PPR蛋白在植物细胞器组分转录后调控中的作用机制
郝媛媛,赵向前,黄福灯,李春寿
综 述
PPR蛋白在植物细胞器组分转录后调控中的作用机制
郝媛媛,赵向前,黄福灯,李春寿
浙江省农业科学院作物与核技术利用研究所,杭州 310021
PPR (pentatricopeptide repeats)蛋白是陆生植物中最大的蛋白家族之一,是一类RNA结合蛋白,参与细胞器基因的转录后加工过程,对细胞器的生物合成和功能具有深远影响。PPR突变后造成叶绿体光合电子传递链和线粒体呼吸链等进程受损,最终通过影响光合作用或者呼吸作用,造成植株生长发育异常,影响产量、育性、籽粒品质等。近年来,关于该蛋白家族成员参与植物生长发育的报道越来越多,但由于该家族庞大,大部分成员的功能还未被解析。本文系统总结了目前PPR蛋白调控细胞器基因转录后加工的分子机理及对细胞器和植株发育的影响,提出PPR家族还未解决的问题,为深入解析该基因家族的功能及育种应用提供理论基础。
PPR蛋白;转录后调控;细胞器代谢;植株生长发育
PPR (pentatricopeptide repeats)蛋白表达基因约在20多年前被发现[1,2],但是对PPR家族的系统研究,是从拟南芥()基因组测序之后才开始。对拟南芥基因组测序发现,将近31%的基因与已知功能的基因不同,而在未知功能的基因中,PPR基因占比6%,约有450个成员构成这个庞大的家族。由于与已知的TPR (tetratricopeptide repeat)蛋白结构域相似,学者们将其命名为PPR蛋白,TPR蛋白是一类蛋白结合型蛋白家族,而PPR蛋白属于RNA结合蛋白,这两种蛋白可以通过保守的氨基酸残基进行区分。PPR蛋白广泛存在于真核细胞中,原核生物只有在少数病原体和共生菌中存在,因此PPR蛋白是一类真核细胞特有的蛋白家族[3]。
近年来,在研究各种类型突变体的过程中发现PPR蛋白在植物不同生长发育阶段发挥重要作用,因此有关植物PPR成员的功能研究越来越多。目前,PPR大部分成员被定位于线粒体和叶绿体,对细胞器基因的转录后调控起重要作用,主要包括RNA剪接、RNA稳定、RNA切割、RNA翻译和RNA编辑[3]。一些PPR基因突变后,下游的RNA底物加工异常,导致产生异常的转录本,进而影响了叶绿体和线粒体的正常发育,从而影响植物光合作用和呼吸作用等重要生物进程,导致植物生长发育受阻。如此,研究庞大的PPR家族成员影响植物生长的分子机理至关重要。
对PPR家族分子机理的研究关键是探寻其所作用的RNA底物及作用方式。植物线粒体及叶绿体的基因组较小,且各基因在基因组的位置已研究的较为清楚,利用成熟的分子生物学手段,可较快速地定位目标RNA,并确定影响植物生长发育的机理。因此,掌握PPR基因对底物的作用机理及作用方式有助于探究并总结PPR家族对植物生长发育的作用。本文以水稻()、拟南芥、玉米()为例,总结了目前多个PPR成员的分子作用机理及不同的PPR突变体对植物形态建成的影响,以期为未来研究庞大的PPR家族成员提供理论依据及研究方向。
1 PPR蛋白分类
PPR蛋白主要分为P类和PLS类,其中P类是由相对保守的35个氨基酸串联重复排列而成,PLS类成员则是由P基序(35个氨基酸)、L (longer)基序(35~36个氨基酸)和S (shorter) (31~32个氨基酸)基序为一个重复单位排列而成[3]。此外,PLS类成员按照其C端的不同氨基酸序列又分为E亚类、E+亚类和DYW亚类。E和E+结构域包含34个氨基酸,这种结构类似于TPR蛋白,DYW结构域包含胞苷脱氨酶序列,其命名来源于3个保守的氨基酸—天冬氨酸(D)、酪氨酸(Y)和色氨酸(W)[3]。
2 PPR蛋白分子作用机理
PPR蛋白作为一种RNA结合蛋白,对mRNA转录后加工起着关键作用。P类成员主要参与RNA的剪接、稳定、切割和翻译过程,PLS类成员主要参与RNA的编辑过程。
2.1 PPR蛋白参与顺式和反式剪接
RNA剪接是指把内含子从前体mRNA (pre- mRNA)上移除,并将外显子连接起来的过程。与细胞核基因不同,叶绿体和线粒体基因的外显子并非连续分布在环状基因组上,而是包含多个多顺反子结构,需要顺式剪切和反式剪切形成多个转录中间产物,并分别去除内含子,才能将外显子连接起来形成成熟mRNA。发生在一条前体mRNA之间的剪接,称为顺式剪接(-splicing),多见于真核生物细胞核基因的剪接;发生在不同前体mRNA之间的剪接,称为反式剪接(-splicing),反式剪接可产生某个成熟mRNA的中间转录物,反式剪接多见于对叶绿体和线粒体基因组的初始转录物进行加工的过程中[4]。
需要剪切的叶绿体和线粒体基因内含子在不同物种中数量不同,被归类为I型内含子和II型内含子,其中大部分是II型内含子[4]。I型内含子由10个结构域构成(P1~P10),作用于RNA折叠,促进剪切和连接;II型内含子由6个双螺旋结构域构成(I~VI),形成复杂的空间结构,构成剪切的活性识别位点,每个结构域都有独特的作用,其剪切方式是经过两步转酯反应完成的,剪切机制类似于细胞核中的剪接体[5]。I型内含子和II型内含子的区别主要是二级结构和第一步剪切步骤的化学反应机制不同[6]。
II型内含子在植物中的分布较普遍,以水稻、玉米和拟南芥为例,拟南芥线粒体基因组9个基因共含有23个内含子,水稻和玉米线粒体基因组8个基因共含有22个内含子,且均为II型内含子。其中需要顺式剪切的内含子有17个(包含拟南芥中的一个内含子),需要反式剪切的内含子有6个[5]。目前,参与RNA剪接的PPR蛋白大多参与、、的顺反式剪切,参与、、RNA剪切的PPR蛋白还未见报道(图1)[7~28]。拟南芥叶绿体基因组有20个II型内含子和1个I型内含子(),分布在18个基因中;水稻和玉米叶绿体基因组有17个II型内含子和1个I型内含子(),分布在16个基因中[29]。这些内含子中只有的一个内含子属于I型内含子,其余均为II型内含子。此外,根据结构特异性和功能差异性,II型内含子可分为IIA组、IIB组、IIC组和IIE/F组[30]。在植物中,依据内含子的二级结构不同,Michel等[31]将叶绿体的II型内含子分为IIA和IIB两组。叶绿体基因只有的第一个内含子需反式剪切,且属于B组内含子,其余所有剪切方式均为顺式剪切。叶绿体中报道参与RNA剪接的PPR蛋白数量相对较少(图2)[32~42]。
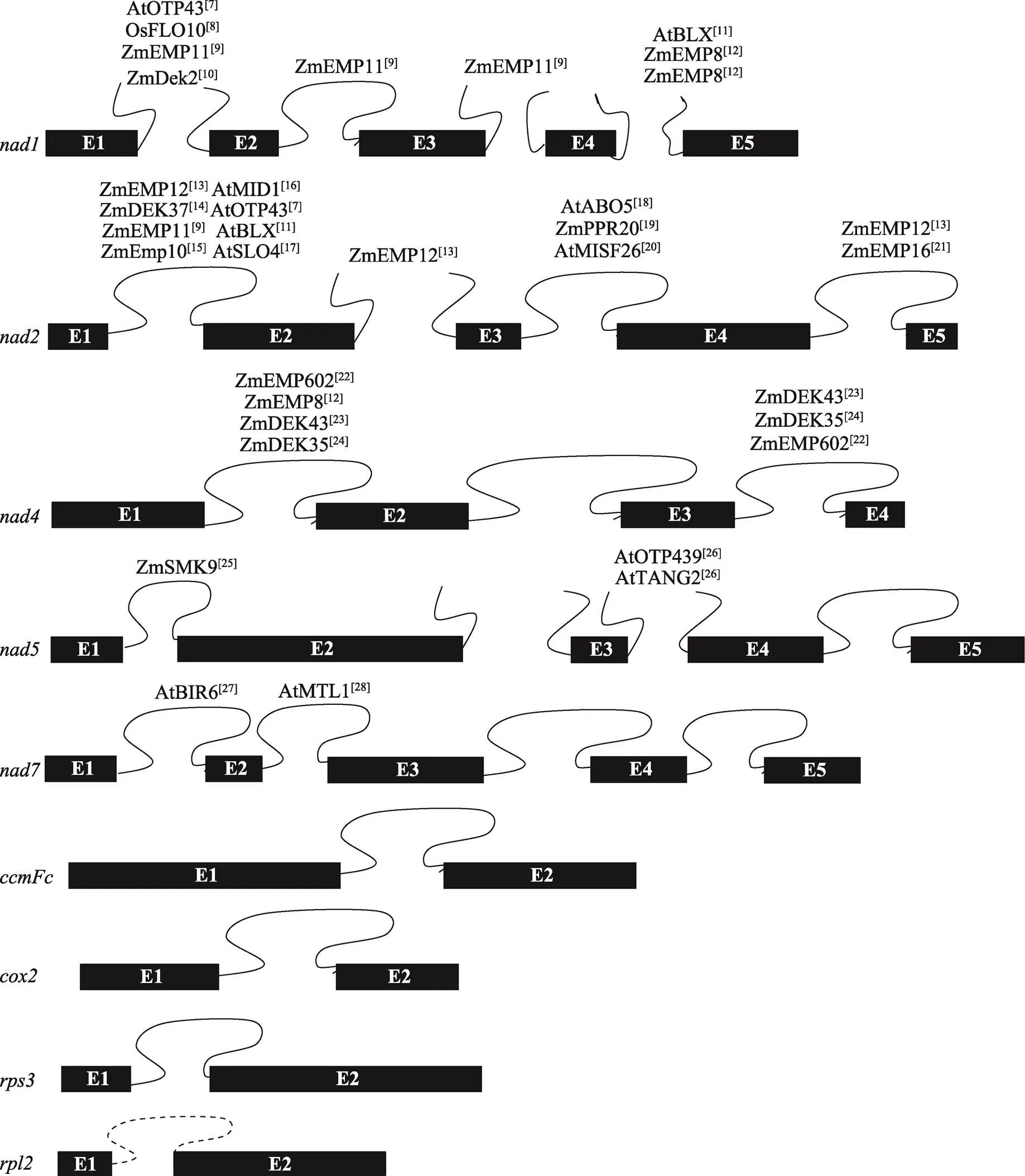
图1 水稻、玉米和拟南芥中参与线粒体内含子剪接的PPR蛋白
水稻和玉米包含22个内含子,拟南芥包含23个内含子,蛋白名称右上角为参考文献。Zm:玉米;Os:水稻;At:拟南芥。图中展示了9个包含内含子的基因结构图,拟南芥比水稻和玉米多了1个内含子(的内含子,虚线标注),黑色方框代表外显子(E),闭合曲线表示顺式剪接的内含子,未闭合的曲线代表反式剪接的内含子。

图2 水稻、玉米和拟南芥中参与叶绿体内含子剪接的PPR蛋白
水稻和玉米包含18个内含子,拟南芥包含21个内含子,蛋白右上角为参考文献。Zm:玉米;Os:水稻;At:拟南芥。是唯一一个I型内含子,其余均为II型内含子,且II型内含子分为两个亚组(Subgroup II A和Subgroup II B)。虚线左侧的内含子为B组内含子,虚线右侧的内含子为A组内含子。图中展示了包含内含子的基因结构图,拟南芥比水稻和玉米多了3个内含子(的1个内含子,的2个内含子,虚线标注)。黑色方框代表外显子(E),闭合曲线表示顺式剪接的内含子,未闭合的曲线代表反式剪接的内含子。
