Molecular Cloning, Characterization and Expression Profile of Myf5 and Myf6 During Growth and Development in the Seriola lalandi
SHI Bao, SUN Ranran, LIU Xuezhou, *, ZHANG Zhengrong, XU Yongjiang, JIANG Yan, and WANG Bin
Molecular Cloning, Characterization and Expression Profile of Myf5 and Myf6 During Growth and Development in the
SHI Bao1), 2), SUN Ranran1), 3), LIU Xuezhou1), 2), *, ZHANG Zhengrong1), XU Yongjiang1), 2), JIANG Yan1), 2), and WANG Bin1), 2)
1) Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture and Rural Affairs,Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071, China 2) Laboratory for Marine Fisheries and Food Production Processes, Pilot National Laboratory for Marine Science and Technology (Qingdao), Qingdao 266237, China 3) College of Fisheries and Life Science, Dalian Ocean University, Dalian 116023, China
Myogenic Regulatory Factors (MRFs) is involved in the muscle growth and differentiation. In this study, the cDNA sequence of yellowtail kingfish MRFs genes were cloned by rapid amplification of cDNA ends (RACE) method; then, the characteristics of these genes and the predicted protein sequences were analyzed by bioinformatics methods, the tissue and embryonic stages differential expression pattern were detected by the quantitative real-time PCR. Our results showed that the yellowtail kingfish (YTK) Myf5 cDNA has a full length of 951bp, encoding 266 amino acids. The yellowtail kingfish Myf6 cDNA has a full length of 1105bp, encoding 250 amino acids. The proteins contain α-helix, β-strand, and loops. The Neighbour-joining tree revealed that YTK Myf5 and Myf6 are closely related to. The yellowtail kingfish Myf5 and Myf6 gene expressed significantly higher in muscle than in other tissues (<0.05). In addition, Myf5 and Myf6 in muscle was significantly expressed in 400g and 500g fish but not in 50g, suggesting that myogenic regulatory factors expression had a great relationship with the fish size. Our results also indicated that Myf5 and Myf6 have different functions during embryonic development, because Myf5 showed highest expression level at the neuroembryo period, but Myf6 had the highest expression level at embryo coverage yolk 70% stage. Myf5 gene showed highest expression at 30d of age, suggesting it played key roles in myogenic period. However, the Myf6 gene was significantly highly expressed at 60d, revealing this gene functioned in the later muscle formation period.
; myogenic regulatory factors; gene cloning; expression
1 Introduction
Fish skeletal muscle is predominantly composed of white muscle, which represents up to 60% of total weight in some fish species, and involved in most fish growth, becoming the edible part of the fish (Zhang., 1996; Almeida., 2010). Therefore, research on fish skeletal muscle is currently a hot topic in field of aquaculture. Muscle growth and differentiation is regulated by the positive regulators of the Myogenic Regulatory Factors (MRFs) family members including myoblast determi- nation protein (MyoD), myogenin (MyoG), Myogenic factor 5 (Myf5) and Myogenic factor 6 (Myf6; which was also named as MRF4) (Braun and Gautel, 2011). MRFs, the transcription factors share a highly conserved central region termed the basic helix loop helix (bHLH) domain,which could be dimerized with ubiquitous E proteins (Edmondson and Olson, 1993). Many studies have identified that MyoD and Myf5 were required for muscle determination (Rudnicki., 1993) and involved in myogenic lineage determination (Tajbakhsh., 1996). MyoG was known as acted as a crucial differentiation factor in the control of myoblast fusion and myofiber maturation (Hasty., 1993; Nabeshima., 1993). Meanwhile, Myf6 played a key role initiating and maintaining the myogenic differentiation program, which functioned to be more complicated (Braun and Arnold, 1995; Zhang., 2015).
MRFs exhibiting functional similarity between higher vertebrates and fish, however, the distinct characteristics of muscle in fish including indeterminate growth, slow and fast muscle type, and contribution of hyperplasia in post-larval growth should be considered (Mommsen, 2001). So far, Myf5 and Myf6 genes had already been identified and characterized in many fish species such as zebrafish () (Chen and Tsai, 2002; Hinits., 2007), striped bass ()(Tan., 2002), common carp (L.) (Kobiyama., 1998), rainbow trout () (Johansen and Overturf, 2005) and snow trout () (Rajesh., 2019). The expression for these genes have been all resolved during early embryonic development, such as(Chen and Tsai, 2002),L.(Cole., 2004) and sea perch () (Ye., 2007), and provided a better understanding of Myf5 and Myf6 function during fish development.
