Effect of UBR5 on the tumor microenvironment and its related mechanisms in cancer*
Guangyu Wang,Sutong Yin,Justice Afrifa,Guihong Rong,Shaofeng Jiang,Haonan Guo (✉),Xianliang Hou (✉)
1 Central Laboratory,Guangxi Health Commission Key Laboratory of Glucose and Lipid Metabolism Disorders,The Second Affiliated Hospital of Guilin Medical University,Guilin 541199,China
2 Reproductive Medical Center,No.924 Hospital of Chinese People's Liberation Army,Guilin 541001,China
3 Department of Medical Laboratory Science,School of Allied Health Sciences,University of Cape Coast,Cape Coast,Ghana
4 Department of Clinical Laboratory,The Affiliated Hospital of Guilin Medical University,Guilin 541001,China
5 Guangxi Key Laboratory of Tumor Immunology and Microenvironmental Regulation,Guilin Medical University,Guilin 541199,China
Abstract Objective UBR5,recently identified as a potential target for cancer therapeutics,is overexpressed in multiple malignant tumors.In addition,it is closely associated with the growth,prognosis,metastasis,and treatment response of multiple types of cancer.Although emerging evidence supports the relationship between UBR5 and cancer,there are limited cancer analyses available.Methods In this study,online databases (TIMER2,GEPIA2,UALCAN,c-BioPortal,STRING) were employed to comprehensively explore expression levels and prognostic values of the UBR5 gene in cancer,using bioinformatic methods.Results We found that various characteristics of the UBR5 gene such as gene expression,survival value,genetic mutation,protein phosphorylation,immune infiltration,and pathway activities in the normal tissue were remarkably different from those in the primary tumor.Furthermore,“protein processing in spliceosome”and“ubiquitin mediated proteolysis”have provided evidence for their potential involvement in the development of cancer.Conclusion Our findings may provide insights for the selection of novel immunotherapeutic targets and prognostic biomarkers for cancer.
Key words:UBR5;cancer;tumor;prognosis;biomarker
Despite its declining incidence in many developed countries,cancer remains the most common cause of death across the globe.More than nineteen million people were diagnosed and over nine million people died as a result of cancer in 2020 alone.Cancer is characterized by a high degree of malignancy,rapid development and poor prognosis[1-3].In the wake of the rapid strides being made by scientists and clinicians to explore novel prognostics,diagnostics and therapeutic options,cancer still remains one of the most elusive diseases in terms of treatment and management.
The humanUBR5gene,which is widely expressed in various cell types,has 59 exons encoding approximately 10 kb of mRNA and > 300 kDa of protein[4].It is highly conserved in metazoans,has unique structural features,and has been implicated in the regulation of the DNA damage response,metabolism,transcription,and apoptosis[5-7].UBR5is a key regulator of cell signaling related to the field of tumor biology.Recent studies have primarily demonstrated thatUBR5plays an important role in the development of many tumors,and its expression may be closely associated with growth and proliferation of malignant tumors[8-9].For example,in breast cancer,UBR5is coamplified with Myelocytomatosis (MYC) to limit MYC-dependent apoptosis by encoding a ubiquitin ligase[10].Also in breast cancer,others have shown that triple-negative breast cancer (TNBC) metastasis and cisplatin resistance may be mediated by elevatedUBR5expression[11-13].Similarly,in colorectal cancer,Xieet al.concluded that an elevatedUBR5levels play an oncogenic role and may be a potential prognostic marker[14].The trends elucidated in the various studies point to the likelihood ofUBR5as an oncogenic mediator in most cancers.However,contrary to other cancers,inactivating mutations have been observed in theUBR5gene,as is the case in approximately 18% of mantle cell lymphoma cases[15].It is therefore clear thatUBR5is a key cell signaling regulator that has been strongly associated with cancer;however,its function as a promoter or inhibitor of tumorigenesis still remains inconclusive.
In this study,we conducted an in-depth and comprehensive bioinformatic analysis of the expression of theUBR5gene and evaluated its potential as a therapeutic target and prognostic biomarker.Findings from this study will provide a better understanding of this gene,and help clinicians select appropriate therapeutic drugs and more accurately prognose long-term outcomes in cancer patients.
Materials and methods
TIMER2
TIMER2 (http://timer.cistrome.org/) is a reliable tool that provides the expression status ofUBR5across The Cancer Genome Atlas (TCGA) datasets from different tumor tissues and adjacent normal tissues.It also provides a robust estimation of immune infiltration levels for TCGA or user-provided tumor profiles using six stateof-the-art algorithms.In this study,the expression status ofUBR5across the TCGA dataset,and the correlation between the infiltration of immune cells andUBR5expression was evaluated[16-18].
