Cortical transcriptome analysis after spinal cord injury reveals the regenerative mechanism of central nervous system in CRMP2 knock-in mice
Ayaka Sugeno , Wenhui Piao Miki Yamazaki , Kiyofumi Takahashi,Koji Arikawa, Hiroko Matsunaga, Masahito Hosokawa , , Daisuke Tominaga, ,Yoshio Goshima, Haruko Takeyama , , , Toshio Ohshima ,
Abstract Recent studies have shown that mutation at Ser522 causes inhibition of collapsin response mediator protein 2 (CRMP2) phosphorylation and induces axon elongation and partial recovery of the lost sensorimotor function after spinal cord injury (SCI). We aimed to reveal the intracellular mechanism in axotomized neurons in the CRMP2 knock-in (CRMP2KI) mouse model by performing transcriptome analysis in mouse sensorimotor cortex using micro-dissection punching system. Prior to that, we analyzed the structural pathophysiology in axotomized or neighboring neurons after SCI and found that somatic atrophy and dendritic spine reduction in sensorimotor cortex were suppressed in CRMP2KI mice. Further analysis of the transcriptome has aided in the identification of four hemoglobin genes Hba-a1, Hba-a2, Hbb-bs, and Hbb-bt that are significantly upregulated in wild-type mice with concomitant upregulation of genes involved in the oxidative phosphorylation and ribosomal pathways after SCI. However, we observed substantial upregulation in channel activity genes and downregulation of genes regulating vesicles, synaptic function, glial cell differentiation in CRMP2KI mice. Moreover, the transcriptome profile of CRMP2KI mice has been discussed wherein energy metabolism and neuronal pathways were found to be differentially regulated. Our results showed that CRMP2KI mice displayed improved SCI pathophysiology not only via microtubule stabilization in neurons, but also possibly via the whole metabolic system in the central nervous system, response changes in glial cells, and synapses. Taken together, we reveal new insights on SCI pathophysiology and the regenerative mechanism of central nervous system by the inhibition of CRMP2 phosphorylation at Ser522. All these experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at Waseda University,Japan (2017-A027 approved on March 21, 2017; 2018-A003 approved on March 25, 2018; 2019-A026 approved on March 25, 2019).
Key Words: CNS regeneration; cortex; CRMP2; hemoglobin; metabolic pathway; spinal cord injury; spine; transcriptome
Introduction
Mammalian central nervous system (CNS) has limited ability to regenerate and recover after injury. Spinal cord injury (SCI)is one such typical injury which is characterized by its semipermanent impairments in sensorimotor functions below the spinal cord lesion. Though new treatment regimen for SCI by stem cell transplantation is gradually developing (Nakamura and Okano, 2013), promising approach for regenerating CNS with intrinsic factors is yet to be revealed.
Collapsin response mediator protein 2 (CRMP2) is one such candidate protein which is abundantly expressed in the adult mammalian brain (Wang and Strittmatter, 1996). CRMP2 is a member of the CRMP family proteins (CRMP1-5) which were identified as downstream molecules from Semaphorin 3A - an axon guidance cue (Schmidt and Strittmatter, 2007).CRMP2 has been reported to bind to microtubules, kinesin or dynein dependent vesicles, transport proteins, and interact with calcium channels (Kawano et al., 2005; Kimura et al., 2005; Wang et al., 2009; Rahajeng et al., 2010; Lin et al., 2011). Through these complex interactions, CRMP2 regulates synaptic transmission (Brittain et al., 2009; Wang et al., 2009), controls microtubule dynamics (Zheng et al.,2018), and modulates vesicle trafficking (Arimura et al., 2009;Rahajeng et al., 2010). The activity of CRMP2 is controlled by phosphorylation and Cdk5 is one of the major upstream kinases that phosphorylates CRMP2 at Ser522. Recently,in a mouse model, inhibition of phosphorylation at Ser522(CRMP2KI) induces axon elongation after SCI along with reduced inflammation and microtubule stabilization at the spinal cord lesion site (Nagai et al., 2016). CRMP2KI mice showed partial recovery in the lost sensorimotor function subsequently and the phosphorylation inhibition of CRMP2 at Ser522 was suggested to have a therapeutic effect on SCI pathophysiology (Nagai et al., 2016).
The pathophysiology of SCI was also observed in the brain sensorimotor cortex. The axotomized neurons and neighborhood neurons significantly shrink and the number of synaptic spines reduces after SCI (Barron et al., 1988; Kim et al., 2006; Carter et al., 2008; Ghosh et al., 2012). These structural change is known to contribute to the rewiring of neuronal circuits within the cortex for compensating the lost sensorimotor functions (Ghosh et al., 2010). The effect of such changes in the cortex on the entire pathophysiology of SCI is not clearly understood and it has also not been investigated in the context of CRMP2 knock-in (CRMP2KI) mouse models.