参与RNA剪接的PPR蛋白突变后,通常会造成成熟(spliced)的mRNA含量减少或缺失,而未剪接(unspliced)的前体mRNA含量增多。根据目前的报道,还没有一个PPR蛋白可参与某个基因所有内含子的剪接,可见一个基因,或者某个特定内含子,是由多个PPR蛋白和其他RNA加工类蛋白共同完成的,且这些RNA加工蛋白的氨基酸序列并不保守,这种现象反映了进化的多样性和剪接过程的复杂性。在进化过程中,不同物种线粒体和叶绿体基因组的内含子发生自然突变,进而招募相应的核编码蛋白(如PPR蛋白)进行剪接加工。若内含子在不同物种中的突变发生在单子叶和双子叶植物进化之后,则同一个内含子的剪接可能需要不同的PPR蛋白;若内含子的突变发生在单双子叶植物进化之前,则作用于内含子的PPR蛋白在不同物种中具有保守性。此外,同一个PPR蛋白也可参与多个基因内含子的剪切过程,这体现了PPR蛋白对下游底物作用机制的复杂性。
2.2 PPR蛋白参与mRNA稳定
PPR 蛋白稳定底物RNA主要是保护底物RNA免受核酸外切酶的非特异性切割。其作用机理是PPR蛋白结合于初始转录物的5ʹ或3ʹ末端,或者结合于多顺反子反式剪切中间转录物的5ʹ或3ʹ末端,作为一种“障碍”,阻挡5ʹ或3ʹ外切酶的非特异性切割,稳定mRNA不被降解。同时,由于所结合位置是固定的、特异性的,所以PPR也界定了转录物的5ʹ或3ʹ末端[43]。因此,PPR蛋白参与mRNA的稳定,既确定了转录物的末端,又保证了转录物免于降解,具有双重作用。由于线粒体和叶绿体基因组由多顺反子组成,因此界定正确的5ʹ或3ʹ末端这一功能对成熟mRNA的形成至关重要。
参与RNA剪接的PPR蛋白功能缺失时,通常会造成成熟的底物RNA转录本减少或缺失,未剪切的中间转录本增加,表明剪接过程受阻。相反,参与mRNA稳定的PPR蛋白功能缺失时,虽然也会阻止mRNA成熟,但是未剪切的转录本并未增多,反而是减少的,由于未剪切的中间转录物被5ʹ或3ʹ外切酶非特异性切割,稳定性降低并缺失,导致不能形成功能正常的成熟mRNA。因此,该分子机理是鉴定PPR蛋白参与底物RNA的剪接或稳定进程的依据。
叶绿体中同时拥有5ʹ和3ʹ核酸外切酶,PPR蛋白可结合于转录物的5ʹ或3ʹ末端,参与mRNA的稳定(图3A)。玉米ZmPPR10可结合于-、-基因间区[44];此外,玉米ZmATP4、ZmCRP1、拟南芥AtHCF152可分别作用于-基因间区、-基因间区、-基因间区[45~47],上述4个蛋白均能同时阻止5ʹ和3ʹ核酸外切酶的非特异性切割。拟南芥AtMRL1、AtPGR3、玉米ZmPPR103分别结合于、、的5ʹUTR,阻止5ʹ核酸外切酶的非特异性切割[48~50]。ZmPPR5较为特殊,它结合于的3ʹ内含子,促进的稳定,并促进剪切[33]。
线粒体基因组在进化过程中经过动态的进化和重排,只有3ʹ核酸外切酶,而无法检测到5ʹ核酸外切酶活性[51]。所以,在线粒体中,参与mRNA稳定的报道多集中于阻止3ʹ核酸外切酶的切割(图3B),而5ʹ端的加工为一种切割作用(见下文)。线粒体的成熟过程经过3个反式剪切过程,所以其前体转录物包含4个中间产物,AtMTSF2和AtPPR19均可结合于第二个前体int1b-exon2-int2-exon3-int3a的内含子3a处,阻止3ʹ核酸外切酶的切割[52,53];AtMTSF1结合于的3ʹUTR,免受3ʹ核酸外切酶的非特异性切割[54];ZmPPR78比较特殊,不参与稳定反式剪切的中间转录物,而是直接稳定成熟的转录物,且突变后,成熟转录本中缺失5ʹATG和3ʹTGA的不完整转录本增多,猜测其可通过阻止3ʹ核酸外切酶的切割稳定3ʹ端,但是由于未能检测到5ʹ核酸外切酶活性,其缩短的5ʹ端转录本还未能解释。由此可见,在线粒体中仍然有未被发现的mRNA加工进程待挖掘[55]。
2.3 PPR蛋白参与mRNA切割
PPR蛋白参与RNA切割包含两种方式:核酸外切酶方式和核酸内切酶方式。较典型的是PPR编码的RF (restorers of fertility)蛋白可以切割育性相关的线粒体RNAs,在三系配套育种工作中发挥重要作用,也是系谱法选育品种的理论依据。细胞质雄性不育(cytoplasmic male sterility, CMS)是一种母系遗传的线粒体遗传缺陷,产生不育的花粉,目前已在200多种高等植物中观察到CMS。与CMS相关的线粒体基因均是以嵌合体的结构分布在线粒体基因组上[57],这种有缺陷的线粒体基因编码的蛋白,使植物特定发育阶段的组织生长受阻从而产生不育的花粉。用包含RF蛋白的恢复系与不育系杂交产生F1杂交种,在F1的植株中携带育性恢复基因,即可恢复植株的雄性育性,其分子机理即PPR蛋白可对线粒体目标RNA正确切割,进而不能产生有毒蛋白,从而恢复F1杂交种的育性。目前大部分的RF蛋白都是PPR蛋白,包括矮牵牛花() RF[58]、油菜() Rfk1[59]、萝卜() Rfo[60]、高粱() PPR13[61]、水稻RF1[62]。以水稻BT-CMS育性恢复基因为例,该基因包含两个位于第10号染色体的PPR基因和。BT-CMS线粒体基因组包含两个拷贝的,分别为和。其中,下游为,ORF79蛋白是一个特异性表达于小孢子的毒性蛋白。的初始转录物为2 kb,Rf-1A参与3ʹ末端和5ʹ端基因间区的切割,加工后分为1 kb和0.4 kb,而BT-CMS中初始转录物不能正确切割,产生毒性蛋白导致不育;Rf-1B降解转录物,使植株不受毒性蛋白ORF79侵害,从而恢复育性,Rf-1A的作用上位于Rf-1B[63]。
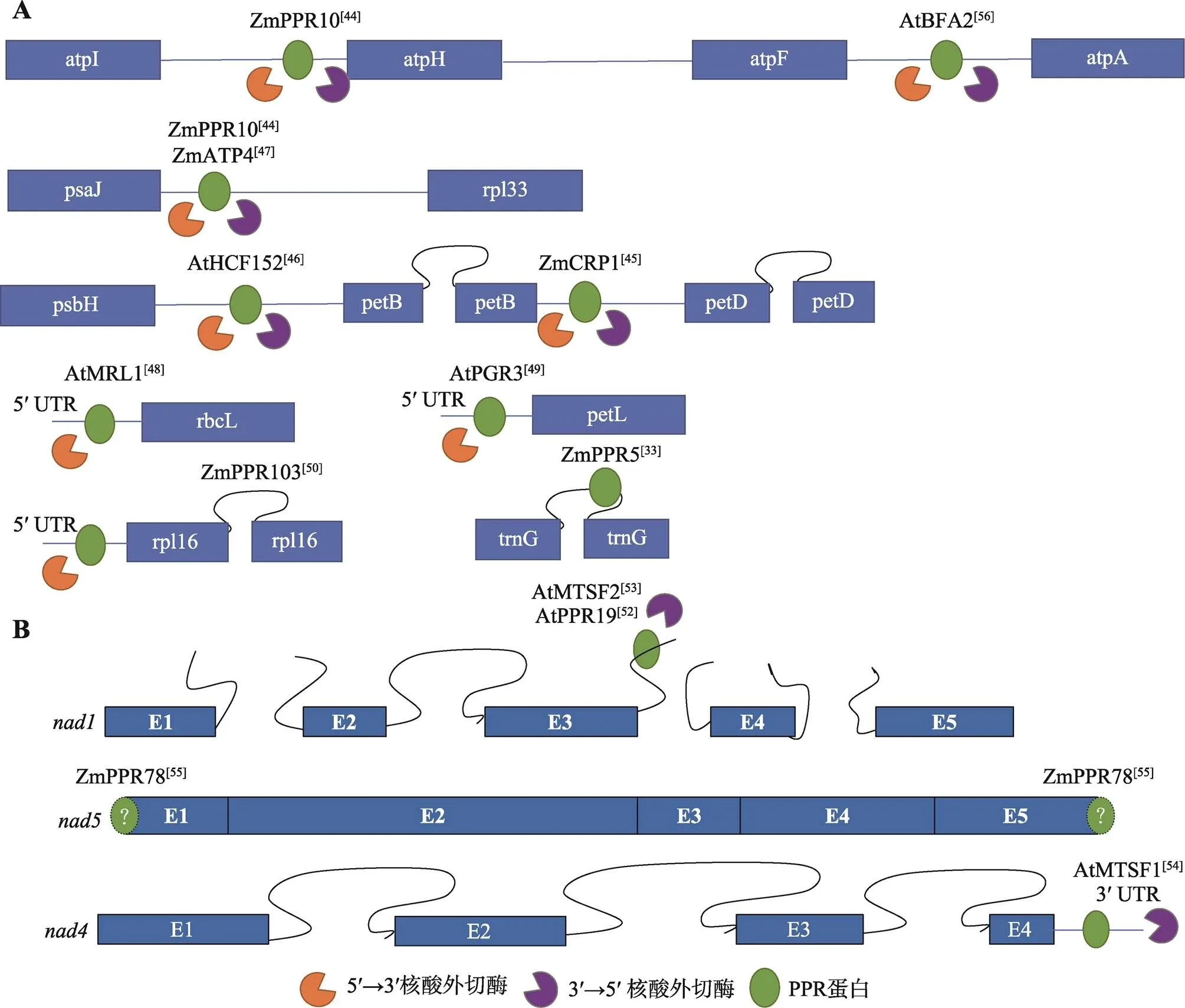
图3 水稻、玉米和拟南芥中参与叶绿体(A)和线粒体(B)转录物稳定的PPR蛋白
蛋白右上角为参考文献。蓝色方框代表外显子,闭合曲线代表顺式剪接的内含子,未闭合的曲线代表反式剪接的内含子,蓝色直线代表基因间区。
PPR蛋白参与5ʹ端切割的具体机制还未研究清楚,Barkan等[43]指出,可能的机制是切割位点的顺式作用元件可与PPR蛋白的结合位点形成一个RNA结构,当PPR蛋白结合于目标位点时,可暴露顺式作用元件,继而通过一种未知机制促进5ʹ端的加工成熟过程。拟南芥中与RF同源的蛋白称之为RFLs (restorers of fertility like),其中RPF1[64]作用于的5ʹUTR区、RPF2[65]作用于和的5'UTR区、RPF3[66]作用于的5ʹUTR区、RPF5[67]作用于和26S RNAs的5ʹUTR区,促进邻近的5ʹ端形成。鉴于线粒体5ʹ→3ʹ核酸外切酶的缺失,这些蛋白功能丧失后,通常形成5ʹ端异常长的转录本。