GEPIA2
GEPIA2 (http://gepia2.cancer-pku.cn/#index) is a tool for analyzing the RNA sequencing expression data of 9,736 tumors and 8,587 normal samples from TCGA[19].GEPIA2 was employed in this study to perform a differentialUBR5expression analysis of tumor and adjacent normal tissue,expression ofUBR5total protein,pathological stage analysis,and correlative prognostic analysis of theUBR5gene.
UALCAN
UALCAN (http://ualcan.path.uab.edu/index.html) is an interactive web resource that provides analysis based on TCGA and MET500 cohort data[20].It allows analysis of relative expression of query genes across tumor and normal samples,as well as in various tumor sub-groups based on individual cancer stages,tumor grade or other clinicopathological features.Protein expression analysis was conducted using Clinical Proteomic Tumor Analysis Consortium (CPTAC) and the available datasets of six tumors were selected in our study.
cBioPortal
The cBioPortal (http://cbioportal.org) is a web resource for exploring,visualizing,and analyzing multidimensional cancer genomics data.This web resource provides the option of querying a single cancer study or querying across multiple cancer studies.It is also possible to view relevant genomic alterations in cancer samples and analyze multidimensional cancer genomics data[21].The alteration frequency,type of alterations ofUBR5and copy number alterations are shown in our study.In addition,we aimed to assess the genetic alterations ofUBR5and its correlation with survival values in cancer patients,using data from TCGA.
STRING
STRING (https://string-db.org/) is a web resource that integrates all known and predicted associations between proteins[22].We conducted a protein-protein interaction network analysis of differentially expressed levels of theUBR5gene,to explore the interactions among them with STRING.
Results
Aberrant expression of UBR5 in patients with cancer
To understand the oncogenic role of humanUBR5,we examined its expression status across the TCGA dataset from different cancer types using the TIMER2 approach.The expression level ofUBR5in the tumor tissues of breast invasive carcinoma (BRCA),cholangiocarcinoma (CHOL),colon adenocarcinoma (COAD),esophageal carcinoma (ESCA),glioblastoma multiforme (GBM),head and neck squamous cell carcinoma (HNSC),liver hepatocellular carcinoma (LIHC),lung adenocarcinoma (LUAD),lung squamous cell carcinoma (LUSC),pheochromocytoma and paraganglioma (PCPG),prostate adenocarcinoma (PRAD),rectum adenocarcinoma (READ),and stomach adenocarcinoma (STAD) was higher than in normal tissues.On the contrary,the expression level of kidney chromophobe (KICH),kidney renal clear cell carcinoma (KIRC),thyroid carcinoma (THCA),and uterine corpus endometrial carcinoma (UCEC) was lower than in normal tissues (Fig.1a).
Based on clinical data extracted from the GTEx dataset,we compared the differential expression level ofUBR5in tumor tissues with that in matched normal tissues of CHOL,lymphoid neoplasm diffuse large B-cell lymphoma (DLBC),pancreatic adenocarcinoma (PAAD),and thymoma (THYM).The results showed a significantly elevatedUBR5expression among the tumor tissues (P< 0.05) compared to that in normal tissues (Fig.1b).
In the CPTAC dataset,we observed significantly higher expression ofUBR5total protein in the primary tissues of breast cancer (P< 0.001),clear cell RCC (P< 0.001),colon cancer (P< 0.001),LUAD (P< 0.001),UCEC (P< 0.001) and ovarian cancer (P< 0.05),than in normal tissues (Fig.1c).
To assess the association betweenUBR5expression and the pathological stages of cancer,the“Pathological Stage Plot”module of GEPIA2 was employed to analyze pathological data from COAD (P< 0.001),ESCA (P< 0.05) and KICH (P< 0.001) patients in the TCGA database (Fig.1d).