In order to clear out such cortex pathology after SCI,we analyzed the structural pathophysiology and mRNA transcription in the cortex of CRMP2KI mice.
Materials and Methods
Animals and surgical procedures
Female wild-type (WT) and CRMP2KI (Crmp2KI/KI) mice(Yamashita et al., 2012) were crossed with YFP-H (thy1-Yellow Fluorescent Protein Mouse line H) or GFP-M (thy1-Green Fluorescent Protein Mouse line M) mice (Feng et al.,2000). Offspring at the age of 6–9 weeks, weighing 22–28 g,were used in the study.The fluorescent signal was used as a landmark to demarcate the region of interest–cortex layer V. Animals were randomly selected for sham or SCI models.For the SCI model, dorsal transection at T7–8 spinal cord was performed as previously described (Hill et al., 2009; Nagai et al., 2015). In brief, mice were anesthetized by inhalation with 2–5% isoflurane (DS Pharma Animal Health, Japan)and a T7–8 laminectomy was performed with 1.5-mm-deep dorsal transection. After surgery, manual bladder evacuation was done every day. All animals could easily access food and water. SCI mice who showed 0–1 Basso Mouse Scale (BMS)score (Basso et al., 2006) at 1 day after SCI were used for the experiments. For RNA sequencing, sham-operated model was made in order for precise analysis. In the condition of shamoperated model, mice were kept under anesthesia for 15–20 min. All animals were group-housed (N= 2 to 4 per cage) in plastic cages and maintained on a 12/12-hour light/dark cycle with the room temperature (RT) maintained at 25 ± 2°C. All these experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at Waseda University, Japan (2017-A027 approved on March 21, 2017; 2018-A003 approved on March 25, 2018; 2019-A026 approved on March 25, 2019).
Soma size analysis by YFP signal
Mice were anesthetized using diethyl ether (Wako, Osaka,Japan) and transcardially perfused with 4% paraformaldehyde(PFA) (nacalai tesque, Kyoto, Japan) in phosphate buffer saline (pH 7.4), one or four weeks after surgery. Brains were dissected out from the animals and post-fixed overnight.They were continuously soaked in PBS, 10% sucrose and 20%sucrose for 12–24 hours and embedded in 2:1 mixture of Tissue Tekoptimal cutting temperature (OCT) compound(Sakura Finetek Japan, Osaka, Japan) and 20% sucrose.Frozen brains were sectioned into 30 µm sections by cryostat(Leica biosystems, Wetzlar, Germany). Slices containing bregma-posterior 0.5 mm, which has been reported to be sensorimotor cortex of the hindlimbs in mice by retrograde tracer (Ghosh et al., 2012), were observed by confocal microscope (FV-1000, Olympus, Tokyo, Japan) in 1.5 µm Z-stacks. Soma size of YFP-positive neurons in the cortex layer V between 500 and 1500 µm from the midline was measured with ImageJ software ver 1.52a (NIH, Bethesda, MD, USA)(Schneider et al., 2012) by encircling the soma.
Golgi-stain and spine analysis
Female mice at the age of 6 weeks were used for spine analysis for enhanced precision. SliceGolgi Kit (Bioenno Tech, LLC, Santa Ana, CA, USA) was used for Golgi staining technique. All the procedures were performed following SliceGolgi Kit instructions except the impregnation period.In brief, dissected brains fixed in fixative solution overnight at 4°C. Hindlimb sensorimotor cortex was sliced (200 µm)by vibratome (Micro Slicer DTK-1000, D.S.K, Kyoto, Japan)and Golgi-stained with impregnation period of 2.5–3 days.Apical dendrites of pyramidal neurons in cortex layer V were selectively observed with bright-field microscope (BX51,Olympus) within 7 days after Golgi-staining. The observed dendrites were selected randomly after confirming proper staining of the dendrites as well as ensuring the dendrites did not overlap with neighboring dendrites for the ease of analysis. All the spines and spine-like protrusions on the apical dendrites were counted with 60× objective lens (UPLFLN 60X,Olympus) and the length of the dendrites was measured with ImageJ software (Schneider et al., 2012).
