水稻黄绿叶突变体ygl3的鉴定与基因功能分析
许子怡,程行,沈奇,赵亚男,汤佳玉,刘喜
水稻黄绿叶突变体的鉴定与基因功能分析
许子怡,程行,沈奇,赵亚男,汤佳玉,刘喜
淮阴师范学院/江苏省环洪泽湖生态农业生物技术重点实验室/江苏省区域现代农业与环境保护协同创新中心,江苏淮安 223300
【】为了丰富和加深人们对植物叶色形成的分子机理认识,对水稻黄绿叶突变体()进行表型鉴定和基因克隆,阐明的分子功能,为解析调控水稻叶色形成的分子机理奠定基础。从水稻中花11 CRISPR-Cas9敲除突变体库中鉴定出2份稳定遗传的等位黄绿叶突变体,命名为与,对突变体的表型进行鉴定,测定野生型和突变体苗期的叶绿素含量,运用透射电镜观察野生型和突变体的叶绿体结构。利用实时荧光定量PCR分析的组织表达模式,并使用BioXM 2.6软件对YGL3及其同源蛋白序列进行比对,采用酵母双杂交方法筛选YGL3的互作蛋白。在苗期,与野生型相比,突变体叶片黄化,叶绿素、类胡萝卜素和总光合色素含量显著降低。透射电镜结果表明,突变体叶绿体形态异常,类囊体片层结构较少,而野生型叶绿体形态正常,类囊体片层结构排列有序。CRISPR-Cas9敲除位点鉴定结果表明,发生单碱基插入,导致蛋白翻译提前终止,该基因编码351个氨基酸的蛋白突变为55个氨基酸的截短蛋白。与野生型相比,的表达水平在突变体中显著下调。qRT-PCR结果表明在水稻根、穗、种子、叶鞘以及叶片中均有表达,且叶片中表达水平最高。编码一个质体定位的尿嘧啶核苷酸激酶。蛋白氨基酸序列比对表明YGL3蛋白在玉米、高粱、拟南芥等物种中均较为保守,与拟南芥同源蛋白氨基酸的同源性为59.4%。qRT-PCR结果表明,叶绿素合成基因(如、和)在突变体中显著下调,而、和等叶绿素合成基因在野生型与突变体之间无显著差异。酵母双杂交系统筛选水稻叶片酵母cDNA文库,发现YGL3与RNA编辑因子MORF8存在互作。水稻黄绿叶突变体的表型是由突变导致,该基因与已报道的水稻黄绿叶基因等位。在叶片中高度表达,同时YGL3与MORF8在酵母中互作。
水稻(L.);黄绿叶;YGL3;CRISPR-Cas9;叶绿体发育
0 引言
【研究意义】水稻(L.)是研究植物功能基因的模式作物,也是世界三分之二人口的口粮。加强水稻分子生物学研究,对水稻分子设计育种具有重要意义。叶绿体是植物主要的光合器官,是脂肪、淀粉、激素等代谢产物的合成场所,对植物产量的获得具有十分重要的作用[1-2]。叶色变异是一种常见的突变性状,突变基因影响叶绿素的合成、降解以及叶绿体的发育。多数水稻叶色变异突变体光合作用效率下降,植株生长发育受阻,造成水稻减产[3-4]。叶色变异通常出现在苗期,也会出现在分蘖期和抽穗期,叶色表型易于识别鉴定。近年来,水稻叶色变异相关基因的鉴定与克隆,丰富了水稻叶色形成的分子机制,为水稻高光效育种提供基因资源。此外,黄化、白条纹、白化转绿等叶色变异表型可以作为形态标记,用于保证杂交稻制种和不育系繁殖的纯度[5-6]。【前人研究进展】水稻叶色变异可以分为条纹、白化、黄化、斑点叶、白化转绿和持绿等。植物叶绿素合成以谷氨酰-tRNA为底物,最终生成叶绿素a和叶绿素b,涉及27个基因编码的15种酶[7]。目前,水稻中已经克隆了14个直接参与叶绿素合成的基因,这些基因突变都会引起叶色异常[8-10]。编码谷氨酰胺tRNA还原酶,其突变体表现出淡绿叶的表型[8]。编码叶绿素合成酶,突变植株表现为叶片黄化[9]。编码镁原卟啉Ⅸ单酯环化酶的催化亚基,参与叶绿素合成[10]。目前,水稻中已鉴定并克隆叶色变异基因超过120个,多数涉及叶绿素的生物合成、降解以及叶绿体的发育[11-12]。此外,质-核信号转导途径受阻[13]、转录后修饰[14]、质体转录复合物形成受阻[15]、叶绿体蛋白酶调控受阻[16-17]、叶绿体蛋白转运受阻[18]、ATP转运蛋白基因[19]、转录因子与表观遗传[20-21]等突变机制也被鉴定出来。【本研究切入点】目前,约120个水稻叶色相关基因已被克隆,但水稻叶色形成调控网络极其复杂,需要鉴定更多的涉及叶色变异的功能基因,而利用CRISPR-Cas9技术创制突变体库,从中筛选鉴定叶色变异突变体,是鉴定叶色控制基因的有效途径。【拟解决的关键问题】本研究从中花11 CRISPR- Cas9突变体库中筛选获得黄绿叶突变体,对突变体进行表型鉴定、光合色素含量测定和叶绿体形态结构观察,并克隆,进行的组织表达分析、蛋白氨基酸序列分析以及互作蛋白的筛选,以期解析调控叶绿体发育的分子机理,为在水稻分子育种上的应用奠定基础。