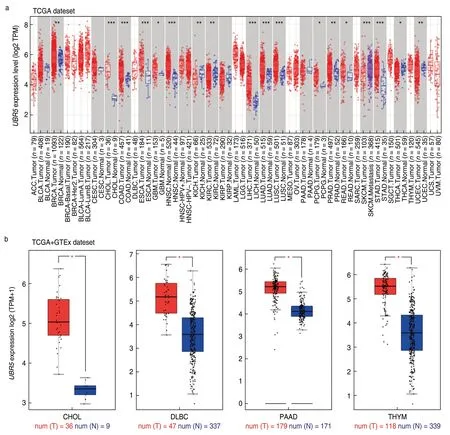
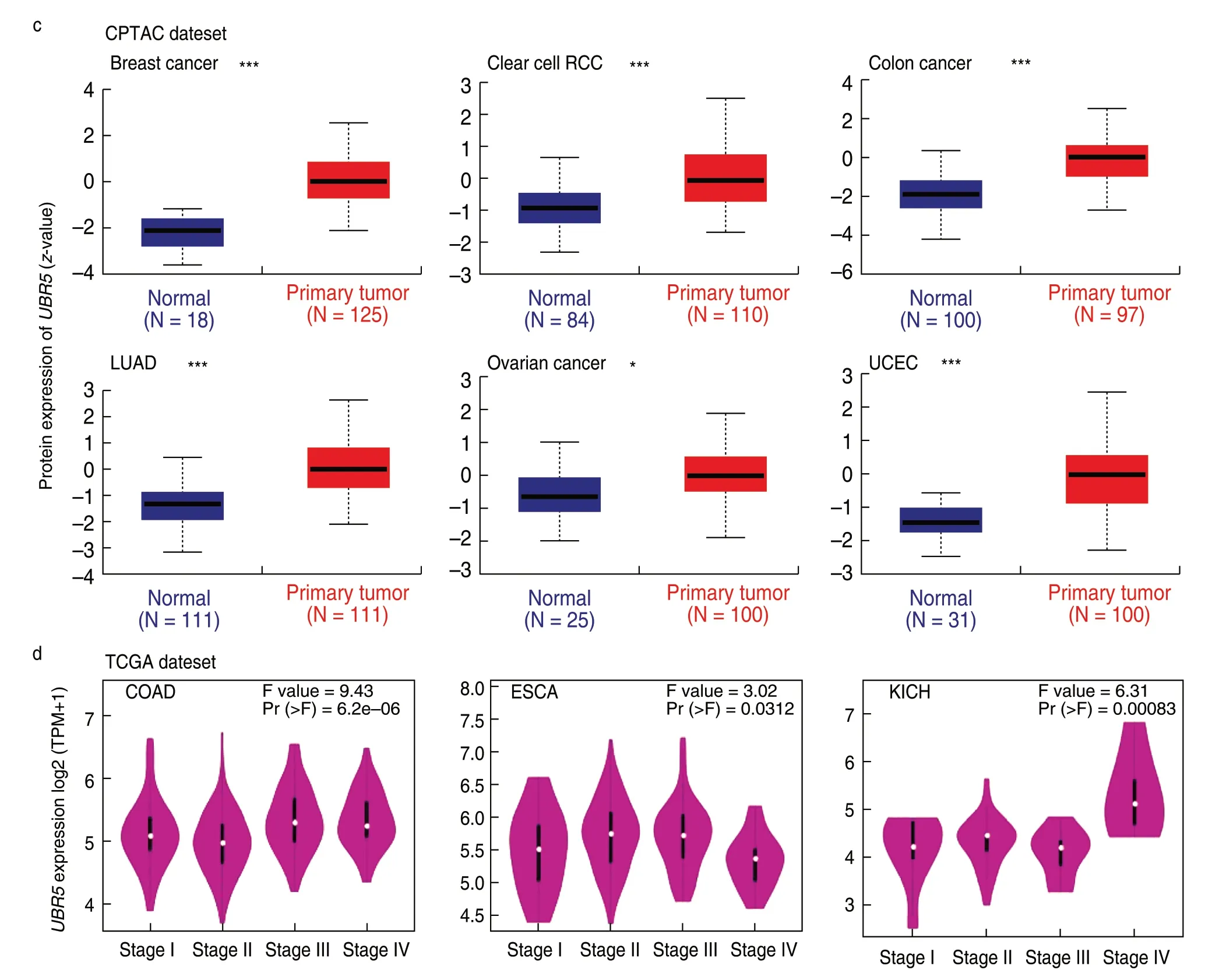
Fig.1 Expression levels of the UBR5 gene in different cancer samples.(a) UBR5 expression status varies in different cancers through TIMER2,*P < 0.05,**P < 0.01,***P < 0.001;(b) Differential expression of UBR5 between the normal tissues and the tumor tissues through GEPIA2,*P < 0.05;(c) Higher expression of UBR5 total protein in the primary tissues through UALCAN,all P < 0.05;(d) Expression levels of UBR5 in different pathological stages through GEPIA2,P < 0.05.
Prognostic value of UBR5 in patients with cancer
Patients were grouped into high-expression and lowexpression groups.We examined the association betweenUBR5expression and the prognosis of patients with cancer using TCGA and GEO datasets.UBR5expression was linked to cancer prognosis:the Fig.2 plot showing overall survival (OS) for BRCA (P=0.041) within the TCGA project indicates that higherUBR5expression is linked to a poor prognosis.Disease-free survival (DFS) for PRAD (P=0.013) and READ (P=0.037) values also supported this conclusion.