除此之外,一些包含DYW结构域的PPR蛋白本身就具有核酸内切酶的活性。CRR2是一个DYW型的PPR蛋白,可作为一种核酸内切酶,切割基因间区,参与的成熟[68],CRR22和CRR28在体外也被证实具有核酸内切酶的活性[69]。此外,PPR蛋白家族的一小部分成员C端包含SMR (small MutS-related)结构域,SMR结构域具有核酸内切酶活性[70]。目前PPR成员中,只发现SOT1的C段SMR结构域具有内切酶的活性,AtSOT1可识别23S–4.5S rRNA前体5ʹ末端13 bp的序列,正确切割前体末端形成成熟的23S及4.5S rRNA[71]。ZmPPR53是AtSOT1的同源蛋白,PPR53直接结合于23S rRNA 5ʹ端上游70 nt的位点,稳定23S rRNA不被5ʹ→3ʹ外切酶切割,也可直接结合于5ʹ端上游66 nt的位点,增加的翻译效率,但是其C端的SMR结构域功能还未能解释清楚[72]。
2.4 PPR蛋白参与mRNA翻译
PPR蛋白参与mRNA的翻译通常结合于底物的5ʹUTR区,促进底物RNA与核糖体的结合,从而激活或者抑制翻译。ZmPPR10[44]、AtPGR3[49]、ZmATP4[47]可分别激活的翻译过程,ZmCRP1[45]可同时激活和的翻译。
叶绿体中,PPR蛋白参与mRNA翻译的经典例子是PPR10。PPR10激活翻译的研究为了解叶绿体PPR介导的翻译激活的潜在机制提供了基础。在PPR10未结合的情况下,PPR10的结合位点和的核糖体结合位点(ribosome binding site, RBS)形成一个RNA发夹结构,PPR10结合后,可使RBS暴露,促进和核糖体结合,激活翻译,同时结合到该位点的PPR10也阻断5ʹ→3ʹ外切核糖核酸酶活性,从而兼具稳定的作用[43]。
线粒体中,PPR蛋白影响翻译的研究较少,且线粒体mRNA的核糖体结合位点不能被清楚认定[73],这就需要UTR特异性结合的PPR和其他蛋白质识别这些结合位点,对线粒体基因表达进行特异性的调节,例如ZmMPPR6结合于线粒体mRNA的5ʹUTR的3ʹ末端,5ʹUTR的RNA二级结构遮挡了起始密码子,从而抑制翻译起始,MPPR6可使这种二级结构松散,从而保证了与核糖体的正确结合,激活翻译[74]。
除了激活翻译外,参与mRNA切割的RF蛋白可抑制底物RNA的翻译。萝卜的不育花器官中成熟毒蛋白ORF138的含量比根中高10倍,而含育性恢复基因Rfo的可育植株中,花器官和其他组织的毒蛋白含量均显著下降。但是,不管在可育或不育的植株中,转录物含量不受影响,这表明Rfo影响了的翻译或翻译后进程,使蛋白表达量在不育和可育植株中含量不同,但是调控翻译的机制还未可知[60]。
2.5 PPR蛋白参与mRNA编辑
RNA编辑是一种发生在转录后核苷酸特异位点的加工修饰现象,包括核苷酸的插入、删除和转换,其中核苷酸的转换包括两种方式,A→I和C→U。A→I的编辑仅由作用于RNA的腺苷脱氨酶(adenosine deaminase acting on RNA, ADAR)完成,主要发生在动物、真菌及细菌中[75];高等植物中RNA编辑主要发生在线粒体与叶绿体中,C→U核苷酸替换的RNA编辑是高等植物中的普遍方式,该方式是由RNA编辑体统筹,通过对初始转录物编码的C残基进行脱氨基来完成的[76]。C→U的改变,(1)一般发生在密码子的第一个或者第二个碱基位点,造成氨基酸的改变;(2)产生新的AUG起始密码子或者消除初始转录物的无义终止密码子,从而形成稳定成熟的mRNA,翻译为有功能的蛋白;(3)编辑后的位点可能产生RNA二级结构,影响mRNA的剪接和稳定[77]。C→U RNA 编辑可以影响细胞器蛋白质的序列、改变调节基序、RNA-蛋白质相互作用或RNA二级结构,在RNA 加工过程中发挥重要作用[77]。目前的研究已确定水稻、玉米、拟南芥中所有的编辑位点,以水稻为例,线粒体共485个编辑位点,分布在36个线粒体基因上(https://www.ncbi. nlm.nih.gov/nucleotide/BA000029);叶绿体共21个编辑位点,分布在11个叶绿体基因上[78],本文列出了目前在水稻、玉米、拟南芥中报道的部分参与RNA编辑的PPR蛋白及对应的编辑位点[79~102](表1)。
RNA编辑体统筹RNA编辑过程,组成成员包括:PPR蛋白、MORFs (multiple organellar rna editing factors)蛋白、ORRM (organelle RNA recognition motif-containing)蛋白、PPO1 (protoporphyrinogen IX oxidase 1)蛋白、OZ1 (organelle zinc finger 1)蛋白[75]。其中PPR蛋白的PLS家族成员就是其中一种,主要作为反式作用因子,结合到编辑位点上游5~20个核苷酸的顺式作用元件上,使编辑位点被辨认出来,招募RNA编辑酶催化C→U的反应[91]。PLS家族成员C端的E1和E2结构域类似于PPR基序,可能通过蛋白互作或者结合RNA底物的方式参与编辑过程[103]。DYW结构域具有脱氨酶活性,可直接参与C→U编辑。
一个PPR蛋白可参与多个底物RNA的多个位点的编辑,同时某个特定RNA的编辑位点,可被不同的PPR蛋白识别。RNA编辑位点的编辑效率并不是100%,平均约有80%。不同组织或不同发育条件下,RNA编辑效率均会受影响[104]。
此外,PPR蛋白可与MORFs蛋白互作共同协作完成RNA的编辑过程。MORFs是叶绿体和线粒体编辑体成员,拟南芥中共10个成员,其中MORF1、3、4、6、7定位于线粒体,MORF2、9定位于叶绿体,MORF5、8双定位于线粒体和叶绿体。不同于PPR蛋白,MORF蛋白参与RNA编辑的过程是非特异性的,一个MORF蛋白即可参与相应细胞器中所有编辑位点的编辑过程。PPR蛋白的P基序或E1、E2结构域与MORF蛋白的N端或者中间区域互作,在不同的编辑位点可装配特异的蛋白复合体。MORF蛋白还可与PPR蛋白的L基序互作,减小PPR蛋白构象上P基序和S基序的距离,增加PLS结构域与底物RNA的亲和性[105]。

表1 水稻、玉米和拟南芥中部分参与叶绿体和线粒体RNA编辑的PPR蛋白
2.6 PPR蛋白通过特异的分子机制识别底物RNA
PPR蛋白的P、L、S基序分别具有保守的氨基酸位点,能区分不同的基序类别,并确定每个PPR重复的起始氨基酸[3]。Cheng等[106]发现,PPR基序识别下游RNA底物具有特异的分子机制。若PPR基序第五位的氨基酸是天冬酰胺(N),则对应的PPR基序结合嘧啶,若为丝氨酸(S)或苏氨酸(T),则结合嘌呤。此外,PPR基序的最后一位氨基酸是天冬氨酸(D),则结合尿嘧啶或鸟嘌呤,若为天冬酰胺(N),则结合胞嘧啶或腺苷酸。综上所述,每个PPR基序的第五位和最后一位氨基酸决定了所结合的RNA碱基。基于此规律,改变拟南芥PPR10对应位置的氨基酸就会改变对应的RNA识别位点[107]。
这种识别方式也适用于PLS型PPR蛋白,该类型蛋白最后一个S基序与编辑位点前的第4位核苷酸结合,该位置允许胞苷脱氨酶活性特异性作用于待编辑的胞苷。位于编辑位点上游5~20个核苷酸的顺式作用元件是PLS型PPR蛋白的识别位点[77]。PPR56可作为PLS型PPR蛋白特异性识别底物的模式蛋白,PPR56第五位与最后一位氨基酸的组合与相对应碱基可归纳为以下规律:T/S+N: A, T/S+D: G, N+N: C/U, N+S: C>U, N+ D: U>C,这与其底物和对应的顺式作用元件相吻合[77]。
