SMARCC1 copy number variation is related to metastatic colon cancer:an investigation based on TCGA data*
Libo Feng,Yu Liu,Dong Xia,Xiaolong Chen (✉)
1 Department of Gastrointestinal Surgery,Southwest Medical University,Luzhou 646000,China
2 Department of Electrocardiogram,Affiliated Hospital of Southwest Medical University,Luzhou 646000,China
Abstract Objective There are no well-defined genetic indicators for distant metastatic illness in patients with colon cancer (CC).The discovery of genetic changes linked to metastatic CC might aid in the development of systemic and local therapeutic approaches.Using The Cancer Genome Atlas (TCGA),we examined the relationship between copy number variation (CNV) of SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily C member 1 (SMARCC1) and distant metastatic illness in patients with CC.Methods Genetic sequencing data of all relevant CC patients and clinical features were collected from TCGA using R.There were 506 CC patients with CNV and clinical outcome data.The CNV of SMARCC1 was examined for its correlation with distant metastatic disease using the TCGA CC dataset (M1 vs.M0).After adjusting for age,sex,T stage,N stage,adjuvant chemotherapy,microsatellite instability (MSI),and surgical margin status,univariate and multivariate logistic regression analyses were performed.Results SMARCC1 CNV was linked to distant metastatic disease (P=0.012 and 0.008 in univariate and multivariate analysis,respectively);positive lymph nodes and margin status were also associated with distal metastases (all P < 0.01).MSI,T stage,N stage,adjuvant treatment,sex,race,and MSI were not associated with metastases (all P > 0.05).Conclusion SMARCC1 CNV is associated with distant metastatic disease in patients with CC.In individuals with CC,such genetic profiles might be utilized therapeutically to support optimal systemic treatment options against local treatments for CC,such as radiation therapy,pending additional confirmation.
Key words:colon cancer (CC);copy number variation (CNV);genetic marker
According to Global Cancer Statistics (Globocan 2018),led by the International Agency for Research on Cancer(IARC) of the World Health Organization (WHO),the worldwide incidence of newly developed colorectal cancer (CRC) in 2018 was approximately 1.85 million,making it the 3rd most common malignant tumor.Approximately 880,000 died of CRC,making it the 2nd most common cause of cancer-related deaths[1].In addition,around 20% of all current patients with CRC have distant metastatic illness[1].Despite advancements in screening and therapy,patients with CRC still have a poor 5-year overall survival rate[1].Some genes linked to the existence and progression of CRC,as well as treatment response indicators,have been identified[2–4];however,genetic markers linked to metastatic CRC are still lacking.Surgical resection,chemotherapy,radiation,and targeted therapy may be applied in the treatment of CRC depending on the stage and location of the original tumor.In individuals with rectal adenocarcinoma,radiation treatment improves local control and overall survival[5].Copy number variation (CNV) is caused by genome rearrangement and refers to an increase or decrease in the copy number of large genome fragments with lengths of more than 1 kb,mainly showing deletion and duplication at the submicroscopic level[6–7].CNV is an important component of the structural variation (SV)of the genome.The mutation rate of CNV sites is much higher than that of single nucleotide polymorphisms(SNPs),which are an important pathogenic factor in human disease[6].
For a variety of malignancies,CNVs have been utilized to assess prognosis and subsequent therapy profiles,such asMYCNamplification in neuroblastoma[8].CNV of genes in primary CRC tumors and their corresponding metastatic disease locations has long been studied,but little is known about how CNVs of genes in the primary tumor of CRC are associated with localized illness and distant metastases[9–14].One key component of the SWI/SNF complex involved in epigenetic control of genome transcription is the SWI/SNF-related,matrix-associated,actin-dependent regulator of chromatin subfamily C member 1 (SMARCC1)[15–18].It is thought to act as a tumor suppressor in a variety of malignancies,including pancreatic and renal carcinomas[19–20].However,its involvement in CRC is unclear and controversial.Using a dataset of 524 individuals with colon cancer (CC) from The Cancer Genome Atlas (TCGA),we evaluated the relationship ofSMARCC1CNV with distant metastatic disease.