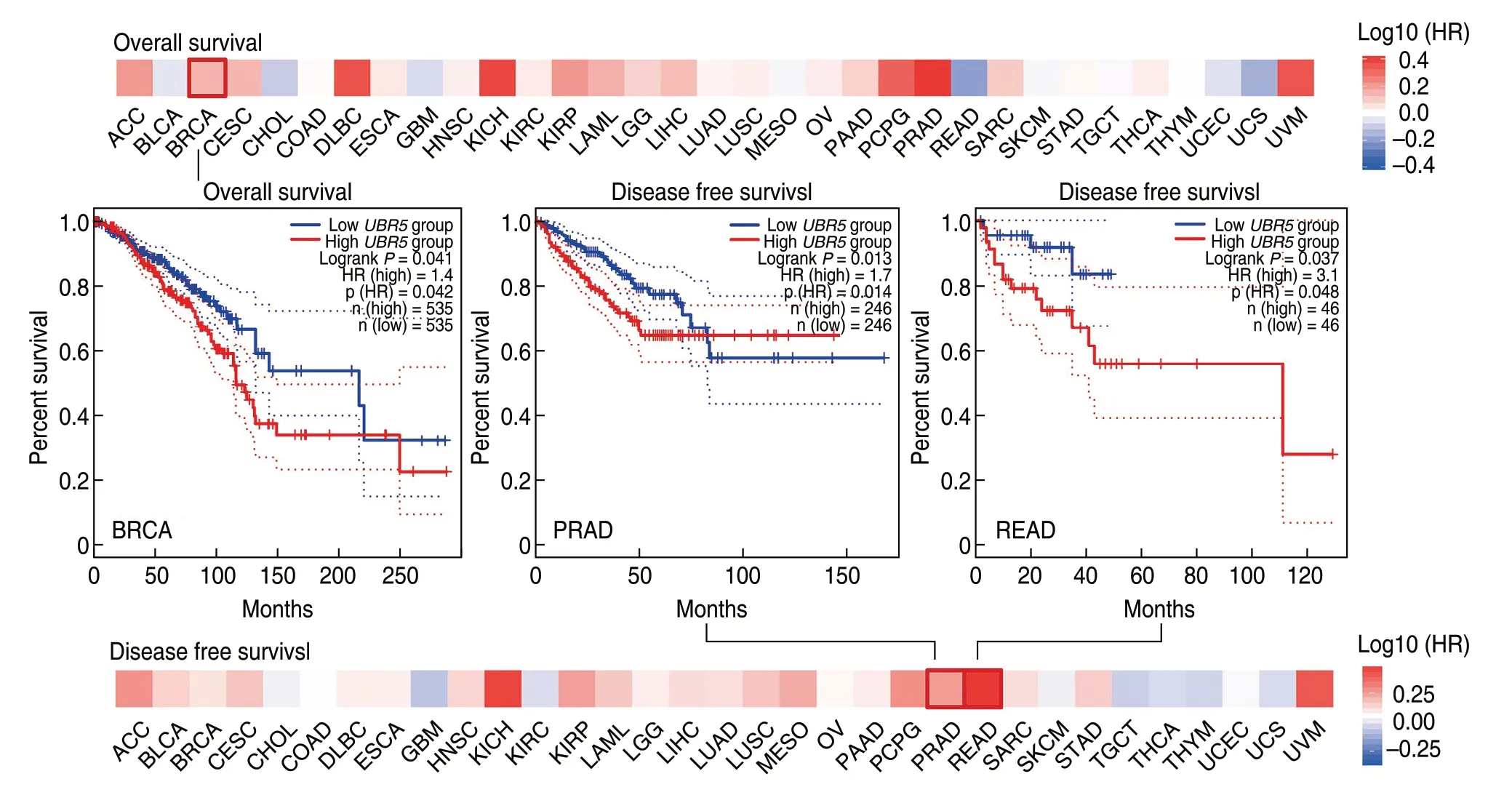
Fig.2 Relationship between UBR5 gene expression level and survival in cancer patients using GEPIA2.Clinical survival curves of BRCA (OS),PRAD (DFS),and READ (DFS) are presented,P < 0.05.