值得注意的是,这种结合机制并不适合所有的PPR蛋白。一些P类家族成员在每个重复之间可能存在多余的氨基酸残基,并不是保守的每35个氨基酸重复紧密相连。而且每个重复第五位的氨基酸和最后一个氨基酸并不是保守地按照上述规律排列,不同的PPR蛋白差异很大,所以还需进一步补充这种分子机制,完善PPR蛋白结合底物RNA的规律。
3 PPR蛋白对细胞器发育的影响
3.1 PPR蛋白影响线粒体和叶绿体发育
几乎所有PPR突变体表现出的表型均是由一种或几种线粒体或叶绿体基因产物的功能缺失。尽管PPR家族成员很多,但不同家族成员之间的功能几乎没有冗余,不同物种中某些同源的PPR蛋白也具有不同的作用底物。PPR突变体的表型主要包括:光合缺陷[35,45,48,68]、叶片发育异常[108]、色素积累[109,110]、生长受阻[8]、胚或胚乳发育异常[7,9,10]、脱落酸信号途径受损[111]和胞质雄性不育[58~62]。
在真核细胞中,线粒体是一种半自主性细胞器,有自身的基因组。在进化过程中,线粒体大部分基因整合到宿主细胞核基因组中,经过转录翻译后,转运到线粒体,参与线粒体代谢和线粒体基因表达调控[112]。陆生植物线粒体基因组包含60~70个基因,这些基因编码的蛋白包括tRNAs、rRNAs、核糖体蛋白、复合体I(NADH脱氢酶)亚基、复合体III(细胞色素C还原酶)亚基、复合体IV(细胞色素C氧化酶)亚基、ATP合酶亚基和细胞色素C合成酶亚基等。
线粒体为植物发育提供了能量,其功能丧失对植物生长有害。线粒体PPR蛋白分子功能的确定有助于阐明RNA加工机制以及氧化磷酸化机制的组装。复合体I嵌入线粒体内膜并介导电子从NADH转移至泛醌[113],是电子进入电子传输链的主要入口点,多个研究表明PPR参与的剪接缺陷能影响复合体I的装配和稳定[13,114,115]。Nad1、Nad2和Nad4蛋白是复合物I的膜臂成分[116],Nad1形成醌结合位点,Nad2是复合物I中质子转移的位点,而Nad4、Nad5形成质子易位子之一,并且在结构上与K或Na+/H+逆向运输蛋白有关。由于需要复合物I中Nad1、Nad2、Nad4、Nad5参与质子转移和醌键结合,PPR蛋白表达受阻而导致的、异常转录都会严重降低复合物I的含量,使活性显著降低[10,12,22,25]。
PPR蛋白ZmEMP8、ZmDEK43、AtPPR19[12,23,52]功能异常时,复合体I亚基功能的缺失还可导致细胞色素途径受损,线粒体氧化磷酸化效率降低,导致植物出现代谢问题[117],诱发交替途径起始[24],导致交替氧化酶基因的转录水平提高。此外,OsNPPR1、OsFlo10[8,118]突变后,由于电子传递效率降低,使得呼吸链产生的ATP含量显著下降[117]。氧化磷酸化中产生的活性氧(reactive oxygen species, ROS)在触发植物细胞程序性死亡(programmed cell death, PCD)中起关键作用[119]。通常,PCD从胚乳的中部开始,然后扩散到外围[120]。PPR突变体由于氧化磷酸化进程的异常,导致产生过量的ROS,使突变体胚乳比野生型更早的发生PCD,这会干扰淀粉和贮藏蛋白的合成[121],所以定位于线粒体的PPR蛋白突变通常会造成胚乳粉质和皱缩的表型。可见,线粒体PPR功能缺陷型突变体,会导致线粒体发育受损,影响线粒体功能。
真核细胞中,叶绿体是光合作用的主要场所,其基因组包含约100个基因左右,这些基因主要参与光合作用、ATP合成和基质中蛋白的转录翻译和降解过程。叶绿体光合作用需要光合电子传递链,由3种复合物PS I、PS II和Cytb6f组成。PPR蛋白HCF152[46]、MRL1[48]、PGR3[49]等突变,通过影响光合电子传递链复合物的成员转录后加工,从而造成叶绿体发育异常;叶绿体ATP合酶是位于类囊体膜上的一个多亚基复合体,它利用光合作用电子传递产生的质子动力将ADP转化为ATP。叶绿体ATP合酶由CFo和CF1模块组成,其中由叶绿体基因参与的亚基产生于两个多顺反子叶绿体转录单位和[122]。PPR蛋白WSL4[32]、ATP4[47]通过影响合酶成员的剪接和翻译,影响叶绿体中ATP的合成过程;此外,在叶绿体基质中存在叶绿体蛋白酶体clp复合体、叶绿体RNA聚合酶复合体以及一些核糖体蛋白[56]。PPR蛋白WSL[42]、CRR2[68]、CLB19[109]的突变影响这些途径叶绿体mRNA的成熟过程。
3.2 PPR蛋白在细胞核中的功能
有一类特殊的PPR蛋白,亚细胞定位于细胞核,包括GRP23、PNM1、SORA1和OsNPPR1。其中GRP23只定位于细胞核,与RNA聚合酶II亚基III互作,突变后致死[123]。PNM1双定位于细胞核和线粒体,其突变体的致死表型只与线粒体定位有关,在细胞核中PNM1可与转录因子TCP互作,调控线粒体氧化磷酸化过程相关的核基因表达[124]。GRP23与细胞核RNA聚合酶II亚基III互作,PNM1与细胞核转录因子TCP8和NAP1互作,说明PPR定位于细胞核可以影响核mRNA的转录和转录后调控进程。此外,TCP8转录因子特异性的转录与线粒体氧化磷酸化途径相关的细胞核基因[125],说明定位于细胞核的PPR蛋白可能通过参与定位于细胞器的核基因的转录后调控过程,参与细胞器的代谢过程。在水稻中,OsNPPR1是定位于细胞核的PPR蛋白,但却影响了线粒体的功能,猜测可能是参与了细胞核中与线粒体发育相关基因的转录后调控而间接影响线粒体发育[118];SOAR1双定位于细胞核和胞质,参与ABA信号途径,作用于ABAR1下游和ABI5的上游[126]。
4 PPR功能展望
本文系统综述了水稻、玉米、拟南芥中PPR蛋白参与转录后调控的分子机理和性状表现。PPR蛋白可参与整个植物生育期的发育过程,影响植株的生长,对植物的正常长成有重要作用。近年来,对PPR成员的功能研究也越来越多。但是,仍有一些重要的科学问题没有解决:(1)庞大的PPR蛋白家族成员是否仅调控100多个细胞器基因的表达;(2) E和E+型的PLS PPR蛋白结合底物后,招募编辑酶的分子机制还未知;(3)萝卜的不育花器官中成熟毒蛋白ORF138的含量极高,而含育性恢复基因Rfo的可育植株中,各组织毒蛋白含量均显著下降。但是,不管在可育或不育的植株中,转录物含量不受影响,虽然参与翻译的过程,但是调控翻译的机制还不清楚;(4)参与RNA切割的线粒体PPR蛋白的切割机制还未知;(5)虽然通过对PPR10的分子机理进行系统的研究,发现了可能存在的PPR基序与底物结合的“密码”[99],但是由于PPR家族各成员之间保守性相差较大,所以“密码”并不适用于所有的PPR成员,是否有更精细明确的结合方式存在还有待探索。
细胞器基因的正常表达,需要PPR家族成员协同工作,研究PPR蛋白可定向控制细胞器基因的表达,为利用生物工程技术,改良作物光合作用和呼吸作用进程提供理论基础。
[1] Manthey GM, Mcewen JE. The product of the nuclear geneis required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondriallocus of., 1995, 14(16): 4031–4043.