Immunostaining
Mice brain sections were prepared as described above.From bregma to bregma posterior 1.0 mm, the brains were sectioned into 15 µm. The slices were washed with PBS for 30 minutes, blocked with 3% horse serum in 0.1% PBSTr (PBS with 0.01% Triton X-100) for 10 minutes, 3% horse serum in 0.01% PBSTr for 30 minutes and incubated with anti-Hbα rabbit monoclonal IgG (1:50, Cat# SN70-09, Invitrogen,Carlsbad, CA, USA) overnight at 4°C. Slides were washed threetimes for 10 minutes each with 0.01% PBSTr the following day and incubated with Alexa Fluor 594 (1:1000, Abcam, Tokyo,Japan) rabbit IgG for 1 hour at room temperature. Following 3 times of 0.01% PBSTr washing, slices were incubated for 30 min with Hoechst 33258 (Fujifilm, Tokyo, Japan). Subsequently,the sections were washed with 0.01% PBSTr three times and mounted with FluoromountTMaqueous mounting medium(MERCK). Images were acquired by confocal laser scanning microscope in 1.5 µm Z-stacks (FV1000, Olympus).
Tissue dissection for RNA sequencing
Female mice at the age of 6 weeks were used for enhanced precision. The mice brains were dissected out 1 week after sham or SCI operation. Mice bodies were laid on ice to prevent degradation of RNA during the dissection process.Fresh brains were temporarily stored in PBS on ice, embedded in SCEM (Super Cryoembedding Medium, SECTION-LAB Co.
Ltd., Hiroshima, Japan) using liquid nitrogen and stored at–80°C until sectioning. Fresh brains were sectioned at 30 µm thickness coronally with Cryostat (CM1860, Leica). The slices were mounted on FrameSlides PPS-Membrane 4.0 µm (Leica)and fixed immediately by air-drying. Dried-mounted slices were stored at room temperature or –20°C until use. Airdrying was conducted every time when the slices were taken out from –20°C freezer in order to prevent RNA degradation by condensation. Micro-dissection punching (Yoda et al.,2017) was performed for sampling cortex layer V, whose pyramidal neurons project axons to the spinal cord. The standard area of punching was determined by the YFP signal of YFP-H lines (Feng et al., 2000). Hindlimb sensorimotor cortex was also selected from micro-dissected samples as described previously (Ghosh et al., 2012). We confirmed the area similarly by Fast Blue tracer as well (Additional Figure 1).
Library construction and RNA sequencing
cDNA libraries were made after Poly (A)+ RNA extraction as per an available protocol with Smart-seq2 (Picelli et al., 2014).RNA quality was confirmed with RIN > 7 using bulk whole brain slice sample. Sequencing libraries were constructed by Nextera XT DNA library prep kit (Illumine, San Diego,CA, USA). Constructed libraries were sequenced by pairedend way with Illumina MiSeq (Illumine). Adapter sequences were trimmed with FLEXBAR (Dodt et al., 2012) and trimmed sequence reads were aligned to the Ensembl mouse reference genome (GRCm38 ver.92) using HISAT2 (Kim et al., 2015) with the default parameters. Annotated gene expression data was normalized as transcripts per million (TPM) value.
Differential gene expression and pathway analysis
The samples with high portion of mitochondrion genes(TPM ≥ 35%) and low number of detected genes (≤ 10,000 genes) were removed from analysis. Genes expressing more than 5 as averaged TPM value and detected in more than 4 samples from each condition (WT + sham, WT + SCI, CRMP2KI+ sham, and CRMP2KI + SCI) were statistically analyzed as shown in statistics section. For pathway enrichment analysis,Gene Ontology (GO, http://www.geneontology.org), Kyoto Encyclopedia of Genes and Genomes (KEGG, http://www.genome.jp/kegg/), Reactome (https://reactome.org/), and Hallmark gene sets from MSigDB (Liberzon et al., 2015)databases from Molecular Signatures Database (MSigDB,https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp)(Liberzon et al., 2011) were used with clusterProfiler (Yu et al., 2012), ReactomePA (Yu and He, 2016), and msigdbr R packages. Fold change (FC) was calculated using averaged TPM value. FC evaluation for finding differentially expressed genes(DEGs) (Top DEGs) was performed by averaging intensitybased z-score. Concretely, genes were separated into several groups by averaged TPM from sham samples and z-score was calculated from the FC values within the group. This procedure was performed repeatedly with different grouping threshold and z-scores were averaged. Genes showing the highest or the lowest averaged z-score were defined to be the top DEGs.
Quantitative real-time PCR validation
cDNA libraries for RT-qPCR validation were constructed with the same procedure as RNA sequencing library construction.SYBR Green PCR Master Mix (Applied Biosystems, Foster City,CA, USA) was used for qPCR. Reactions were carried out in triplicates with 10 ng of cDNA and 200 nM of each primer in a final volume of 20 µL. The reaction protocol was 95°C for 1 minute; 40 amplification cycles at 95°C for 15 seconds, 50°C for 30 seconds, 72°C for 1 minute. Relative quantification of gene expression was analyzed with ∆∆Ct method and the internal control ACTB, whose mRNA expression did not change statistically in the RNA sequencing experiments. Genes and primers used for validation are as follows: ACTB-F (TTG TGT AAG GTA AGG TGT GC), ACTB-R (GGT TGA GGT GTT GAG GCA G), Hba-F (ATG CCC ACA AGC TGC GT), Hba-R (GTG CTC ACA GAG GCA AGG AA), Hbb-F (GCC CAG CAC AAT CAC GA), and Hbb-R (TGC CTT TAA CGA TGG CCT GA). In order to calculate relative expression ratio of Hb genes, ACTB was used as a housekeeping control for qPCR analysis.