Materials and methods
Patient data
In July 2021,a cohort of 524 samples (506 cases were determined after data integration) from original tumor specimens were chosen from the TCGA database using R based on the availability of metastatic staging and copy number data.The results presented here are based on data downloaded from the TCGA Research Network (http://cancergenome.nih.gov/) on July 22nd.
Clinicopathological data
Clinical data on age,sex,race/ethnicity,and staging were obtained from the TCGA data portal.The TCGA data portal also included pathologic information such as tumor size,tumor location,resection margins,lymph node status,and metastatic status upon diagnosis.
CNV analysis
The TCGA level 3 CNV data for colorectal cancer were obtained from the TCGA data portal.Each sample was processed and normalized using TCGA level 3 CNV(Affymetrix Genome-Wide SNP Array 6.0).The study was conducted using mean copy number estimations of regions that overlapped the whole genome.The gene was classified using the Genomic Identification of Significant Targets in Cancer (GISTIC) algorithm.CNV was characterized as a copy number ≥1 or ≤1.
Statistical analysis
The univariate relationship ofSMARCC1CNV with variables in the CC set was investigated using the chisquared test or Fisher’s exact test,as applicable.A logistic regression model was used to perform a univariate study of metastatic disease (M1vs.M0) using predictors and variables.A logistic regression model was used to conduct a multivariate analysis of metastatic disease(M1vs.M0) by inputting all factors in the model and utilizing a backward variable selection technique with an alpha level of removal of 0.1.The model was modified to includeSMARCC1CNV,chemotherapy,N stage,and T stage.R (version 4.1.0) was used for all analyses,and the threshold for significance was set at 0.05.Following the discovery of a link betweenSMARCC1CNV and distant metastatic disease in the colorectal cohort,SMARCC1CNV data were analyzed to determine whether the CNV represented a gain or a loss,and the majority of the CNVs were homozygous deletions,indicating copy number loss.
Results
A total of 506 individuals with CC were included in the CC copy number analysis group (Table 1).The median age was 68 years (range,31–90 years).
Pathological characteristics
Patients with distant metastatic disease were identified in 68 (13.4%) of the CC copy number analysis cohort,whereas 204 (40.3%) had positive nodal disease.
Genetic analysis for all patients
There were 22 samples with CNV ofSMARCC1,with homozygous deletions resulting in a reduction in copy number.SMARCC1CNV was shown to have a statistically significant relationship with distant metastatic disease (P=0.017) in the univariate analysis.SMARCC1CNV was shown to have no link to positive nodal disease,positive resection margins,sex,race,or T stage (Table 2).
The occurrence of distant metastatic disease was observed to be related to the existence ofSMARCC1CNV in the univariate analysis of metastatic disease (Table 2;P=0.012).Distant metastases were also connected with N1-2 nodal disease (P< 0.001),R1-2 (P< 0.001) resection,Adjuvant treatment (P< 0.001),but had no retaltion with MSI (P=0.993),T3 or T4 advanced local disease(P=0.984),adjuvant chemotherapy (P=0.985),sex (P=0.530),and race (P=0.500).
On multivariate analysis (Table 2),the presence ofSMARCC1CNV was associated with distant metastatic disease (P=0.008),as well as N1-2 nodal disease (P<0.001).The data for race and MSI were removed from the multivariate model to determine the amount of NA data.