Yellowtail kingfish (, here after referred to as YTK) is distributed in most oceanic areas, including the coasts of Australia, New Zealand, Japan, China, the United States, Chile. (Ai., 2020). YTK is well- received by consumers because of its characteristics of balanced nutrition, because of the increased demand for YTK in global, this species has recently been designated as one of the important species for the diversification of aquaculture industry in many countries (Palomino., 2014; Candebat., 2020; Dettleff., 2020).
To gain more insight into the potential function of MRFs, we have cloned Myf5 and Myf6 genes in YTK. Subsequently, we tried to explore tissue distribution of both Myf5 and Myf6 genes, and quantify their transcriptional changes during fish embryonic and juvenile development, as well as in different fish size. Our data will undoubtedly expand our knowledge about fish Myf5 and Myf6 genes, which may provide some novel information for fish muscle growth.
2 Material and Methods
2.1 Experimental Fish and Sample Processing
YTK were taken from Dalian Fugu Aquatic Products Co., Ltd., and reared under standard conditions (18–25℃; salinity at 28–30ppt; pH 8.0–8.3), feeding with 2%–5% of the body weight of the fish. One year old YTK (body weight 0.5–0.55kg, body length 38–40cm), 2 year old YTK (body weight 2–2.5kg, body length 50–52cm), 3 year old YTK (7–7.5kg, body length 93–95cm) were chosen as samples. After MS-222 (260mgL−1) anesthesia, rapid dissection, 11 tissues including brain, pituitary, liver, muscles, spleen, kidney, gills, head, stomach, intestinal and head kidney were collected. Tissues were put into liquid nitrogen for quick freezing, and then transferred to −80℃ for storage.
According to the division of embryonic stages of YTK (Xu., 2019), microscope (NIKON MSZ800, Japan) was used to observe the embryonic development, record the time sequence of development, and collect embryo samples at 18 developmental stages, from fertilized eggs to newly hatched larvae. At the same time, samples of 14 growth and development stages including hatchling juveniles (1d to 60d old) were collected. All of the samples were quickly frozen in liquid nitrogen, used for gene expression analysis during embryonic and juvenile juvenile development. All experimental methods were performed following the guidelines for the care and use of experimental animals of China (China’s national standard: GB/ T358922018).
2.2 Total RNA Extraction and First Strand cDNA Synthesis
RNAiso Plus (TaKara, Japan) was used to extract total RNA from muscle tissue, and the integrity of RNA was detected by 1.2% agarose gel electrophoresis. The RNA concentration was measured using Nanodrop2000 (Ther- mo, USA). The PrimeScript RT reagent Kit reverse transcription kit (TaKara, Japan) was used to synthesize the first strand of cDNA for amplification of conserved regions. The first strands of 5’ RACE and 3’ RACE cDNA were synthesized by SMARTerTM RACE cDNA Amplification Kit (Clontech, USA) for subsequent full-length RACE experiments.

Table 1 Nucleotide sequences of primers used for PCR amplification of S. lalandi
The primers used for gene cloning are shown in Table 1. Amplification of intermediate fragments using muscle cDNA as template, PCR reaction system (25μL): 2μL 10 ×PCR buffer, 2μL MyoD1-F, 2μL MyoD1-R, 0.5μL Taq enzyme, 2.5μL dNTP mixture, 2μL template, and 14μL ddH2O. The PCR product was detected by 1.2% agarose gel electrophoresis. The target band was recovered by EZNA™ Gel Extraction Kit Manual (Omega, USA), and quickly connected to the pEASY-T1 vector, and then transferred to Transl T1 competent cells (whole-type gold, China) were cultured in a 37℃ incubator overnight, and positive clones were picked and sent to Biotech (Shanghai) Co., Ltd. for sequencing. The full length gene sequences were obtained using nested PCR reaction (primers are shown in Table 1).