Statistical analysis
For the comparison of soma size distribution analysis,Kolmogorov-Smirnov test was performed with GraphPad Prism software version 6.0b (GraphPad Software Inc., San Diego, CA, USA). Statistical differences of spine analysis were calculated with an unpaired two-tailed Student’st-test. DEGs were defined using Wilcoxon test. AdjustedP-values were calculated with Benjamini-Hochberg method. Genes and pathways withP-value < 0.05 were defined as significantly changed or enriched.
Results
CRMP2KI suppresses somatic atrophy in sensorimotor cortex after spinal cord injury
A previous study demonstrated that neurons of the corticospinal tract showed somatic atrophy after SCI in rat and mice (Barron et al., 1988; Carter et al., 2008). In mice, cortical layer V neurons undergo severe atrophy at 4 weeks after SCI(Carter et al., 2008). In order to check whether these changes occur upon the inhibition of phosphorylation of CRMP2, we examined the size of the sensorimotor neurons in cortex layer V observing YFP signal. In WT mice, sensorimotor neurons showed severe somatic atrophy at 1 week after SCI and this atrophy continued until 4 weeks after SCI (Figure 1AandB).However, in CRMP2KI mice, sensorimotor neurons shrank more mildly at 1 week after SCI and recovered to their original size at 4 weeks after SCI (Figure 1AandB). Between WT and CRMP2KI mice, significant difference was observed for both 1 week and 4 weeks after SCI (P< 0.05;Figure 1C).
CRMP2KI suppresses postsynaptic spine reduction in sensorimotor cortex after spinal cord injury
Neurons of the corticospinal tract have fewer post-synaptic spines after SCI (Ghosh et al., 2012; Kim et al., 2006). In order to reveal the effect of phosphorylation inhibition of CRMP2 on synaptic pathology, we analyzed the spine density. The location of analysis was apical dendrites of the pyramidal neurons. The number of spines decreased 1 week after SCI in WT mice (Figure 2). However, CRMP2KI mice did not show any changes in spine density (Figure 2B). CRMP2KI mice had originally fewer spines on the pyramidal neurons in cortex layer V at 7 weeks.
Cortical transcriptome profiles in all comparisons
We found the structural pathologies in sensorimotor cortex were suppressed in synaptic and somatic levels by the phosphorylation inhibition of CRMP2. However,the intracellular response in axotomized neurons is not fully understood. To reveal the detailed pathophysiology in axotomized neurons of the cortex, we performed RNA sequencing for micro-dissected cortex layer V (Figure 3A),1 week after SCI in WT and CRMP2KI mice. For the precise detection of DEGs, we performed statistical analysis for each pair of groups instead of multiple comparisons. We identified 967 genes in WT mice and 811 genes in CRMP2KI mice as DEGs after SCI. The mutation effect of CRMP2 phosphorylation inhibition was analyzed using the sham model samples of WT and CRMP2KI mice. A total of 1674 DEGs were found in CRMP2KI mice compared with WT mice. PCA analysis was performed with all DEGs’ TPM data and micro-dissected samples were separated well reflecting experimental conditions (Figure 3B). We found that the number of overlapped DEGs did not account for the high percentage of whole DEGs (Figure 3C).
Differential gene expression by the phosphorylation inhibition of CRMP2
We analyzed the mutation effect of CRMP2 phosphorylation inhibition. We detected 1674 DEGs. Among them, 1176 genes were upregulated, while 498 genes were downregulated(Additional Figure 2). Pathway enrichment analysis revealed that many pathways related to intracellular metabolism and neuronal functions were differentially expressed in CRMP2KI sensorimotor cortex (Table 1andAdditional Table 1and2). Further, genes involved in the cytosolic or mitochondrial translation and oxidative phosphorylation, proteasome,spliceosome pathways were significantly upregulated (P<0.05,q< 0.05), and fatty acid metabolism, lysosome, neuronal system, synaptic pathway genes were downregulated (P<0.05,q< 0.2). In addition to the oxidative phosphorylation and ribosomal pathways in mitochondria, genes regulating mitochondrion organization were also upregulated (Additional Figure 2C). GO analysis also displayed that neuronal specific structural or functional terms such as axon, and vesicles were enriched among downregulated DEGs (Additional Figure 2D). This suggests that the inhibition of phosphorylation of CRMP2 might dynamically change the intracellular metabolic system and neuronal functions. We also analyzed the top 20 significant DEGs (Additional Tables 3and4). Various genes whose function is restricted to cytoskeletal protein binding like Nrcam, Map1b, and Vapa were greatly downregulated.