Genomic alterations of UBR5 in cancer
The cBioPortal was used to determine the genetic alteration status ofUBR5in cancer,based on TCGA datasets.As shown in Fig.3a,the highest alteration frequency was found in the bladder urothelial carcinoma tumor with“Amplification”as the primary type,whereas PCPG exhibited the lowest alteration among all of the cancer samples queried.It is noted that all uveal melanoma cases with genetic alterations showed copy number amplifications ofUBR5.As shown in Fig.3b,519 mutations were identified in patients.Out of the alterations,377 missense mutations,126 truncating mutations,4 in-frame mutations and 12 fusion mutations were detected.Missense mutations ofUBR5were identified as the main type of genetic alteration,and E2121Kfs*28/E2121Rfs*13/K2120Rfs*13 alteration was predicted to induce a frame shift mutation of theUBR5gene.The 3D structure of theUBR5protein can be observed in Fig.3c.In addition,genetic alterations have been found in patients with different types of cancer,which is related to survival prognoses.As shown in Fig.3d,STAD cases with alteredUBR5showed better prognosis in progression-free (P=0.0492),but not overall (P=0.920),disease-specific (P=0.392) and disease-free (P=0.362) survival,compared with cases withoutUBR5alteration.UCEC cases with alteredUBR5showed better prognosis in disease-specific (P=0.0492),but not overall (P=0.0643),disease-free (P=0.761) and progression-free survival (P=0.148),compared with cases withoutUBR5alteration.
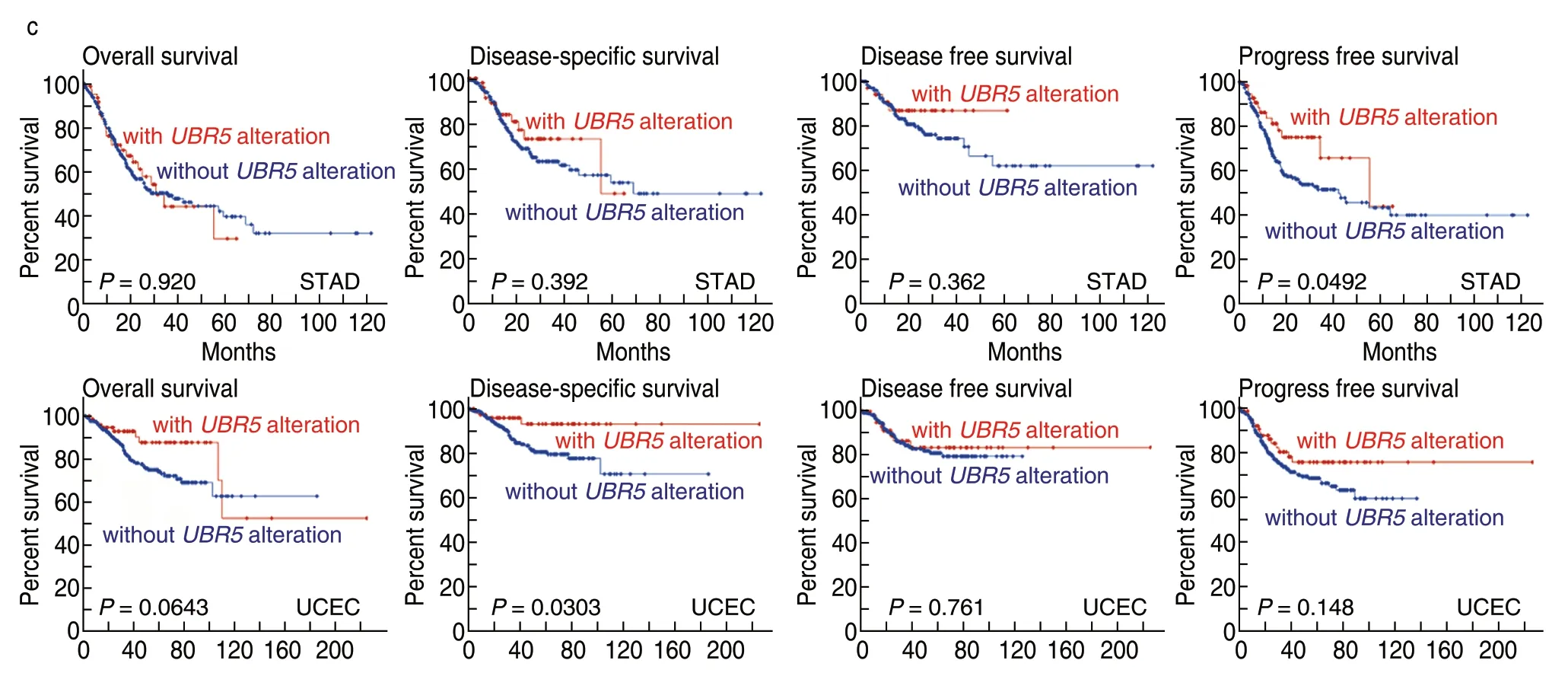
Fig.3 Genetic features of mutations of UBR5 in different tumors (cBioPortal).(a) Alteration frequency in different tumor samples;(b) Sites and case number of UBR5 genetic alterations;(c) 3D structure of the UBR5 protein;(d) Clinical survival curve of STAD and UCEC.