[2] Coffin JW, Dhillon R, Ritzel RG, Nargang FE. Thenuclear gene encodes a protein with a region of homology to thePET309 protein and is required in a post-transcriptional step for the expression of the mitochondrially encoded COXI protein., 1997, 32(4): 273–280.
[3] Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, Lecharny A, Le Ret M, Martin-Magniette ML, Mireau H, Peeters N, Renou JP, Szurek B, Taconnat L, Small I. Genome-wide analysis ofpentatricopeptide repeat proteins reveals their essential role in organelle biogenesis., 2004, 16(8): 2089–2103.
[4] Bonen L.- and-splicing of group II introns in plant mitochondria., 2008, 8(1): 26–34.
[5] Brown GG, des Francs-Small CC, Ostersetzer-Biran O. Group II intron splicing factors in plant mitochondria., 2014, 5: 35.
[6] Huang WH, Zhu YJ, Wu WJ, Li X, Zhang DL, Yin P, Huang JR. The pentatricopeptide repeat protein SOT5/ EMB2279 is required for plastidandintron splicing., 2018, 177(2): 684–697.
[7] De Longevialle AF, Meyer EH, Andrés C, Taylor NL, Lurin C, Millar AH, Small ID. The pentatricopeptide repeat geneis required for-splicing of the mitochondrialintron 1 in., 2007, 19(10): 3256–3265.
[8] Wu MM, Ren YL, Cai MH, Wang YL, Zhu SS, Zhu JP, Hao YY, Teng X, Zhu XP, Jing RN, Zhang H, Zhong MS, Wang YF, Lei CL, Zhang X, Guo XP, Cheng ZJ, Lin QB, Wang J, Jiang L, Bao YQ, Wang YH, Wan JM. Riceencodes a pentatricopeptide repeat protein that is essential for the- splicing of mitochondrialintron 1 and endosperm development., 2019, 223(2): 736–750.
[9] Ren XM, Pan ZY, Zhao HL, Zhao JL, Cai MJ, Li J, Zhang ZX, Qiu FZ. EMPTY PERICARP11 serves as a factor for splicing of mitochondrialintron and is required to ensure proper seed development in maize., 2017, 68(16): 4571–4581.
[10] Qi WW, Yang Y, Feng XZ, Zhang ML, Song RT. Mitochondrial function and maize kernel development requires Dek2, a pentatricopeptide repeat protein involved inmRNA splicing., 2017, 205(1): 239– 249.
[11] Sun Y, Huang JY, Zhong S, Gu HY, He S, Qu LJ. Novel DYW-type pentatricopeptide repeat (PPR) protein BLX controls mitochondrial RNA editing and splicing essential for early seed development of., 2018, 45(3): 155–168.
[12] Sun F, Zhang XY, Shen Y, Wang HC, Liu R, Wang XM, Gao DH, Yang YZ, Liu YW, Tan BC. The pentatricopeptide repeat protein EMPTY PERICARP8 is required for the splicing of three mitochondrial introns and seed development in maize., 2018, 95(5): 919–932.
[13] Sun F, Xiu ZH, Jiang RC, Liu YW, Zhang XY, Yang YZ, Li XJ, Zhang X, Wang Y, Tan BC. The mitochondrial pentatricopeptide repeat protein EMP12 is involved in the splicing of threeintrons and seed development in maize., 2019, 70(3): 963–972.
[14] Dai DW, Luan SC, Chen XZ, Wang Q, Feng Y, Zhu CG, Qi WW, Song RT. Maize Dek37 encodes a P-type PPR protein that affects-splicing of mitochondrialintron 1 and seed development., 2018, 208(3): 1069–1082.
[15] Cai MJ, Li SZ, Sun F, Sun Q, Zhao HL, Ren XM, Zhao YX, Tan BC, Zhang ZX, Qiu FZ.encodes a mitochondrial PPR protein that affects the-splicing ofintron 1 and seed development in maize., 2017, 91(1): 132–144.