2.3 Protein Sequence Alignment and Phylogenetic Analysis
Expasy online database prediction (www.expasy.org/ tools/protparam.html) was used to predict the structure, molecular weight and isoelectric point of related genes. The software DNAMAN6.0, SignalP4.1 (http://www.cbs. dtu.dk/services/SignalIP) was used for amino acid sequence derivation, splicing and homology analysis to predict the signal peptide position. PSORT II software (https://psort.hgc.jp/form2.html) was used for subcellular localization analysis. NCBI database (https://www.ncbi. nlm.nih.gov/Structure/cdd/wrpsb.cgi) was used for domain prediction, and ClustalW online software (http:// www.genome.jp/tools-bin/clustalw) was chosen to analyze amino acid sequence alignment, MEGA 6 software was used for system evolution analysis (the self-expansion value was 1000). A neighbour-joining tree was constructed based on the deduced amino acid sequences of other reported species using MEGA 6.0 software.
2.4 Expression Analysis of Myf5 and Myf6 by Real-Time Quantitative PCR
According to the Myf5 and Myf6 sequences and 18S rRNA (EU047719.1), the real-time quantitative PCR primers of YTK Myf5 and Myf6 genes were designed by the Premier 5.0 software (shown in Table 1). Total RNA was extracted from different tissues, embryos and juveniles of different developmental stages with RNAiso Plus (TaKara, Japan) according to the manufacturer instructions. Total RNA was first treated with DNase I (Invitrogen, CA, USA) to remove any genomic DNA contamination. The cDNA was synthesized using the Takara PrimerScript™ First Strand cDNA Synthesis kit (TaKaRa, China) according to the manufacturer’s instructions. Then, the mRNA expression levels of Myf5 and Myf6 genes were evaluated using a relative quantitative real-time RT-PCR assay with SYBR Premix Ex TaqTMII (TaKaRa, China). The real-time quantitative PCR reaction condi- tions were shown as follows: pre-denatured at 95℃ for 30 s, denatured at 95℃ for 5s, and annealed at 60℃ for 20s, 72℃ for 30s, 40 cycles. The relative expression of Myf5 and Myf6 mRNA of different tissues was calculated by 2−△△Ctmethod.

Fig.1 The full-length cDNA sequence of S. lalandiMyf5 gene and deduced amino acid sequence. The underline represents Basic region, and the box represents HLH region.
2.5 Statistical Analysis
The mean ± standard deviation (Mean±SD) is used to represent the obtained experimental data. For comparison between multiple sets of data, SPSS 19.0 statistical software is used for data analysis (one-way ANOVA, Duncan comparative analysis,<0.05).
3 Result
3.1 Gene Identification and Sequences Analysis
The full length of Myf5 cDNA obtained 951bp (GenBank accession number: MK101226), including 15bp5’ UTR, 801bp ORF and 330bp 3’UTR, encoding 266 ami- no acids, the protein predicted molecular weight 28749.81 and isoelectric point 6.0. There are two distinct domains in Myf5, one is the Basic region (27–98), and the other is its HLH domain (99–150) (Fig.1). The full length of Myf6 cDNA obtained 1105bp (GenBank accession number: MK036016), including 166bp 5’UTR, 753bp ORF and 186bp 3’UTR, encoding 250 amino acids, the protein predicted molecular weight 27323.58, and iso- electric point 6.82, Myf6 also contained two distinct domains, the HLH domain (102–153) and its extended stru- cture basic region (2–101, Fig.2), the protein predicted molecular weight 26586.50, and isoelectric point 4.91.

Fig.2 The full-length cDNA sequence of S. lalandiMyf6 gene and deduced amino acid sequence. The underline represents Basic region, and the box represents HLH region.
3.2 Phylogenetic Analysis of Myf5 and Myf6
The multiple alignment results of amino acid sequences of YTK Myf5 and Myf6 of different species are shown in the figure below (Fig.3). The deduced amino acid sequence with other fish species showed well conserved re- gions across the sequence (Fig.3). The Neighbour-joining tree constructed based on multiple sequence alignment revealed that YTK is closely related to(Fig.4). Phylogenetic analysis showed that YTK Myf5 and Myf6 shared high amino acid identity (>90%) with, while only 52.24%–62.10% identity with mammals.

Fig.3 Comparison of deduced amino acids equences of Myf5 and Myf6 of S. lalandiwith other published Myf5 and Myf6. The Basic region is indicated by single line. The predicted bHLH domain is indicated by double line. Identical amino acid residues are represented by stars.
3.3 Tissue Distribution Using Real-Time PCR(qRT-PCR)
The qRT-PCR showed that Myf5 and Myf6 were expressed highest in muscle tissues, indicating that Myf5 and Myf6 mainly played roles in muscles (Fig.5A and 5B). In addition, Myf5 is highly expressed in gill and stomach, but Myf6 is highly expressed in head and stomach (Figs.5A and 5B). Both of the Myf5 and Myf6 genes expressed in kidney are low (Fig.5A and 5B).