Protein phosphorylation of UBR5 in patients with cancer
We also investigatedUBR5phosphorylation levels using the CPTAC dataset.Clear cell clear cell renal cell carcinoma (RCC),ovarian cancer,LUAD,UCEC and breast cancer were analyzed.The analysis ofUBR5phosphoprotein expression level is presented in Fig.4a.The clinical data showed a higher phosphorylation level of the S327 locus in all primary tumor tissues compared with that seen in normal tissues (Fig.4b-f,allP< 0.05),followed by a lower phosphorylation level of the S636 locus for colon cancer (Fig.4f,P=1.2e-06),LUAD (Fig.4d,P=1.8e-05),colon cancer (Fig.4f,P=1.3e-14) and the S1549 locus for ovarian cancer (Fig.4c,P=5.2e-03),
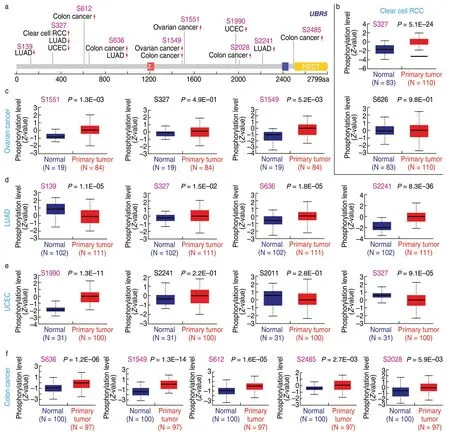
Fig.4 Phosphorylation analysis of different tumors.(a) Analysis of UBR5 phosphoprotein expression level based on the CPTAC dataset,S139,S327,S612,S626,S1549,S1551,S1990,S2028,S2241,and S2485;The box plots for different cancers,including clear cell RCC (b),ovarian cancer (c),LUAD (d),UCEC (e) and breast cancer (f),all P < 0.05.
Immune cell infiltration of UBR5 in patients with cancer
Next,we used the TIMER2,TIDE,XCELL,MCPCOUNTER and EPIC algorithms to assess the correlations ofUBR5expression with immune infiltration levels.Heat map of different expressedUBR5gene are further presented in Fig.5a.We found a significant negative correlation betweenUBR5expression and the estimated infiltration value of cancer-associated fibroblasts for Testicular Germ Cell Tumor (TGCT).(Fig.5b,cor=-0.242,P=3,11e-03)

Fig.5 Correlation analysis of UBR5 in the tumor microenvironment and immune infiltration.(a) Correlation heat map of differentially expressed UBR5 gene (TIMER2);(b) The correlation between differentially expressed UBR5 gene and immune cell infiltration (TIMER2).
Enrichment analysis of UBR5 -related partners
In an attempt to investigate the potential enrichment of particular molecular mechanisms in tumorigenesis,we attempted to screen out targetingUBR5-binding proteins andUBR5expression-related genes using STRING and GEPIA2.Fig.6a showed the interaction network of 50UBR5-binding proteins supported by experimental evidence.There were significant positive correlations between the expression level ofUBR5and that of cell division cycle and apoptosis regulator 1 (CCAR1) (R=0.56),Arginine/serine-rich coiled-coil 2 (RSRC2) (R=0.52),Suppressor of mek1 (SMEK1) (R=0.52),Ubiquitin specific peptidase 7 (USP7) (R=0.52) and Zinc finger protein 7 (ZNF7) (R=0.68) genes (allP< 0.001;Fig.6b).As shown in Fig.6c,the heatmap also revealed that the above-mentioned genes were positively correlated withUBR5in the majority of types of tumor.An intersection analysis of 50UBR5-binding proteins and 100UBR5expression-related genes showed one common member,namely,SRSF1 (Fig.6d).In addition,the KEGG data suggested that“spliceosome”and“ubiquitin mediated proteolysis”pathways were involved in cancer progression.(Fig.6e).