[16] Zhao P, Wang F, Li N, Shi DQ, Yang WC. Pentatricopeptide repeat protein MID1 modulatesintron 1 splicing anddevelopment., 2020, 10(1): 2008.
[17] Weissenberger S, Soll J, Carrie C. The PPR protein SLOW GROWTH 4 is involved in editing ofand affects the splicing ofintron 1., 2017, 93(4–5): 355–368.
[18] Liu Y, He JN, Chen ZZ, Ren XZ, Hong XH, Gong ZZ., encoding a pentatricopeptide repeat protein required for-splicing of mitochondrialintron 3, is involved in the abscisic acid response in., 2010, 63(5): 749–765.
[19] Yang YZ, Ding S, Wang Y, Wang HC, Liu XY, Sun F, Xu CH, Liu BH, Tan BC. PPR20 is required for the-splicing of mitochondrialintron 3 and seed development in maize., 2020, 61(2): 370–380.
[20] Wang CD, Aubé F, Quadrado M, Dargel-Graffin C, Mireau H. Three new pentatricopeptide repeat proteins facilitate the splicing of mitochondrial transcripts and complex I biogenesis in., 2018, 69(21): 5131–5140.
[21] Xiu ZH, Sun F, Shen Y, Zhang XY, Jiang RC, Bonnard G, Zhang JH, Tan BC. EMPTY PERICARP16 is required for mitochondrialintron 4-splicing, complex I assembly and seed development in maize., 2016, 85(4): 507–519.
[22] Ren ZJ, Fan KJ, Fang T, Zhang JJ, Yang L, Wang JH, Wang GY, Liu YJ. Maizeencodes a P-type PPR protein that is essential for seed development., 2019, 60(8): 1734–1746.
[23] Ren RC, Wang LL, Zhang L, Zhao YJ, Wu JW, Wei YM, Zhang XS, Zhao XY. DEK43 is a P-type pentatricopeptide repeat (PPR) protein responsible for the-splicing ofin maize mitochondria., 2020, 62(3): 299–313.
[24] Chen XZ, Feng F, Qi WW, Xu LM, Yao DS, Wang Q, Song RT. Dek35 encodes a PPR protein that affects-splicing of mitochondrialintron 1 and seed development in maize., 2017, 10(3): 427– 441.
[25] Pan ZY, Liu M, Xiao ZY, Ren XM, Zhao HL, Gong DM, Liang K, Tan ZD, Shao YQ, Qiu FZ. ZmSMK9, a pentatricopeptide repeat protein, is involved in the-splicing of, kernel development and plant architecture in maize., 2019, 288: 110205.
[26] Des Francs-Small CC, De Longevialle AF, Li YH, Lowe E, Tanz SK, Smith C, Bevan MW, Small I. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartitetranscript encoding a subunit of mitochondrial complex I., 2014, 165(4): 1409–1416.
[27] Koprivova A, Des Francs-Small CC, Calder G, Mugford ST, Tanz S, Lee BR, Zechmann B, Small I, Kopriva S. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrialtranscripts., 2010, 285(42): 32192–32199.
[28] Haïli N, Planchard N, Arnal N, Quadrado M, Vrielynck N, Dahan J, Des Francs-Small CC, Mireau H. The MTL1 pentatricopeptide repeat protein is required for both translation and splicing of the mitochondrialmRNA in., 2016, 170(1): 354–366.
[29] De Longevialle AF, Small ID, Lurin C. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles., 2010, 3(4): 691–705.
[30] PYLE AM. Group II Intron Self-Splicing., 2016, 45(1): 183–205.
[31] MICHEL F, UMESONO K, OZEKI H. Comparative and functional anatomy of group II catalytic introns—a review., 1989, 82(1): 5–30.
[32] Wang Y, Ren YL, Zhou KN, Liu LL, Wang JJ, Xu Y, Zhang H, Zhang L, Feng ZM, Wang LW, Ma WW, Wang YL, Guo XP, Zhang X, Lei CL, Cheng ZJ, Wan JM.encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development., 2017, 8: 1116.
[33] Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts., 2008, 28(17): 5337–5347.
[34] Wang XM, Yang ZP, Zhang Y, Zhou W, Zhang AH, Lu CM. Pentatricopeptide repeat protein PHOTOSYSTEM I BIOGENESIS FACTOR2 is required for splicing of., 2020, 62(11): 1741–1761.
[35] Khrouchtchova A, Monde RA, Barkan A. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts., 2012, 18(6): 1197–1209.
[36] de Longevialle AF, Hendrickson L, Taylor NL, Delannoy E, Lurin C, Badger M, Millar AH, Small I. The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the-splicing of plastidintron 2 in., 2008, 56(1): 157–168.
[37] Chateigner-Boutin AL, Des Francs-Small CC, Delannoy E, Kahlau S, Tanz SK, De Longevialle AF, Fujii S, Small I. OTP70 is a pentatricopeptide repeat protein of the E subgroup involved in splicing of the plastid transcript., 2011, 65(4): 532–542.
[38] Schmitz-Linneweber C, Williams-Carrier RE, Williams- Voelker PM, Kroeger TS, Vichas A, Barkan A. A pentatricopeptide repeat protein facilitates the-splicing of the maize chloroplastpre-mRNA., 2006, 18(10): 2650–2663.
[39] Aryamanesh N, Ruwe H, Sanglard LVP, Eshraghi L, Bussell JD, Howell KA, Small I, Des Francs-Small CC. The pentatricopeptide repeat protein EMB2654 Is essential for-splicing of a chloroplast small ribosomal subunit transcript., 2017, 173(2): 1164– 1176.
[40] Lee K, Park SJ, des Francs-Small CC, Whitby M, Small I, Kang H. The coordinated action of PPR4 and EMB2654 on each intron half mediates-splicing oftranscripts in plant chloroplasts., 2019, 100(6): 1193–1207.
[41] Liu X, Lan J, Huang YS, Cao PH, Zhou CL, Ren YK, He NQ, Liu SJ, Tian YL, Nguyen T, Jiang L, Wan JM. Corrigendum: WSL5, a pentatricopeptide repeat protein, is essential for chloroplast biogenesis in rice under cold stress., 2018, 69(18): 4495.
[42] Tan JJ, Tan ZH, Wu FQ, Sheng PK, Heng YQ, Wang XH, Ren YL, Wang JL, Guo XP, Zhang X, Cheng ZJ, Jiang L, Liu XM, Wang HY, Wan JM. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice., 2014, 7(8): 1329–1349.
[43] Barkan A, Small I. Pentatricopeptide repeat proteins in plants., 2014, 65: 415–442.
[44] Pfalz J, Bayraktar OA, Prikryl J, Barkan A. Site-specific binding of a PPR protein defines and stabilizes 5' and 3' mRNA termini in chloroplasts., 2009, 28(14): 2042–2052.
[45] Barkan A, Walker M, Nolasco M, Johnson D. A nuclear mutation in maize blocks the processing and translation of several chloroplast mRNAs and provides evidence for the differential translation of alternative mRNA forms., 1994, 13(13): 3170–3181.
[46] Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. HCF152, anRNA binding pentatricopeptide repeat protein involved in the processing of chloroplastRNAs., 2003, 15(6): 1480–1495.
[47] Zoschke R, Kroeger T, Belcher S, Schöttler MA, Barkan A, Schmitz-Linneweber C. The pentatricopeptide repeat-SMR protein ATP4 promotes translation of the chloroplastmRNA., 2012, 72(4): 547– 558.
[48] Johnson X, Wostrikoff K, Finazzi G, Kuras R, Schwarz C, Bujaldon S, Nickelsen J, Stern DB, Wollman FA, Vallon O. MRL1, a conserved pentatricopeptide repeat protein, is required for stabilization ofmRNA inand., 2010, 22(1): 234–248.
[49] Cai WH, Okuda K, Peng LW, Shikanai T. PROTON GRADIENT REGULATION 3 recognizes multiple targets with limited similarity and mediates translation and RNA stabilization in plastids., 2011, 67(2): 318–327.
[50] Hammani K, Takenaka M, Miranda R, Barkan A. A PPR protein in the PLS subfamily stabilizes the 5'-end of processedmRNAs in maize chloroplasts., 2016, 44(9): 4278–4288.
[51] Ruwe H, Wang GW, Gusewski S, Schmitz-Linneweber C. Systematic analysis of plant mitochondrial and chloroplast small RNAs suggests organelle-specific mRNA stabilization mechanisms., 2016, 44(15): 7406–7417.
[52] Lee K, Han JH, Park YI, des Francs-Small CC, Small I, Kang H. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization oftranscripts and is crucial for mitochondrial function anddevelopment., 2017, 215(1): 202–216.
