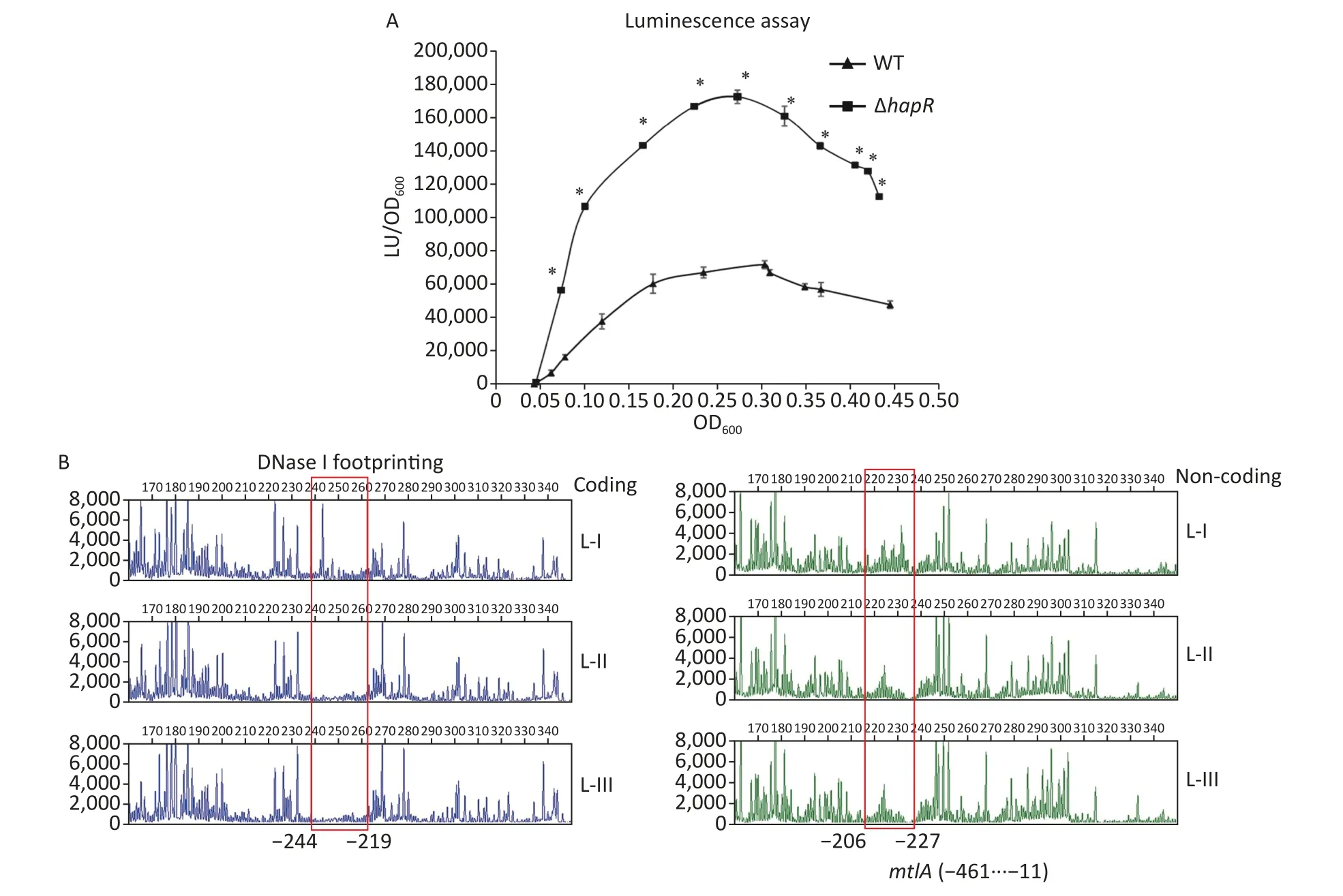
Figure 3.Negative regulation of mtlA by HapR.(A) luminescence assay.The promoter DNA region of mtlA was cloned into a pBBRlux vector.Then,it was transferred into V.cholerae strains to determine the luminescence activity of each strain using Infinite® 200 Pro NanoQuant.The lux activity was calculated as light units/OD600 (LU/OD600).The assay was performed in at least three independent bacterial cultures,with values expressed as the mean ± standard deviation.Paired Student’s t-test was used to calculate the statistical significance,with P < 0.01 considered as significant.The asterisks indicate the statistical significance between ΔhapR and WT.(B) DNase I footprinting assay.Fluorescently labeled DNA probes were incubated with increasing quantities of His-HapR (Lanes I,II,and III contain 0,2.31,and 6.92 pmol,respectively) and were digested with DNase I.The results were analyzed with an ABI 3500XL DNA analyzer.Footprint regions are boxed and marked with positions.The negative numbers represent the nucleotide positions upstream of the first nucleotide of the translation start site of mtlA.WT,wild type.
Biographical note of the first author:ZHANG Yi Quan,male,born in 1985,PhD,Associate Professor,majoring in gene regulation of pathogenic bacteria.
Supplementary Table S1. Oligonucleotide primers used in this study
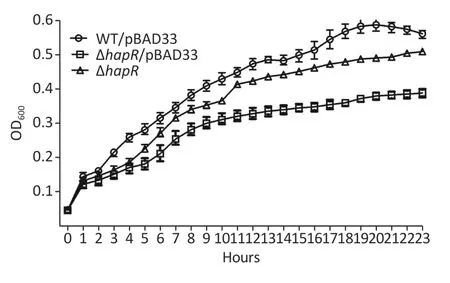
Supplementary Figure S1. Growth curves of the Vibrio cholerae strains in mannitol fermentation media. WT/pBAD24,ΔhapR/pBAD24,and C-ΔhapR were statically cultured in mannitol fermentation media at 37°C,and the OD600 values for each strain were monitored with a 1-h interval.The experiments were performed at least twice,with three biological replicates for each strain per time.

Supplementary Figure S2. Structural organization of the mtlADR promoter.The transcription and translation starts are marked with bent arrows.The Shine-Dalgarno (SD) box and -10/-35 elements are enclosed in boxes.The MQSR box-like sequence is enclosed in a yellow shadowed box.The Fur,CRP,and HapR binding sites (references in[5,9] and the current study) are underlined with solid,dotted,and break point lines,respectively.
Received:July 17,2021;
Accepted:October 19,2021
 Biomedical and Environmental Sciences2022年1期
Biomedical and Environmental Sciences2022年1期
- Biomedical and Environmental Sciences的其它文章
- Physical Activity Guidelines for Chinese (2021)
- Association of Residential Greenness with the Prevalence of Metabolic Syndrome in a Rural Chinese Population:the Henan Rural Cohort Study*
- SlRT1 Mediates Sestrin1-lnduced lmprovement in Hepatic lnsulin Resistance*
- An Analysis on Failure Reasons of Quit Smoking among Smokers in Beijing Communities
——Based on a Cross-Sectional Study* - Factors Associated with Diagnostic Delay of Pulmonary Tuberculosis in China*
- Application of Oral Fluid in Measles lgG Antibody Detection for Medical Staff *