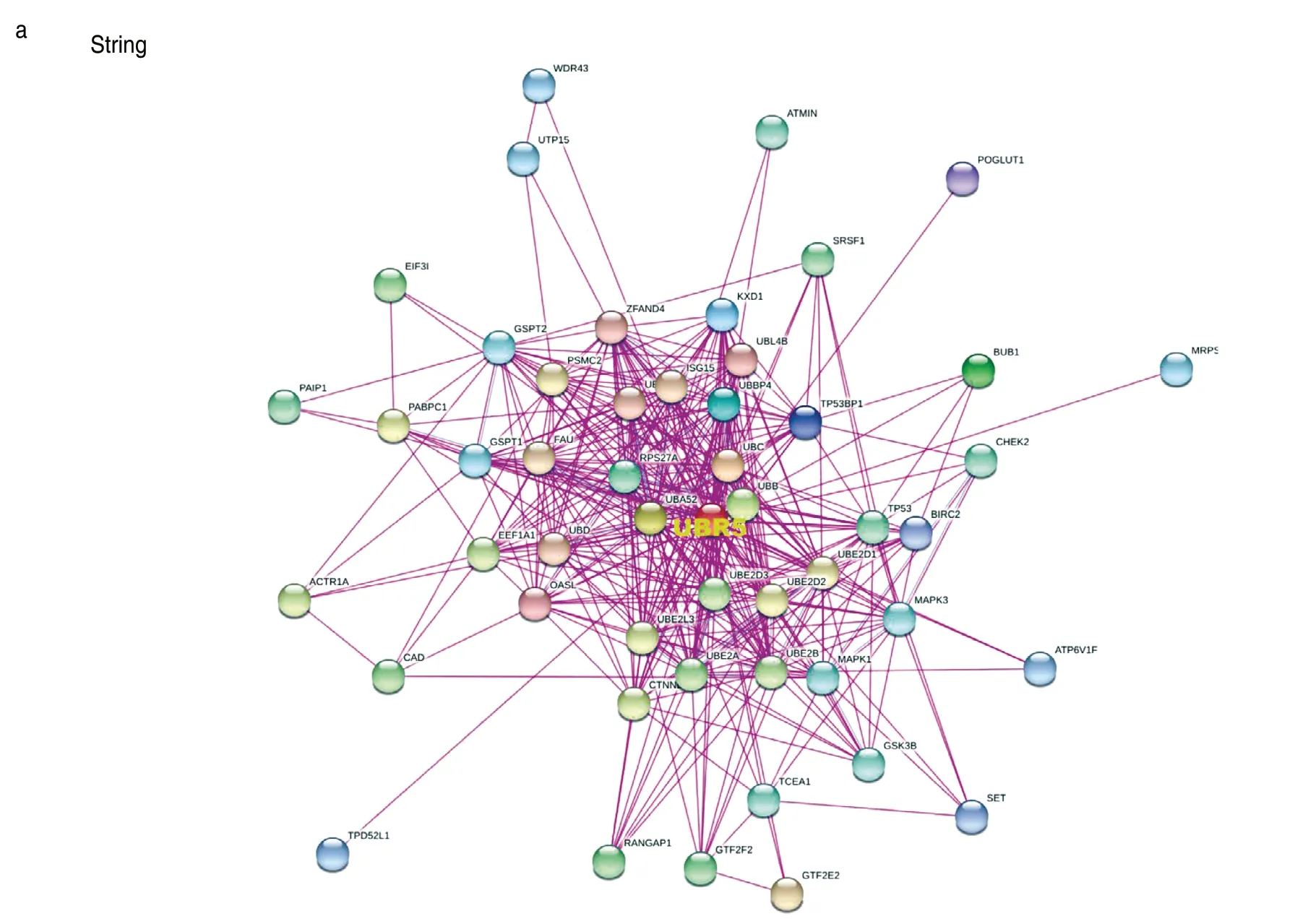
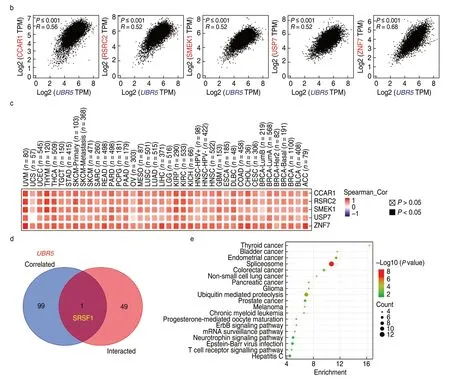
Fig.6 UBR5-related gene enrichment analysis.(a) Interaction network of 50 UBR5-binding proteins through STRING tool;(b) UBR5 expression level was positively correlated with that of CCAR1,RSRC2,SMEK1,USP7 and ZNF7 genes;(c) Correlation heat map of the differentially expressed UBR5 gene;(d) One common member named SRSF1 was observed through intersection analysis;(e) UBR5 expression-related genes for enrichment analysis.