Table 1 Sample characteristics of patients with colon cancer included in copy number analysis (n=506)
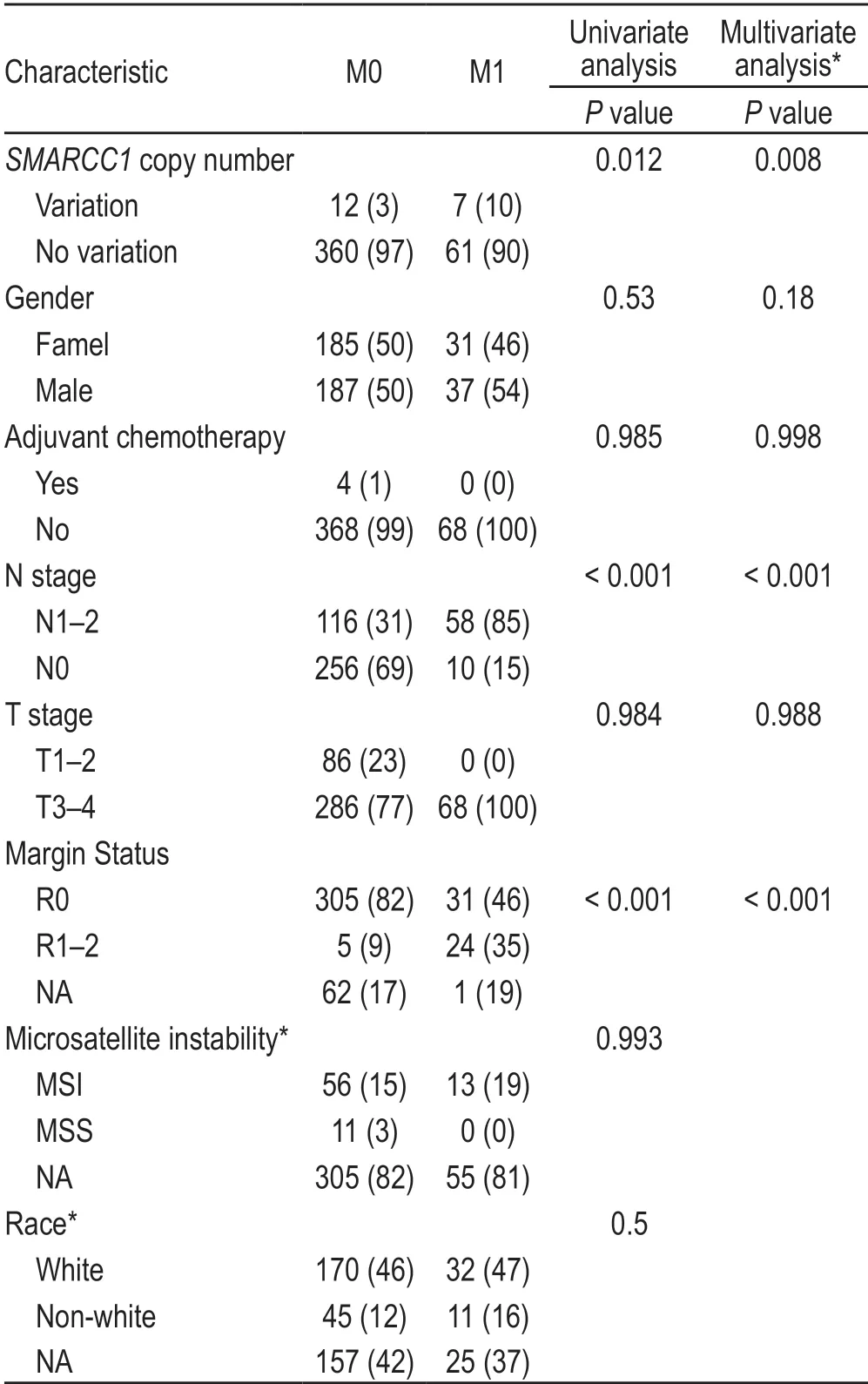
Table 2 Univariate and multivariate analysis for presence of distant metastatic disease (n,%)
Discussion
An examination of data of 506 individuals using the TCGA revealed thatSMARCC1CNV was independently associated with distant metastatic disease in CC.We usedmultivariate analysis,which included age,sex,surgical margin status,T stage,N stage,MSI,and adjuvant chemotherapy,to corroborate our findings.According to our data,SMARCC1CNV is strongly linked with distant metastatic disease in individuals with all types of CC.Furthermore,our findings imply that CNV ofSMARCC1might be a valuable genetic marker for personalizing CC treatment and identifying individuals who would benefit from more aggressive treatment methods.