3.4 Expression Levels of Myf5 and Myf6 in YTK in Different Tissues in Various Sizes
Our results showed that Myf5 mRNA had no significant difference in the 50g weight fish in brain, pituitary, liver and muscle. The highest expression level of Myf5 was shown in the 400g weight fish (<0.05; Fig.6A) in both of pituitary and muscle. In addition, the expression of Myf5 gene in the pituitary is significantly higher than that in brain, liver and muscle in 500g fish samples (<0.05; Fig.6A). Meanwhile, for Myf6 gene, there was no significant change in the expression level of YTK Myf6 mRNA in the brain (<0.05; Fig.6B). The expression level of Myf6 gene in 400g fish was also significantly higher in the pituitary and muscle than that in the brain and liver, which consistent with the Myf5 gene results. The highest expression level of Myf6 gene in the 500g stage of the body weight was seen in muscle (<0.05; Fig.6B).
3.5 Expression Levels of Myf5 and Myf6 in YTK During Embryogenesis
The gene expression level of Myf5 began to increase from middle gastrula embryo stage, reached its highest level at neural embryo stage, and then began to decrease (<0.05; Fig.7A). However, the expression level of Myf6 was higher at fertilized egg stage, and sharply decreased at embryo encompassing yolk 50% stage, then reached the highest value at yolk 70% stage, and then decreased from yolk 100% stage (<0.05; Fig.7B).

Fig.4 Phylogenetic tree for Myf5 and Myf6 amino acid sequences of mammals and teleosts. The tree was generated by MEGA 6.0 software using the neighbor-joining method, following Clustal X. Scale bar indicates an evolutionary distance of 0.01 amino acid substitution per position in the sequence. Bootstrap values are indicated (1000 replicates). GenBank ID for Myf5 mammalians: Homo sapiens NP_005584.2, Bos taurus AAA51415.1, Mus musculus NP_032682.1; for birds: Gallus gallus CAA51712.1; for amphibian: Xenopus laevis CAA40062.1, Notophthalmus viridescens CAA58045.1; for fish: Takifugu rubripes AAR20811.1, Oncorhynchus mykiss AAV30214.1, Paralichthys olivaceus ABI96686.1, Morone saxatilis AAL66387.1, Epinephelus coioides AMR58947.1, Cyprinus carpio BAA33566.1, Seriola lalandi XP_0232688 83.1, Danio rerio AAI65074.1, Seriola dumerili XP_022605279.1, Larimichthys crocea XP_019132416.2. GenBank ID for Myf6 mammalians: Homo sapiens CAG46563.1, Bos taurus NP_861527.1, Mus musculus NP_032683.1; for birds: Gallus gallus NP_001025917.1; for fish: Seriola lalandi XP_023268934.1, Seriola dumerili XP_022605203.1, Paralichthys olivaceus 961011.1, Takifugu rubripes NM_001032771.1, Epinephelus coioides HM190248.1, Cyprinus carpio GU 339054.1, Tetraodon nigroviridis AY576806.1, Monopterus albus AIS22055.1, Trachidermus fasciatus AFP28938.1.

Fig.5 The expression levels ofS. lalandiMyf5 (A) and Myf6 (B)mRNA in different tissues. BR, brain; P, pituitary; L, liver; M, muscle; SP, spleen; K, kidney; G, gill; H, head; ST, stomach; I, intestinal; HK, head kidney.
3.6 Expression Levels of Myf5 and Myf6 During Development
Our results showed that the expression level of Myf5 mRNA increased after hatching out of the membrane. Then increased significantly after 20d, peaked at 30d of age, then significantly decreased and maintained a lower expression level (<0.05; Fig.8A). The expression level of Myf6 mRNA was low at 0–40d, and began to increase significantly at 45d, and maintained a high expression until it reached a peak at 60d (<0.05; Fig.8B).

Fig.6 The expression levels ofS. lalandiMyf5 (A) and Myf6 (B)mRNA in brain, pituitary, liver and muscle in different size.