Table 1 |Enriched biological pathways of differentially expressed genes after SCI in WT and CRMP2KI mice
Differential gene expression after spinal cord injury in WT and CRMP2KI mice
We detected 967 DEGs after SCI in WT mice. Of them, 886 DEGs showed upregulation. Pathway analysis revealed that cytosolic or mitochondrial translation and oxidative phosphorylation pathway genes were significantly enriched among the upregulated genes (Table 1andAdditional table 5). From the limited number of 81 downregulated genes,we could not detect any enriched pathways. Interestingly,hemoglobin genes Hba-a1, Hba-a2, Hbb-bs, and Hbb-bt were remarkably upregulated and became the top 4 upregulated genes (FC > 6.9) after SCI (Table 2).
On the other hand, in CRMP2KI mice, we detected a total of 811 DEGs in which 224 were upregulated; while 587 were downregulated after SCI (Additional Figure 3). Upregulated cluster consisted mainly of channel and transporter activity genes (Table 1andAdditional figure 3C). From the downregulated genes, neuronal system and cytosolic translation pathway genes were specifically enriched.
Genes regulating synaptic activity and organization of axon,myelin sheath, vesicles, and synaptic membranes were all downregulated (Figure 4AandAdditional Table 6). Notably,gliogenesis, glial cell differentiation, and developmental genes were also enriched in the downregulated DEGs after SCI in CRMP2KI mice (Additional Table 6). Hemoglobin genes were not differentially expressed in CRMP2KI mice because of the broad distribution of TPM values.
As summarized inTable 1, oxidative phosphorylation pathway genes were upregulated in both CRMP2KI mutation and SCI.In order to compare these DEGs expression level in other condition, heatmap was generated for DEGs after SCI in WT mice and pathway analysis was performed for several clusters(Figure 4B). We found that oxidative phosphorylation pathway and mitochondrial component genes were expressed at a high level in both CRMP2KI + sham and CRMP2KI + SCI condition compared with WT + SCI condition.
Hemoglobin expression in sensorimotor cortex after SCI
In transcriptome analysis, four hemoglobin genes were remarkably upregulated after spinal cord injury in WT mice(Table 2). These hemoglobin expressions were validated with RT-qPCR at first. cDNA libraries were prepared from microdissected sensorimotor cortex. Both Hba and Hbb genes were significantly upregulated after SCI in WT mice (Figure 5A).However, there was no significant difference between sham and SCI in CRMP2KI mice. These results were consistent with the transcriptome analysis obtained from RNA sequencing.
Next, we performed immunostaining of Hbα in WT mice to analyze if this hemoglobin upregulation could occur at the protein level in the sensorimotor cortex after SCI. The ratio of Hbα positive cells increased after SCI but not significantly (P=0.063) unlike our expectation (Figure 5B).
Discussion
The inhibition of CRMP2 phosphorylation has been proven to be effective for the diagnosis of SCI by this study. Previous study showed that the phosphorylation inhibition of CRMP2 suppresses glial scar formation and inflammation at the spinal cord lesion, which leads to axonal regeneration (Nagai et al., 2016). In this study, we found that spine reduction and somatic atrophy were suppressed in the sensorimotor cortex in CRMP2KI mice. Spine reduction is considered to regulate the rewiring of neuronal circuits as compensation for lost sensorimotor function and brain activity map changed in rat SCI model (Ghosh et al., 2010, 2012; Zhang et al., 2015).We show that CRMP2KI mice have the same spine density as the originally fewer density, 1 week after SCI. According to the previous study (Jin et al., 2016), fewer spines were also observed in CA1 pyramidal neurons of CRMP2KI mice at the age of 5 weeks. Focusing on the effect of SCI, the phosphorylation inhibition of CRMP2 suppressed further spine reduction by SCI. Although it is necessary to take into account that there might be relevance between suppression of spine reduction after SCI and the original fewer spines in CRMP2KI mice, the phosphorylation inhibition of CRMP2 suppressed further post-synaptic spine reduction by SCI from the primary number. This supports the idea that it could be possible in CRMP2KI mice, there is only a mild alteration of electrical activity in the somatosensory cortex,with concomitant suppression of somatic atrophy and spine reduction. Simultaneously, somatic atrophy gave an indication of neuronal stress after SCI. Significant somatic atrophy is observed 4 weeks after SCI in WT mice, but somatic size in CRMP2KI regained the original size on the same time scale.This result suggests that phosphorylation inhibition of CRMP2 contribute to suppressing the somatic atrophy of axotomized neurons and facilitating the recovery toward original size after SCI. This size recovery is also observed in the mice treated with ChABC which have retrograde neuroprotective effect after SCI (Carter et al., 2008). This gave us the possibility that the phosphorylation inhibition of CRMP2 also have similar neuroprotective effect in axotomized neurons.Considering the sensorimotor structural pathology after SCI, the phosphorylation inhibition of CRMP2 contributes to suppressing SCI pathology not only at the spinal cord lesion site; but also in the intracellular response of CNS neurons.