When various genetic markers,such as mutations,gene expression,methylation status,and CNV,were included in the TCGA analysis,no indication of a substantial genetic relationship with distant metastatic illness was found[21].Our work is unusual in that it uses a larger database with more samples of distant metastatic illness to determine a CNV of a single gene that is independently linked with distant metastatic disease.LowSMARCC1expression was linked to the development of extrahepatic metastases in a study of 30 individuals with CC[22].In line with these findings,the majority ofSMARCC1CNVs in our population were homozygous deletions,suggesting that the gene was downregulated.SMARCC1CNV is linked to more aggressive CC,as evidenced by the relationship with nodal disease and distant metastasis in our cohort.
The new cohort included a large number of samples of both colectomy and metastatic illness,allowing for a more statistically powerful correlation analysis.Nodal disease was linked to distant metastasis and increases with advanced T stage.The molecular route of tumorigenesis is another element that influences the development of distant metastatic diseases.Hereditary non-polyposis CC is linked to tumors that initially present in the left colon with a reduced risk of metastasis,as well as MSI.Several studies have found that distinct molecular and clinical characteristics exist in various parts of the colon[2,9,23–24];thus,we adjusted for MSI when looking for a link betweenSMARCC1CNV and distant metastatic illness.SMARCC1CNV was not found to be associated with MSI/MSS on univariate analysis (P=0.879),andSMARCC1was independently associated with distant metastatic disease (P=0.018) in the multivariate analysis,despite the expected trend between distant metastatic disease in the CC cohort and microsatellite stability (MSS) (P=0.993) in the multivariate analysis.These data show that the link betweenSMARCC1CNV and distant metastatic illness is not based on genetic differences between rightand left-sided primary malignancies.
Multiple studies have demonstrated that the number of lymph nodes sampled after CC surgery is related to the outcome in stage II CC,suggesting that further therapy might prevent the disease from spreading[25–27].With increased evidence of tumor dissemination in the early stages of CC,additional indications,such asSMARCC1CNV,may be beneficial in directing therapy toward a more aggressive regimen aimed at avoiding tumor spread.Future research with a longitudinal design will be required to elucidateSMARCC1’s role as a molecular marker that affects therapy,allowing for greater association with the development of advanced illness with distant metastases.
We note that,in addition to the retrospective approach,this study has limitations.The data came from individuals who had their original tumor resected;thus,there were no cases of advanced,unresectable illness in the study.Some clinical information,such as race/ethnicity,was not recorded,and some CNVs may be linked more with various nationalities/backgrounds[28].Only the capacity to compare genetic changes with the occurrence of metastatic illness was possible in the retrospective analysis.The lack of a prospective component following genetic analysis restricts the application of the findings and prevents the identification of variables linked to development of distant metastasis.Although prior studies have found somatic alterations and CNV in the genomes of several cancer types,no previous study has linkedSMARCC1CNV at the original location to distant metastatic illness.The link might have an effect on therapeutic options and could even be used as a marker for targeted molecular therapy.
Conclusions
We found a statistically significant association betweenSMARCC1CNV and distant metastatic disease in CC and rectal adenocarcinoma in our sample.SMARCC1CNV may be utilized clinically to support optimal systemic treatment methods against more aggressive local treatments in patients with CC,including radiation therapy for rectal adenocarcinoma,pending additional confirmation in longitudinal investigations.
Conflicts of interest
The authors indicated no potential conflicts of interest.
 Oncology and Translational Medicine2021年5期
Oncology and Translational Medicine2021年5期
- Oncology and Translational Medicine的其它文章
- Special IgD-λ type multiple myeloma based on bone marrow cell morphology:A case report
- Evaluation of endocrine therapy combined with intensity modulated radiation therapy in patients with advanced prostate cancer
- CircBAGE2 (hsa_circ_0061259) regulates CCND1 and PDCD10 expression by functioning as an miR-103a-3p ‘sponge’ to alter the proliferation and apoptosis of prostate cancer cells*
- Ultrasound-guided No Touch liver pedicle microwave ablation in hepatocellular carcinoma treatment
- Cone beam computed tomography-guided differences among registration methods for lung cancer and the effects of tumor position,treatment model,and tumor size on positioning errors*
- Identification of microRNA-21 as a valuable diagnostic marker of oral squamous cell carcinoma and potential target*