[53] Wang CD, Aubé F, Planchard N, Quadrado M, Dargel- Graffin C, Nogué F, Mireau H. The pentatricopeptide repeat protein MTSF2 stabilizes aprecursor transcript and defines the 3' end of its 5' -half intron., 2017, 45(10): 6119–6134.
[54] Haïli N, Arnal N, Quadrado M, Amiar S, Tcherkez G, Dahan J, Briozzo P, des Francs-Small CC, Vrielynck N, Mireau H. The pentatricopeptide repeat MTSF1 protein stabilizes themRNA inmitochondria., 2013, 41(13): 6650–6663.
[55] Zhang YF, Suzuki M, Sun F, Tan BC. The Mitochondrion- Targeted PENTATRICOPEPTIDE REPEAT78 protein is required formature mRNA stability and seed development in maize., 2017, 10(10): 1321– 1333.
[56] Zhang L, Zhou W, Che LP, Rochaix JD, Lu CM, Li WJ, Peng LW. PPR Protein BFA2 is essential for the accumulation of thetranscript in chloroplasts., 2019, 10: 446.
[57] Hanson MR, Bentolila S. Interactions of mitochondrial and nuclear genes that affect male gametophyte development., 2004, 16Suppl(Suppl): S154– S169.
[58] Dahan J, Mireau H. The Rf and Rf-like PPR in higher plants, a fast-evolving subclass of PPR genes., 2013, 10(9): 1469–1476.
[59] Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants., 2002, 99(16): 10887–10892.
[60] Brown GG, Formanová N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang JF, Cheung WY, Landry BS. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats., 2003, 35(2): 262–272.
[61] Klein RR, Klein PE, Mullet JE, Minx P, Rooney WL, Schertz KF. Fertility restorer locusof sorghum (L) encodes a pentatricopeptide repeat protein not present in the colinear region of rice chromosome 12., 2005, 111(6): 994–1012.
[62] Komori T, Ohta S, Murai N, Takakura Y, Kuraya Y, Suzuki S, Hiei Y, Imaseki H, Nitta N. Map-based cloning of a fertility restorer gene,, in rice (L)., 2004, 37(3): 315–325.
[63] Wang ZH, Zou YJ, Li XY, Zhang QY, Chen LT, Wu H, Su DH, Chen YL, Guo JX, Luo D, Long YM, Zhong Y, Liu YG. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing., 2006, 18(3): 676–687.
[64] Hölzle A, Jonietz C, Törjek O, Altmann T, Binder S, Forner J. A RESTORER OF FERTILITY-like PPR gene is required for 5'-end processing of themRNA in mitochondria of., 2011, 65(5): 737–744.
[65] Jonietz C, Forner J, Hölzle A, Thuss S, Binder S. RNA PROCESSING FACTOR2 is required for 5' end processing ofandmRNAs in mitochondria of., 2010, 22(2): 443–453.
[66] Jonietz C, Forner J, Hildebrandt T, Binder S. RNA PROCESSING FACTOR3 is crucial for the accumulation of maturetranscripts in mitochondria ofaccession Columbia., 2011, 157(3): 1430–1439.
[67] Hauler A, Jonietz C, Stoll B, Stoll K, Braun HP, Binder S. RNA processing factor 5 is required for efficient 5' cleavage at a processing site conserved in RNAs of three different mitochondrial genes in., 2013, 74(4): 593–604.
[68] Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplastin., 2003, 36(4): 541–549.
[69] Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage inchloroplasts., 2009, 21(1): 146–156.
[70] Liu S, Melonek J, Boykin L M, Small I, Howell KA. PPR-SMRs: ancient proteins with enigmatic functions., 2013, 10(9): 1501–1510.
[71] Zhou W, Lu QT, Li QW, Wang L, Ding SH, Zhang AH, Wen XG, Zhang LX, Lu CM. PPR-SMR protein SOT1 has RNA endonuclease activity., 2017, 114(8): E1554–E1563.
[72] Zoschke R, Watkins KP, Miranda RG, Barkan A. The PPR-SMR protein PPR53 enhances the stability and translation of specific chloroplast RNAs in maize., 2016, 85(5): 594–606.
[73] Hazle T, Bonen L. Comparative analysis of sequences preceding protein-coding mitochondrial genes in flowering plants., 2007, 24(5): 1101–1112.
[74] Manavski N, Guyon V, Meurer J, Wienand U, Brettschneider R. An essential pentatricopeptide repeat protein facilitates 5' maturation and translation initiation ofmRNA in maize mitochondria., 2012, 24(7): 3087–3105.
[75] Yan JJ, Zhang QX, Yin P. RNA editing machinery in plant organelles., 2018, 61(2): 162– 169.
[76] Walbot V. RNA editing fixes problems in plant mitochondrial transcripts., 1991, 7(2): 37–39.
[77] Small ID, Schallenberg-Rudinger M, Takenaka M, Mireau H, Ostersetzer-Biran O. Plant organellar RNA editing: what 30 years of research has revealed., 2020, 101(5): 1040–1056.
[78] Corneille S, Lutz K, Maliga P. Conservation of RNA editing between rice and maize plastids: are most editing events dispensable?, 2000, 264(4): 419–424.
[79] Li XJ, Zhang YF, Hou MM, Sun F, Shen Y, Xiu ZH, Wang XM, Chen ZL, Sun SSM, Small I, Tan BC.encodes a pentatricopeptide repeat protein required for mitochondrialtranscript editing and seed development in maize () and rice ()., 2014, 79(5): 797–809.
[80] Wang HC, Sayyed A, Liu XY, Yang YZ, Sun F, Wang Y, Wang MD, Tan BC. SMALL KERNEL4 is required for mitochondrialtranscript editing and seed development in maize., 2020, 62(6): 777–792.
[81] Liu YJ, Xiu ZH, Meeley R, Tan BC.encodes a pentatricopeptide repeat protein that is required for mitochondrial RNA editing and seed development in maize., 2013, 25(3): 868– 883.
[82] Sun F, Wang XM, Bonnard G, Shen Y, Xiu ZH, Li XJ, Gao DH, Zhang ZH, Tan BC.encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required forediting, mitochondrial function and seed development in maize., 2015, 84(2): 283–295.
[83] Yang YZ, Ding S, Wang HC, Sun F, Huang WL, Song S, Xu CH, Tan BC. The pentatricopeptide repeat protein EMP9 is required for mitochondrialandtranscript editing, mitochondrial complex biogenesis and seed development in maize., 2017, 214(2): 782–795.
[84] Li XL, Huang WL, Yang HH, Jiang RC, Sun F, Wang HC, Zhao J, Xu CH, Tan BC. EMP18 functions in mitochondrialandtranscript editing and is essential to seed development in maize., 2019, 221(2): 896–907.
[85] Qi WW, Tian ZR, Lu L, Chen XZ, Chen XZ, Zhang W, Song RT. Editing of mitochondrial transcriptsandby Dek10 is essential for mitochondrial function and maize plant development., 2017, 205(4): 1489–1501.
[86] Li XJ, Gu W, Sun SL, Chen ZL, Chen J, Song WB, Zhao HM, Lai JS.encodes a PPR protein required for seed development in maize., 2018, 60(1): 45–64.
[87] Ding S, Liu XY, Wang HC, Wang Y, Tang JJ, Yang YZ, Tan BC. SMK6 mediates the C-to-U editing at multiple sites in maize mitochondria., 2019, 240: 152992.
[88] Wang Y, Liu XY, Yang YZ, Huang J, Sun F, Lin JS, Gu ZQ, Sayyed A, Xu CH, Tan BC.encodes a novel PPR-DYW protein that is required for mitochondrial RNA editing at multiple sites, complexes I and V biogenesis, and seed development in maize., 2019, 15(8): e1008305.
[89] Doniwa Y, Ueda M, Ueta M, Wada A, Kadowaki KI, Tsutsumi N. The involvement of a PPR protein of the P subfamily in partial RNA editing of anmitochondrial transcript., 2010, 454(1–2): 39–46.
[90] Boussardon C, Salone V, Avon A, Berthomé R, Hammani K, Okuda K, Shikanai T, Small I, Lurin C. Two interacting proteins are necessary for the editing of thesite inplastids., 2012, 24(9): 3684–3694.
[91] Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing., 2007, 104(19): 8178–8183.
[92] Sung TY, Tseng CC, Hsieh MH. The SLO1 PPR protein is required for RNA editing at multiple sites with similar upstream sequences inmitochondria., 2010, 63(3): 499–511.
[93] Verbitskiy D, van der Merwe JA, Zehrmann A, Härtel B, Takenaka M. The E-class PPR protein MEF3 ofcan also function in mitochondrial RNA editing with an additional DYW domain., 2012, 53(2): 358–367.