Fig.7 The expression levels ofS. lalandiMyf5 (A) and Myf6 (B)mRNA in different embryonic stages. (1), Fertilized egg; (2), 2 cells egg; (3), 4 cell egg; (4), 8 cell egg; (5), 16 cell egg; (6), 32 cell egg; (7), multi-cell stage; (8), mulberry embryo; (9), high blastocyst; (10), low blastocyst; (11), early gastrula embryo; (12), middle gastrula embryo; (13), late gastrula embryo; (14), neural embryo stage; (15), embryo encompassing yolk 50%; (16), embryo encompassing yolk 70%; (17), embryo encompassing yolk 100%; (18), newly hatched larva.
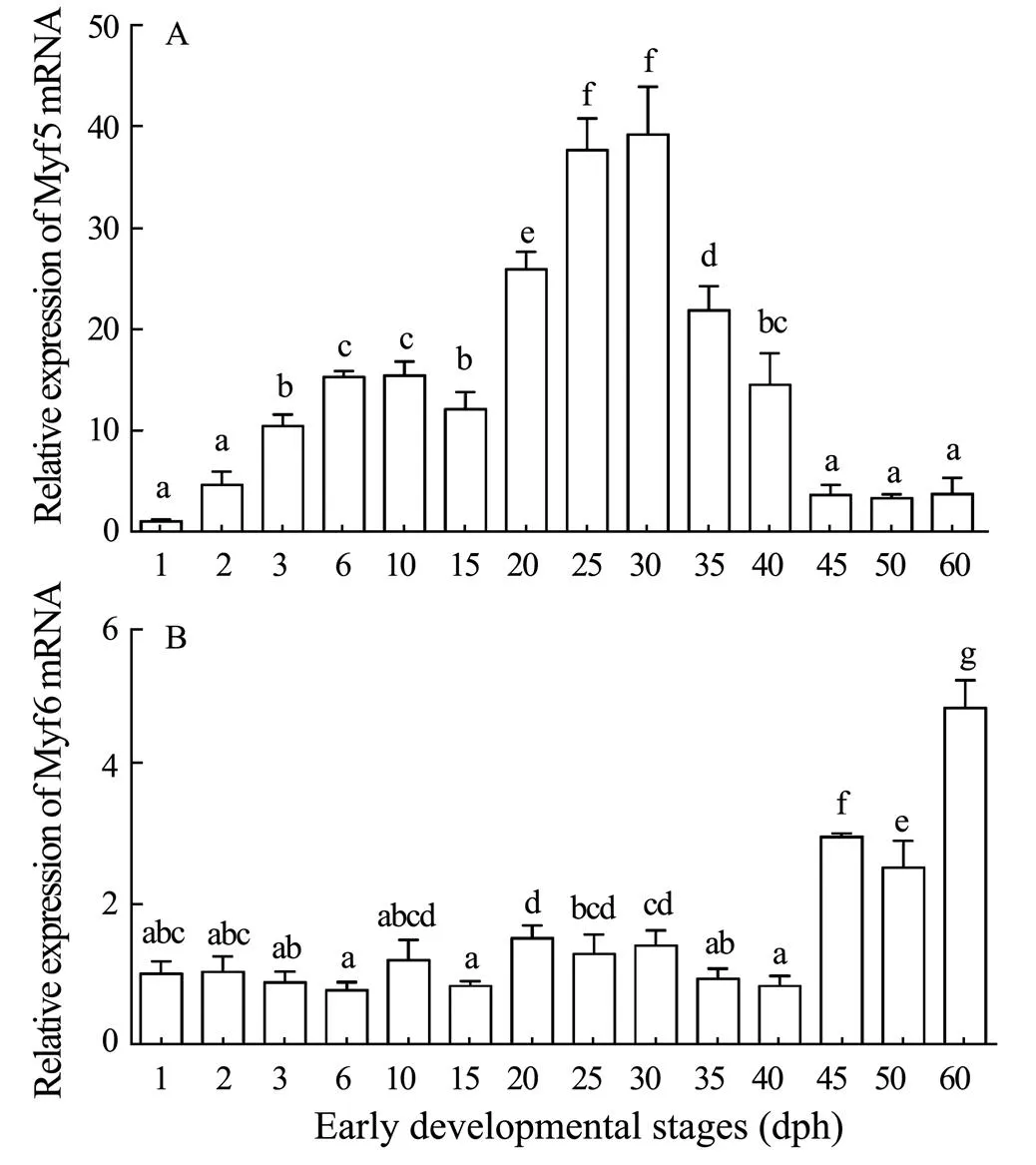
Fig.8 The expression levels of S. lalandiMyf5 (A) and Myf6 (B) mRNAin larval and juvenile development stages.