Figure 1|Constitutively activated CRMP2 suppresses soma atrophy in corticospinal neurons after SCI.

Figure 2|Constitutively activated CRMP2 suppresses spine decrease in CST neurons after SCI.

Figure 3|RNA sequencing analysis of micro-dissected samples.
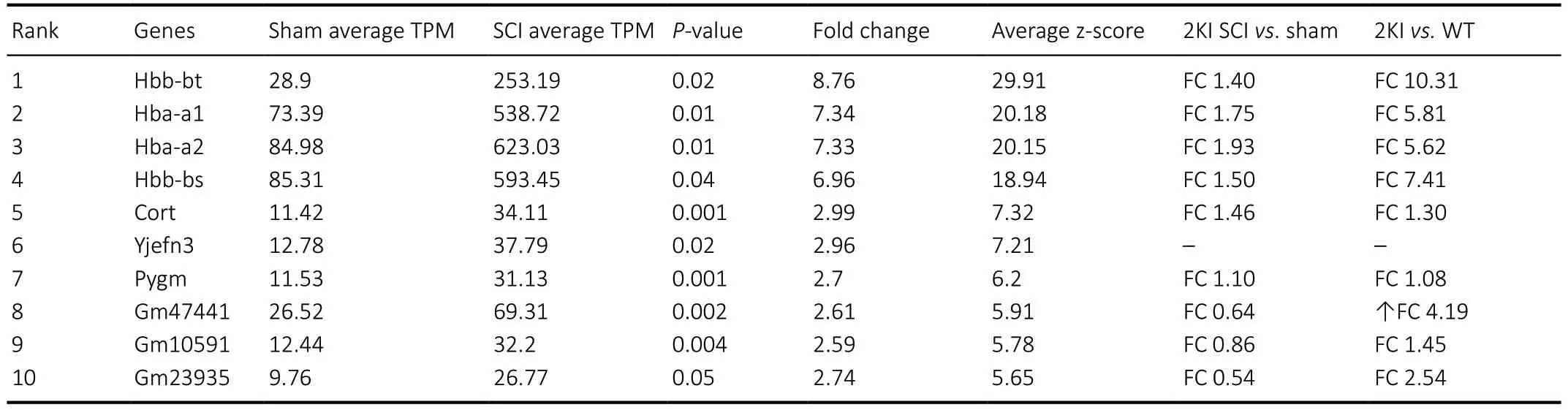
Table 2 |Top 10 significantly upregulated DEGs after spinal cord injury in WT mice

Figure 4|Differential gene expression analysis after SCI.

Figure 5|Hemoglobin (Hb) was upregulated after SCI and increased at protein level.
We also performed RNA sequencing analysis in microdissected sensorimotor cortex to reveal the detailed intracellular response in axotomized neurons after SCI in WT and CRMP2KI mice. Each cortical transcriptome profile has different characteristics when subjected to SCI, CRMP2 mutation and both. We observed that genes involved in the cytosolic, mitochondrial translation and oxidative phosphorylation pathways were significantly upregulated after SCI. Previous studies showed that genes involved in similar pathways like translation, RNA splicing, and oxidative phosphorylation were differentially expressed within a week after SCI in the cortex (Jaerve et al., 2012; Baek et al., 2017).This showed that our transcriptome analysis with microdissected cortex is consistent with the previous studies.One of these studies also suggested that impaired oxidative phosphorylation and mitochondrial dysfunction occur after SCI (Baek et al., 2017). We also found upregulation of mitochondrion component genes. Oxidative phosphorylation pathway takes place in mitochondria and impaired oxidative phosphorylation induces oxidative stress by generating reactive oxygen species (ROS) in cells generally. ROS and oxidative stress have been considered to be a hallmark of SCI and progressive SCI pathology at spinal cord lesion sites (Xu et al., 2005; Jia et al., 2012). In addition, one transcriptome analysis in cortex layer V also detected differential expression in response to oxidative stress (Kruse et al., 2011). Our findings regarding upregulated genes and enriched pathways after SCI in WT also suggests the oxidative stress as well.Notably, we further detected 4 hemoglobin (Hb) genes remarkably increased after SCI in WT mice, which was also validated by qPCR. Recently, Hb was found to be expressed in CNS tissues, including cortical neurons. Also, some studies on neurodegenerative diseases such as Alzheimer’s disease,Multiple Sclerosis, and Parkinson’s disease suggested the relevance between Hb and their pathophysiology (Biagioli et al., 2009; Richter et al., 2009; Schelshorn et al., 2009; Brown et al., 2016; Freed and Chakrabarti, 2016; Altinoz et al.,2019). In the field of SCI studies, this is the first report that indicates the involvement of Hb in SCI pathophysiology. The role of Hb in nervous system has not been fully understood and continuous studies were needed for clear explanation.However Hb is suggested to be involved in O2homeostasis and mitochondrial homeostasis by buffering protons by other studies (Biagioli et al., 2009; Freed and Chakrabarti, 2016;Altinoz et al., 2019). Though we could not detect statistically significant upregulation of Hb at the protein level, Hb transcription upregulation and simultaneously upregulated Hb functions, oxidative phosphorylation gave us some idea that the involvement of oxidative stress or impairment of mitochondrial homeostasis in axotomized neurons after SCI.