Discussion
It is understood thatUBR5is a tumor-related gene that affects the biological behavior of tumors in many aspects,such as cell cycle regulation,apoptosis regulation,tumor suppressor gene regulation,invasion and metastasis regulation[23].Some studies have reported a correlation between theUBR5gene,tumor microenvironment,and cancer immunotherapy,suggesting that the gene may modulate tumor progression and provide an immunotherapeutic effect[24-25].However,the prognostic value and the biological function of theUBR5gene in cancer has not been well-characterized.With further investigation into this gene,knowledge regarding its regulatory mechanism in cancer will become increasingly clear,which will aid in the molecular diagnosis and targeted therapy of cancer,as well as improving prognostic assessments in cancer patients.Thus,we present a comprehensive overview of theUBR5gene based on data from TCGA,CPTAC and GEO databases.
We first explored expression of theUBR5gene and its correlation with the pathological cancer stage.We found that 17 genes were differentially expressed in cancerous tissues compared with the corresponding control tissues (higher expression of BRCA,CHOL,COAD,ESCA,GBM,HNSC,LIHC,LUAD,LUSC,PCPG,PRAD,READ and STAD;lower expression of KICH,KIRC,THCA and UCEC).UBR5gene expression in cancerous tissues was further confirmed in studies from GTEx and CPTAC datasets.These data demonstrate that differentially expression ofUBR5may play a significant role in cancer.
Furthermore,in BRCA,PRAD and READ patients,high expression ofUBR5were significantly associated with a poor prognosis.Zhanget al.found thatUBR5was overexpressed in gallbladder cancer tumor tissues and was significantly associated with tumor size,histological and tumor differentiation[26].Yanget al.revealed that high expression ofUBR5was associated with poor overall and disease-free survival in patients with gastric cancer[27].This analysis demonstrates thatUBR5may be an important biomarker for predicting the prognosis of patients with cancer.
Since theUBR5gene was significantly differentially expressed in cancer tissues,we explored its molecular characteristics.There were frequent genetic alterations in theUBR5gene expressed in cancer tissues,with mutation and amplification being the most common.It has been reported that theUBR5gene is localized to chromosome 8q22[28].Mutation and amplification occur frequently in this region in many types of cancer,including breast cancer,esophageal cancer and mammary ductal carcinoma[29-31].Tumorigenesis and the progression of cancer are complex and multi-faceted,and genetic alteration plays an important role in these processes.We found a low to high correlation of prognoses with the differential expression of theUBR5gene,suggesting thatUBR5plays a synergistic role in tumorigenesis and the progression of cancer.
We then focused on protein phosphorylation ofUBR5in patients with cancer using UALCAN.Phosphorylation is a formidable regulator of many proteins involved in essential intracellular processes.Studies have reported on the possible role of phosphorylation in both protein function and the progression of specific cancers[32].Phosphorylation may provide key information about the derangements and serve as major targets for therapeutics,which is a rapidly growing area of cancer research[33].The results showed that S327,S636 and S1549 all exhibited a higher phosphorylation level ofUBR5.Bethardet al.revealed thatUBR5has 477 potential phosphorylation sites.However,few studies have specifically targeted the identification of these phosphorylation sites[34].Further laboratory studies to evaluate the potential role ofUBR5phosphorylation in tumorigenesis are needed.
We also found a negative correlation betweenUBR5expression and immune infiltration of cancer-associated fibroblasts.Evidence indicates that cancer development is a complex process that involves interactions between tumor cells,stromal fibroblasts,and immune cells.Tumor-infiltrating immune cells play a role in the promotion or inhibition of tumor growth[35-37].This analysis demonstrates the role ofUBR5in the tumor microenvironment and the promotion or inhibition in different types of cancer.
Additionally,analysis of“protein processing in spliceosome”and“ubiquitin mediated proteolysis”pathways may bring novel insights into the potential association ofUBR5with etiology or pathogenesis of cancers[38].
In conclusion,we hope these results will be a helpful guide to aid in helping diagnose cancer and to assist in the design of new immunotherapeutic drugs.
Conflicts of interest
The authors indicated no potential conflicts of interest.
Ethics approval and consent to participate
Ethics approval is not applicable because this study did not involve human or animal testing.
 Oncology and Translational Medicine2021年6期
Oncology and Translational Medicine2021年6期
- Oncology and Translational Medicine的其它文章
- Autophagy-related lncRNA and its related mechanism in colon adenocarcinoma
- GFPT2 pan-cancer analysis and its prognostic and tumor microenvironment associations*
- Malnutrition as a predictor of prolonged length of hospital stay in patients with gynecologic malignancy:A comparative analysis*
- Effect of radiotherapy on tumor markers and serum immune-associated cells in patients with esophageal cancer*
- Correlation analysis of breast fibroadenoma and the intestinal flora based on 16S rRNA sequencing*
- Development and validation of a tumor microenvironment-related prognostic signature in lung adenocarcinoma and immune infiltration analysis*