[94] Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage inchloroplasts., 2009, 21(1): 146–156.
[95] Okuda K, Hammani K, Tanz SK, Peng LW, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastidandtranscripts., 2010, 61(2): 339–349.
[96] Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. A study of newchloroplast RNA editing mutants reveals general features of editing factors and their target sites., 2009, 21(11): 3686–3699.
[97] Zehrmann A, Van Der Merwe J, Verbitskiy D, Härtel B, Brennicke A, Takenaka M. The DYW-class PPR protein MEF7 is required for RNA editing at four sites in mitochondria of., 2012, 9(2): 155–161.
[98] Takenaka M. MEF9, an E-subclass pentatricopeptide repeat protein, is required for an RNA editing event in thetranscript in mitochondria of., 2010, 152(2): 939–947.
[99] Verbitskiy D, Härtel B, Zehrmann A, Brennicke A, Takenaka M. The DYW-E-PPR protein MEF14 is required for RNA editing at site-1895 in mitochondria of., 2011, 585(4): 700–704.
[100] Kim SR, Yang JI, Moon S, Ryu CH, An K, Kim KM, Yim J, An G. Rice OGR1 encodes a pentatricopeptide repeat-DYW protein and is essential for RNA editing in mitochondria., 2009, 59(5): 738–749.
[101] Toda T, Fujii S, Noguchi K, Kazama T, Toriyama K. Rice MPR25 encodes a pentatricopeptide repeat protein and is essential for RNA editing oftranscripts in mitochondria., 2012, 72(3): 450–460.
[102] Zhang ZG, Cui XA, Wang YW, Wu JX, Gu XF, Lu TG. The RNA editing factor WSP1 is essential for chloroplast development in rice., 2017, 10(1): 86–98.
[103] Rugen N, Straube H, Franken LE, Braun HP, Eubel H. Complexome profiling reveals association of PPR proteins with ribosomes in the mitochondria of plants., 2019, 18(7): 1345–1362.
[104] Chateigner-Boutin AL, Hanson MR. Developmental co-variation of RNA editing extent of plastid editing sites exhibiting similar-elements., 2003, 31(10): 2586–2594.
[105] Yan JJ, Zhang QX, Guan ZY, Wang Q, Li L, Ruan FY, Lin RC, Zou TT, Yin P. MORF9 increases the RNA-binding activity of PLS-type pentatricopeptide repeat protein in plastid RNA editing., 2017, 3: 17037.
[106] Cheng SF, Gutmann B, Zhong X, Ye YT, Fisher MF, Bai FQ, Castleden I, Song Y, Song B, Huang JY, Liu X, Xu X, Lim BL, Bond CS, Yiu SM, Small I. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants., 2016, 85(4): 532–547.
[107] Barkan A, Rojas M, Fujii S, Yap A, Chong YS, Bond CS, Small I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins., 2012, 8(8): e1002910.
[108] Petricka JJ, Clay NK, Nelson TM. Vein patterning screens and the defectively organized tributaries mutants in., 2008, 56(2): 251–263.
[109] Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A, Andrés C, De La Luz Gutiérrez-Nava M, Cantero A, Delannoy E, Jiménez LF, Lurin C, Small I, León P. CLB19, a pentatricopeptide repeat protein required for editing ofandchloroplast transcripts., 2008, 56(4): 590–602.
[110] Zhou WB, Cheng YX, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang JR. The Arabidopsis geneencoding a DYW protein is required for editing oftranscripts and the rapid development of chloroplasts during early growth., 2009, 58(1): 82–96.
[111] Mei C, Jiang SC, Lu YF, Wu FQ, Yu YT, Liang S, Feng XJ, Comeras SP, Lu K, Wu Z, Wang XF, Zhang DP.pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signalling., 2014, 65(18): 5317–5330.
[112] Millar AH, Whelan J, Soole KL, Day DA. Organization and regulation of mitochondrial respiration in plants., 2011, 62: 79–104.
[113] Lee CP, Taylor NL, Millar AH. Recent advances in the composition and heterogeneity of themitochondrial proteome., 2013, 4: 4.
[114] Zhu CG, Jin GP, Fang P, Zhang Y, Feng XZ, Tang YP, Qi WW, Song RT. Maize pentatricopeptide repeat protein DEK41 affects-splicing of mitochondrialintron 3 and is required for normal seed development., 2019, 70(15): 3795–3808.
[115] Ren RC, Lu XD, Zhao YJ, Wei YM, Wang LL, Zhang L, Zhang WT, Zhang CY, Zhang XS, Zhao XY. Pentatricopeptide repeat protein DEK40 is required for mitochondrial function and kernel development in maize., 2019, 70(21): 6163–6179.
[116] Hirst J. Mitochondrial complex I., 2013, 82: 551–575.
[117] Noctor G, De Paepe R, Foyer CH. Mitochondrial redox biology and homeostasis in plants., 2007, 12(3): 125–134.
[118] Hao YY, Wang YL, Wu MM, Zhu XP, Teng X, Sun YL, Zhu JP, Zhang YY, Jing RN, Lei J, Li JF, Bao XH, Wang CM, Wang YH, Wan JM. The nuclear-localized PPR protein OsNPPR1 is important for mitochondrial function and endosperm development in rice., 2019, 70(18): 4705–4720.
[119] Wu J, Sun YF, Zhao YN, Zhang J, Luo L, Li M, Wang JL, Yu H, Liu GF, Yang LS, Xiong GS, Zhou JM, Zuo JR, Wang YH, Li JY. Deficient plastidic fatty acid synthesis triggers cell death by modulating mitochondrial reactive oxygen species., 2015, 25(5): 621– 633.
[120] Young TE, Gallie DR. Regulation of programmed cell death in maize endosperm by abscisic acid., 2000, 42(2): 397–414.
[121] Young TE, Gallie DR, Demason DA. Ethylene-mediated programmed cell death during maize endosperm development of wild-type andgenotypes., 1997, 115(2): 737–751.
[122] Hahn A, Vonck J, Mills DJ, Meier T, Kühlbrandt W. Structure, mechanism, and regulation of the chloroplast ATP synthase., 2018, 360(6389): eaat4318.
[123] Ding YH, Liu NY, Tang ZS, Liu J, Yang WC.is essential for early embryogenesis and encodes a novel nuclear PPR motif protein that interacts with RNA polymerase II subunit III., 2006, 18(4): 815–830.
[124] Hammani K, Gobert A, Hleibieh K, Choulier L, Small I, Giegé P. Andual-localized pentatricopeptide repeat protein interacts with nuclear proteins involved in gene expression regulation., 2011, 23(2): 730–740.
[125] Martín-Trillo M, Cubas P. TCP genes: a family snapshot ten years later., 2010, 15(1): 31–39.
[126] Mei C, Jiang SC, Lu YF, Wu FQ, Yu YT, Liang S, Feng XJ, Comeras SP, Lu K, Wu Z, Wang XF, Zhang DP.pentatricopeptide repeat protein SOAR1 plays a critical role in abscisic acid signalling., 2014, 65(18): 5317–5330.
The role of PPR proteins in posttranscriptional regulation of organelle components in plants
Yuanyuan Hao, Xiangqian Zhao, Fudeng Huang, Chunshou Li
Pentatricopeptide repeat (PPR) proteins constitute one of the largest protein families in land plants. They are sequence-specific RNA-binding proteins and play key roles in posttranscriptional processes within organelles. Their combined actions have profound effects on chloroplast photosynthetic electron transport chain and mitochondrial respiratory chain, affecting photosynthesis and respiration respectively, and ultimately on yield, fertility, and grain quality. Over the past decade, much has been learned about the molecular functions of these proteins on plant growth and development. However, due to the large size of this protein family, the functions of most membersremain largely unknown.Here, we summarize the molecular mechanisms of PPR proteins functions on organelle genes, and effects on development of organelles and plants. Problems that need to be resolved are also identified. This article will provide a theoretical basis for understanding the functions of PPR protein family and genetic improvements of grain yield and quality.
PPR proteins; post-transcriptional regulation; organelle metabolism; plant growth and development
2021-06-30;
2021-08-20
国家自然科学基金项目(编号:32001524)资助[Supported by the National Natural Science Foundation of China (No. 32001524)]
郝媛媛,博士,助理研究员,研究方向:稻米品质的改良和分子机理。E-mail: 499085663@qq.com
李春寿,学士,研究员,研究方向:籼型杂交稻的选育。E-mail: lichunshou@126.com
10.16288/j.yczz.21-233
2021/10/26 17:08:41
URI: https://kns.cnki.net/kcms/detail/11.1913.R.20211026.1131.002.html
(责任编委: 宋任涛)