In this study, we reveal for the first time the mRNA transcription profile in sensorimotor cortex in CRMP2KI in sham-operated and SCI condition. Completely different pathways were up- or down-regulated after SCI. All the enriched pathways except the pathways involved in translation were related to typical functions of nervous system and most of the enriched pathways were downregulated after SCI.The point to be emphasized is that genes regulating synaptic compartments and functions were strongly downregulated simultaneously with the suppression of spine reduction after SCI. In addition, similar synaptic pathways were downregulated by the genetic modification of CRMP2 originally. This means axotomized neurons in CRMP2KI have quite lower mRNAs regulating synaptic functions. Although further studies are needed for clarifying the concrete mechanism, it is likely that downregulation of synaptic genes contributes to the suppression of spine reduction after SCI in CRMP2KI and might contribute to the improvement of SCI pathophysiology as well. We also found an interesting observation that glial cell differentiation and myelin sheath pathways were downregulated after SCI in CRMP2KI mice. Besides, some studies showed that microglial activation appeared in cortex(Wu et al., 2014a, b; Jure and Labombarda, 2017). We could not detect the pathway of microglial activation after SCI in WT mice, but downregulation of glial cells differentiation pathway in CRMP2KI indicates that there might be some changes in the response of glial cells including microglia, astrocytes and oligodendrocytes upon the phosphorylation inhibition of CRMP2. Indeed, the glial scar formation and inflammation at spinal cord lesion was suppressed in CRMP2KI mice (Nagai et al., 2016). Our transcriptome analysis in sensorimotor cortex indirectly supports the different response after SCI in glial cells in CRMP2KI. In addition, axon elongation was also confirmed after SCI in CRMP2KI (Nagai et al., 2016) mice.The downregulation of genes regulating myelin sheath could be involved in suppressed retrograde axon degeneration in CRMP2KI mice. To summarize, the downregulation of these pathways in CRMP2KI may be directly or indirectly involved in pathophysiology of SCI, spine reduction, glial scar formation,and axon degeneration.
It is also important that we found several metabolic intracellular pathways and synaptic compartments are differentially expressed in CRMP2KI mice. Genes involved in translation, oxidative phosphorylation and proteasome pathways were upregulated and fatty acid metabolism,lysosome pathways, and synaptic compartments were downregulated. These changes in the metabolic pathways might come from protein-protein interactions of CRMP2.Previously, CRMP2 interactome analysis indicated that CRMP2 interacts with ten proteins that regulate energy metabolism and nine proteins that regulate protein metabolism including ribosomal subunit (Martins-de-Souza et al., 2015). The interactions between these proteins and mutated CRMP2 is not clear, but the phosphorylation inhibition of CRMP2 could change these interaction levels as well and affect the metabolic system overall. Our transcriptome results are partly consistent with and partly contradict the previous proteome study interestingly. The protein levels of molecules regulating oxidative phosphorylation, proteasome and fatty acid metabolism pathways were actually decreased in CRMP2KI mice (Nakamura et al., 2018). It is possible that the phosphorylation inhibition of CRMP2 broadly induce the improvements in SCI pathophysiology via intracellular metabolic pathways. In addition, a previous phosphoproteome analysis in CRMP2KI mice indicated proteins regulating synaptic functions had different phosphorylation profile as well (Nakamura et al., 2018). Our results that genes involved in regulating synaptic compartments were differentially expressed in CRMP2KI mice might be caused by this alteration of phosphorylation in synaptic proteins. We also found that genes regulating oxidative phosphorylation and mitochondrial components were even more upregulated in both CRMP2KI condition than SCI in WT mice. It is possible that the phosphorylation inhibition of CRMP2 might already have similar condition as observed after SCI in WT mice.