4 Discussion
In the present study, we identified and characterized two important myogenic regulators of muscle growth, Myf5 and Myf6 in yellowtail kingfish (.). In addition, we have elucidated the transcriptional regulation of the two genes in different stages and tissues. It has been found that the sequence of Myf5 and Myf6 were similar to most cyprinids fish species (Fig.3), suggesting a good conservation in the size and constitution of the protein (Rajesh., 2019). In this study, the cDNA coding regions of the Myf5 and Myf6 have been determined in the YTK. YTK Myf5 and Myf6 shared high amino acid identity (>90%) with greater amberjack(), while only 52.24–62.10% identity with mammal such as mouse and human Myf5 and Myf6. The polyadenylation signal (AATAAA) is located in the region 18bp upstream of the poly (A) tail of the Myf6 sequence, but lost in Myf5 sequence (Figs.1 and 2). The deduced Myf5 and Myf6 protein in YTK had very similar functional domain (helix-loop-helix domain (bHLH)) with other species Myf5 and Myf6 (Fig.3). In addition, both of the two proteins contained a myogenic basic domain and a specific myogenic determination factor 5 domain in C terminal structure. Phylogenetic analysis of Myf5 and Myf6 with other teleosts indicated that both of Myf5 and Myf6 cluster with(Fig.3), especially.
In addition, the tissue-specific expression pattern revealed that YTK Myf5 and Myf6 mRNAs were highest in muscle. This was also observed in(Codina., 2008),(Zhu., 2014),(Chu., 2014) and(Lin., 2016). Meanwhile, the results of real- time fluorescence quantification showed that these two genes are also expressed in other tissues, including brain, muscle, spleen, kidney, heart and kidney, as reported by other studies (Zhang.,2006; Zhu., 2016) demonstrated that the expression in the eyes could be attributed to the presence of a small group of extraocular muscles. However, comparing younger fish and elder fish, it has been found that only 400g YTK and 500g YTK showed higher expression of Myf5 and Myf6 in muscle, respectively (Fig.6). Generally, YTK is considered as fast growers (Bowyer., 2012), fish apparently indicated size dependent differential growth in terms of absolute weight gain (Rajesh., 2019). Our results showed that expression of myogenic regulatory factors had a great relationship with the size of fish, so it is particularly important to analyze the expression level in growth and development stages.
Studies ofgene expression during development showed that Myf5 increased at the early gastrointestinal embryo stage, which increased gene expression began after the multi-cell stage (Ye., 2007). Our results indicated thathad the highest Myf5 expression during the neuroembryo period, which was similar to the results of Chinese perch () (Zhu., 2016). In our study, Myf6 had the highest ex- pression level at embryo coverage yolk 70% stage, it was different from theresult which Myf6 was reached higher level at muscular effect stage, but sustained lower level at other stages (Zhu., 2016). These results indicated that Myf5 and Myf6 have different functions during the embryonic development. It has been well known that Myf5 is participated in establishment and maintenance of muscle progenitor lineage, therefore, higher level of Myf5 expression after neurula stage confirmed this (Emerson, 1990). In this study, the Myf6 expressed at embryo coverage yolk 70% stage suggested Myf6 was mainly involved in terminal differentiation myotubes (Hinits., 2007). During juvenile stage, it has been found that the expression level of Myf5 gene was highest at 30d of age (metamorphosis period of YTK). It has been suggested that Myf5 gene played key roles in myogenic period, when requiring a large amount of skeletal muscle formation (Rudnicki., 1993; Sabourin and Rudnicki, 2000). However, the expression level of Myf6 gene was the highest at 60d, indicating that the gene functions in the late stage of muscle formation.
5 Conclusions
We have cloned and characterized Myf5 and Myf6 of the yellowtail kingfish () and confirmed that it is played key roles in myogenesis. Our alinment suggested that Myf5 and Myf6 shared an analogous structure in highly conserved bHLH domains with other vertebrates. Tissue expression patterns revealed a peak level of these two genes mRNA in muscle. For different size, 400g YTK and 500g YTK showed higher expression of Myf5 and Myf6 in muscle, respectively. Spatiotemporal expression patterns indicated the different levels of two genes mRNA during embryo and juvenile development.Myf5 had the highest expression during the neuroembryo period and 30d of age, however, Myf6 had the highest expression level at embryo coverage yolk 70% and 60d stage. Our data presents a first step in elucidating the potential biological role of yellowtail kingfish Myf5 and Myf6. It furthermore provides information that Myf5 and Myf6 played different roles during embryo and juvenile development.
Acknowledgements
The study is supported by the National Key Research and Development Program (Nos. 2018YFD0901204 and 2019YFD0900503), the National Natural Science Foundation of China (No. 31772829), the Laboratory for Marine Fisheries Science and Food Production Processes, Qingdao National Laboratory for Marine Science and Technology, P. R. China (No. 2017-3A01), Central Public- interest Scientific Institution Basal Research, CAFS & Key Laboratory of Sustainable Development of Marine Fisheries, Ministry of Agriculture and Rural Affairs, P. R. China (No. 2019HY-XKQ01), the Central Public-interest Scientific Institution Basal Research Fund, YSFRI, CAFS (No. 20603022021004), the Central Public-interest Scientific Institution Basal Research fund, CAFS (No. 47) and the China Agriculture Research System (No. CARS-47).
Ai, Q., Sang, L., Tan, H., Huang, X., and Li, C., 2021. Genetic and morphological differences between yellowtail kingfish () from the Bohai Sea, China and the Southern Ocean, Australia., 6 (3): 260-266, DOI: 10.1016/j.aaf.2020.03.004.
Almeida, F. L. A. D., Pessotti, N. S., Pinhal, D., Padovani, C. R., Natália de Jesus Leito, and Carvalho, R. F., 2010. Quantitative expression of myogenic regulatory factors MyoD and myogenin in pacu () skeletal muscle during growth., 41 (8): 997-1004.
Bowyer, J. N., Qin, J. G., Smullen, R. P., and Stone, D. A. J., 2012. Replacement of fish oil by poultry oil and canola oil in yellowtail kingfish () at optimal and suboptimal temperatures., 356-357: 211-222.
Braun, T., and Arnold, H. H., 1995. Inactivation of Myf-6 and Myf-5 genes in mice leads to alterations in skeletal muscle development., 14 (6): 1176-1186.
Braun, T., and Gautel, M., 2011. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis., 12 (8): 349- 361.
Candebat, C. L., Booth, M., Codabaccus, M. B., and Pirozzi, I., 2020. Dietary methionine spares the requirement for taurine in juvenile Yellowtail Kingfish ()., 522: 735090.
Chen, Y. H., and Tsai, H. J., 2002. Treatment with Myf5-mor- pholino results in somite patterning and brain formation defects in zebrafish., 70 (8): 447-456.
Chu, W., Li, Y., and Wu, P., 2014. Characterization and expression analysis of myogenin gene in white muscle of Chinese mandarin fish,., 7: 71-76.
Codina, M., Bian, Y. H., Gutiérrez, J., and Du, S. J., 2008. Cloning and characterization of myogenin from seabream () and analysis of promoter muscle specificity., 3 (1): 128-139.
Cole, N. J., Hall, T. E., and Martin, C. I., 2004. Temperature and the expression of myogenic regulatory factors (MRFs) and myosin heavy chain isoforms during embryogenesis in the common carpL., 207: 4239-4248.
Dettleff, P., Hernandez, E., Patridge, G., Cruz, F. L., and Mar- tínez, V., 2020. Understanding the population structure and reproductive behavior of hatchery-produced yellowtail kingfish ()., 522: 734948.
Edmondson, D. G., and Olson, E. N., 1993. Helix-loop-helix proteins as regulators of muscle-specific transcription., 268: 755-758.
Emerson, C. P., 1990. Myogenesis and developmental control genes., 2 (6): 1065-1075.
Hasty, P., Bradley, A., Morris, J. H., Edmondson, D. G., Venuti, J. M., Olson, E. N.,., 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene.,364: 501-506.
Hinits, Y., Osborn, D. P. S., Carvajal, J. J., Rigby, P. W. J., and Hughes, S. M., 2007. Mrf4 (myf6) is dynamically expressed in differentiated zebrafish skeletal muscle., 7 (7): 738-745.
Johansen, K. A., and Overturf, K., 2005. Quantitative expression analysis of genes affecting muscle growth during development of rainbow trout ()., 7: 576-587.