In conclusion, our study showed many possibilities of SCI pathophysiology in axotomized neurons in WT and CRMP2KI mice. Further, the transcriptome profile of CRMP2KI was revealed in detail. We confirmed that CRMP2KI suppressed the structural pathophysiology of spine reduction and somatic atrophy after SCI in cortex. In addition, translation and oxidative phosphorylation pathways were upregulated in cortex after SCI. Interestingly, fatty acid metabolism, lysosome,and neuronal system relating pathways were downregulated originally by phosphorylation inhibition of CRMP2 though translation, oxidative phosphorylation, and proteasome pathways were increased. After SCI in this CRMP2 mutation,neuronal system, synaptic, translation, glial cell differentiation,and myelin sheath pathways were decreased though only channel activity function was upregulated. This transcriptome analysis indicated (1) CRMP2KI seems to influence the whole metabolic system in CNS such as oxidative phosphorylation,fatty acid metabolism, and proteasome pathways, (2) these metabolic systems might contribute to the SCI pathophysiology in axotomized neurons as well and (3) response to SCI in CRMP2KI mice seems to involve synaptic activity and glial cell differentiation in addition to the putatively effective pathways such as microtubules stabilization. Hb involvement in SCI pathology is also suggested for the first time in this study.However in this study we could not cut in the functional level. In spite of the suggestions from mRNA transcriptome,locations that each protein works and protein interaction after SCI are not directly revealed. Cell-to-cell interaction in cortex is also unclear. At this point, our methods have some limitations.
The intracellular mechanisms underlying for axon elongation and functional recovery in CRMP2KI or Hb functions in axotomized neurons are still unclear in detail. In order to clarify the concrete mechanism on CNS regeneration, the complicated SCI pathophysiology and pathophysiological phenomena in CRMP2KI need to be further investigated.
Acknowledgments:We thank Sachiyo Aburatani, Hisashi Anbutsu and Yoriko Uotani for support in OIL, and support by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under Grant Number JP20am0101104.
Author contributions:AS, WP, MY, and KT performed experiments.AS, DT, HM, KA, and MH analyzed RNA sequencing data. TO and YG developed CRMP2KI mice. HT and TO supervised the overall project. AS and TO wrote the manuscript.
Conflicts of interest:The authors declare no conflict of interest.
Financial support:This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from The Ministry of Education, Culture,Sports, Science and Technology (No. 26430043; to T.O)
Institutional review board statement:All these experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at Waseda University, Japan (2017-A027 approved on March 21, 2017; 2018-A003 approved on March 25, 2018;2019-A026 approved on March 25, 2019).
Copyright license agreement:The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement:Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check:Checked twice by iThenticate.
Peer review:Externally peer reviewed.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 4.0 License, which allows others to remix,tweak, and build upon the work non-commercially, as long as appropriatecredit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Figure 1:Fast blue tracer positive region of sensorimotor cortex.
Additional Figure 2:Transcriptome analysis between WT and CRMP2KI mice.
Additional Figure 3:Transcriptome analysis after spinal cord injury in CRMP2KI.
Additional Table 1:Enriched pathways of upregulated DEGs by the phosphorylation deficient of CRMP2 (P < 0.05, q < 0.05).
Additional Table 2:Enriched pathways of downregulated DEGs by the phosphorylation deficient of CRMP2 (P < 0.05, q < 0.2).
Additional Table 3:Top20 significantly upregulated DEGs by the phosphorylation deficient of CRMP2.
Additional Table 4:Top20 significantly downregulated DEGs by the phosphorylation deficient of CRMP2.
Additional Table 5:Enriched pathways of upregulated DEGs after spinal cord injury in WT mice (P < 0.05, q < 0.05).
Additional Table 6:Enriched pathways of downregulated DEGs after spinal cord injury in CRMP2KI mice (P < 0.05, q < 0.05).
- 中国神经再生研究(英文版)的其它文章
- Clusterin: a multifaceted protein in the brain
- Neuroprotective effect of immunomodulatory peptides in rats with traumatic spinal cord injury
- Transcranial pulse current stimulation improves the locomotor function in a rat model of stroke
- Comparative transcriptomic analysis of rat versus mouse cerebral cortex after traumatic brain injury
- Delayed atomoxetine or fluoxetine treatment coupled with limited voluntary running promotes motor recovery in mice after ischemic stroke
- Extremely low frequency electromagnetic fields promote cognitive function and hippocampal neurogenesis of rats with cerebral ischemia