Kobiyama, A., Nihei, Y., Hirayama, Y., Kikuchi, K., and Watabe, S., 1998. Molecular cloning and developmental expression patterns of the MyoD and MEF2 families of muscle transcription factors in the carp., 20 (201): 2801-2813.
Lin, Y., Zhou, J., Li, R., and Wang, S., 2016. MRF gene family in: Molecular cloning, tissue expression, and mRNA expression in muscle development., 16: 461-467.
Mommsen, T. P., 2001. Paradigms of growth in fish., 129: 207-219.
Nabeshima, Y., Hanaoka, K., Hayasaka, M., and Watabe, S., 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect., 364: 532-535.
Palomino, J., Herrera, G., Dettleff, P., and Klein, W. H., 2014. Growth differentiation factor 9 and bone morphogenetic protein 15 expression in previtellogenic oocytes and during early embryonic development of Yellow-tail Kingfish., 47: 60.
Rajesh, M., Kamalam, B. S., Ciji, A., Cruz, L. D., and Victor Martínez., 2019. Molecular characterisation and transcriptional regulation of muscle growth regulatory factors myogenin and myogenic factor 6 in the Trans-Himalayan cyprinid fish.y, 231: 188-200.
Rudnicki, M. A., Schnegelsberg, P. N. J., Stead, R. H., and Klein, W. H., 1993. MyoD or Myf-5 is required for the formation of skeletal muscle.,75: 1351-1359.
Sabourin, L. A., and Rudnicki, M. A., 2000. The molecular regulation of myogenesis., 57: 16-25.
Tajbakhsh, S., Rocancourt, D., and Buckingham, M., 1996. Muscle progenitor cells failing to respond to positional cues adopt non-myogenic fates in myf-5 null mice., 384: 266-270.
Tan, X., Hoang, L., and Du, S., 2002. Characterization of muscle-regulatory genes, Myf5 and Myogenin, from striped bass and promoter analysis of muscle-specific expression.,4: 537-545.
Xu, Y., Zhang, Z., Liu, X., Lin, Y., and Zhou, J., 2019. Morphometric characteristics of the embryonic and postembryonic development of yellowtail kingfish,., 26: 172-182 (in Chinese with English abstract).
Ye, H., Chen, S., and Xu, J., 2007. Molecular cloning and characterization of the Myf5 gene in sea perch ()., 147: 452-459.
Zhang, G., Swank, D. M., and Rome, L. C., 1996. Quantitative distribution of muscle fiber types in the scup., 229: 71-81.
Zhang, R., Ludwig, A., Zhang, C., Zhou, X., Lin, Y., and Liu, L., 2015. Local adaptation of(Cyprinidae) on the Tibetan Plateau.,5: 09780.
Zhang, Y., Tan, X., Zhang, P., Zhang, C., Li, Y., and Liu, Y., 2006. Characterization of muscle-regulatory gene, MyoD, from flounder () and analysis of its expression patterns during embryogenesis.,8: 139-148.
Zhu, K., Chen, L., Zhao, J., Wang, H., Wang, W., and Li, Z., 2014. Molecular characterization and expression patterns ofin compensatory growth of., 170: 10-17.
Zhu, X., Li, Y. L., Liu, L., Wang, J. H., Li, H. H., and Wu, P., 2016. Molecular characterization of Myf5 and comparative expression patterns of myogenic regulatory factors in.,20: 1-10.
November 26, 2020;
April 19, 2021;
June 11, 2021
© Ocean University of China, Science Press and Springer-Verlag GmbH Germany 2021
. E-mail: liuxz@ysfri.ac.cn
(Edited by Ji Dechun)
 Journal of Ocean University of China2021年6期
Journal of Ocean University of China2021年6期
- Journal of Ocean University of China的其它文章
- Meshless Method with Domain Decomposition for Submerged Porous Breakwaters in Waves
- Facial Features of an Air Gun Array Wavelet in the Time-Frequency Domain Based on Marine Vertical Cables
- Magma Evolution Processes in the Southern Okinawa Trough:Insights from Melt Inclusions
- Summery Intra-Tidal Variations of Suspended Sediment Transportation–Topographical Response and Dynamical Mechanism in the Aoshan Bay and Surrounding Area, Shandong Peninsula
- High-Resolution Geochemical Records in the Inner Shelf Mud Wedge of the East China Sea and Their Indication to the Holocene Monsoon Climatic Changes and Events
- Geological Guided Tomography Inversion Based on Fault Constraint and Its Application