1 材料与方法
1.1 试验材料
从中花11 CRISPR-Cas9突变体库中鉴定到2份叶片黄化突变体与。2个黄绿叶突变体经多代自交,黄绿叶能够稳定遗传。
1.2 光合色素含量测定
选取生长一致的野生型和突变体各10株,参考Porra等[22]方法测定苗期叶片的光合色素含量。称取0.2 g叶片,剪碎放入含有5 mL 95%无水乙醇的离心管中,摇匀黑暗处理48h,每隔12 h摇匀一次,设置3次生物重复。用双通道紫外分光光度计(TU1901,北京)测定在663、645和470 nm波长下的吸光值。
1.3 透射电镜观察
选取二叶期的野生型和突变体叶片,置于含有2.5%戊二醛固定液的离心管中,抽真空1 h,室温放置24 h。再用四氧化锇固定,而后经过脱水、置换、包埋和切片,以JEM-1200Ex型透射电镜观察拍照。
1.4 CRISPR-Cas9敲除位点鉴定
利用Primer Premier 5软件设计CRISPR-Cas9敲除位点测序引物(表1)。分别取50 mg野生型和突变体叶片剪碎,加入30 μL DNA提取液(北京聚合美生物科技有限公司;MF848),用200 μL枪头将叶片捣碎,置于Eppendorf Mastercycler nexusX2梯度PCR仪中95℃5 min,然后离心1 min。吸取上清液2 μL作为模板进行PCR扩增;PCR反应体系为DNA 2 μL、引物2 μL、10×Buffer 25 μL、dNTP(10 mmol·L-1)10 μL、KOD 0.1 μL和ddH2O 10.9 μL。PCR反应程序为94℃2 min;98℃10 s,55℃ 30 s,68℃ 30 s,33个循环;68℃5 min,4℃5 min。PCR产物进行测序,利用BioXM 2.6软件进行DNA序列比对,确定突变位点。
1.5 氨基酸序列比对
利用YGL3蛋白氨基酸序列在NCBI(http://www. ncbi.nlm.nih.gov/)中查找不同物种中YGL3的同源蛋白,下载不同物种的氨基酸序列,使用BioXM 2.6软件对氨基酸序列进行比对分析。
1.6 相关基因的表达分析
利用康为世纪RNApure Plant Kit(Cat.#CW0559)提取中花11不同组织和突变体RNA,使用PrimeScriptRT Reagent Kit(TaKaRa)反转录合成cDNA。荧光定量PCR反应体系为8.8 μL模板、1.2 μL引物和10 μL SYBR Green,在Bio-rad CFX96荧光定量PCR仪上进行扩增,引物序列参考刘喜等[23]。
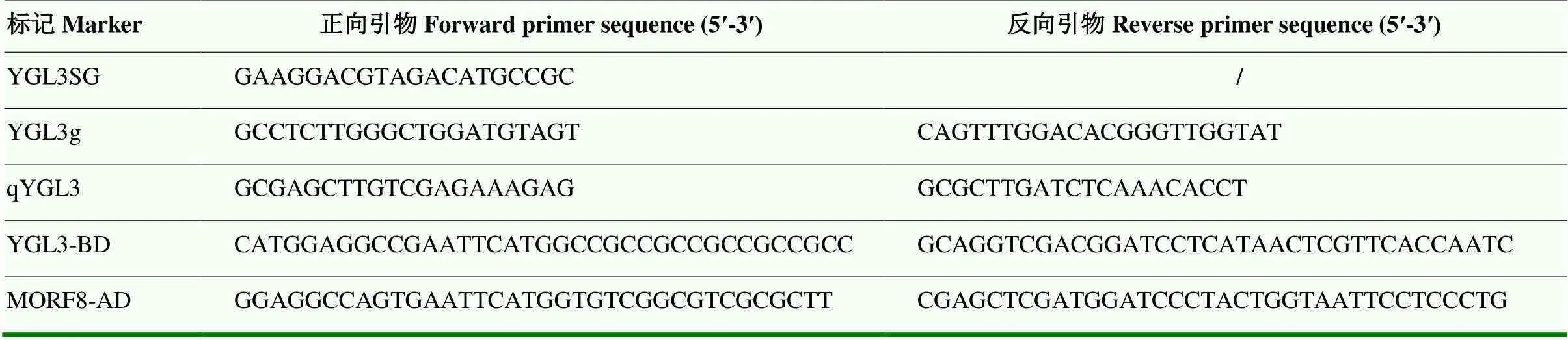
表1 本研究所用引物序列
1.7 酵母双杂交
将的全长CDS克隆融合到pGBKT7载体中,将的全长CDS克隆融合到pGADT7载体中,采用Clontech酵母转化试剂盒(Cat.#630489)将不同组合的质粒共转化酵母菌AH109,涂布于二缺培养基(SD/-Leu/-Trp,DDO),30℃生化培养箱培养2—3 d。然后,挑取二缺培养基上的3个单克隆置于四缺培养基(SD/-Leu/-Trp/-His/-Ade,QDO),30℃生化培养箱培养5 d。酵母转化步骤参照Clontech说明书。
2 结果
2.1 突变体表型考察及CRISPR-Cas9敲除位点的鉴定
从中花11 CRISPR-Cas9突变体库中筛选获得2份稳定遗传黄绿叶突变体与。与野生型相比,苗期突变体与均表现出黄叶表型(图1-A)。光合色素含量测定结果表明,苗期突变体的叶绿素a、叶绿素b、总色素含量和类胡萝卜素含量均显著低于野生型(图1-B),表明突变体黄叶表型主要由光合色素含量降低导致。CRISPR-Cas9编辑靶标位点测序发现在野生型和突变体间存在单碱基插入,导致编码的蛋白翻译提前终止,即是功能缺失型突变体(图1-C—图1-D)。利用qRT-PCR分析在野生型和突变体中的表达水平,结果表明,与野生型相比,在突变体中表达水平显著降低(图1-E)。
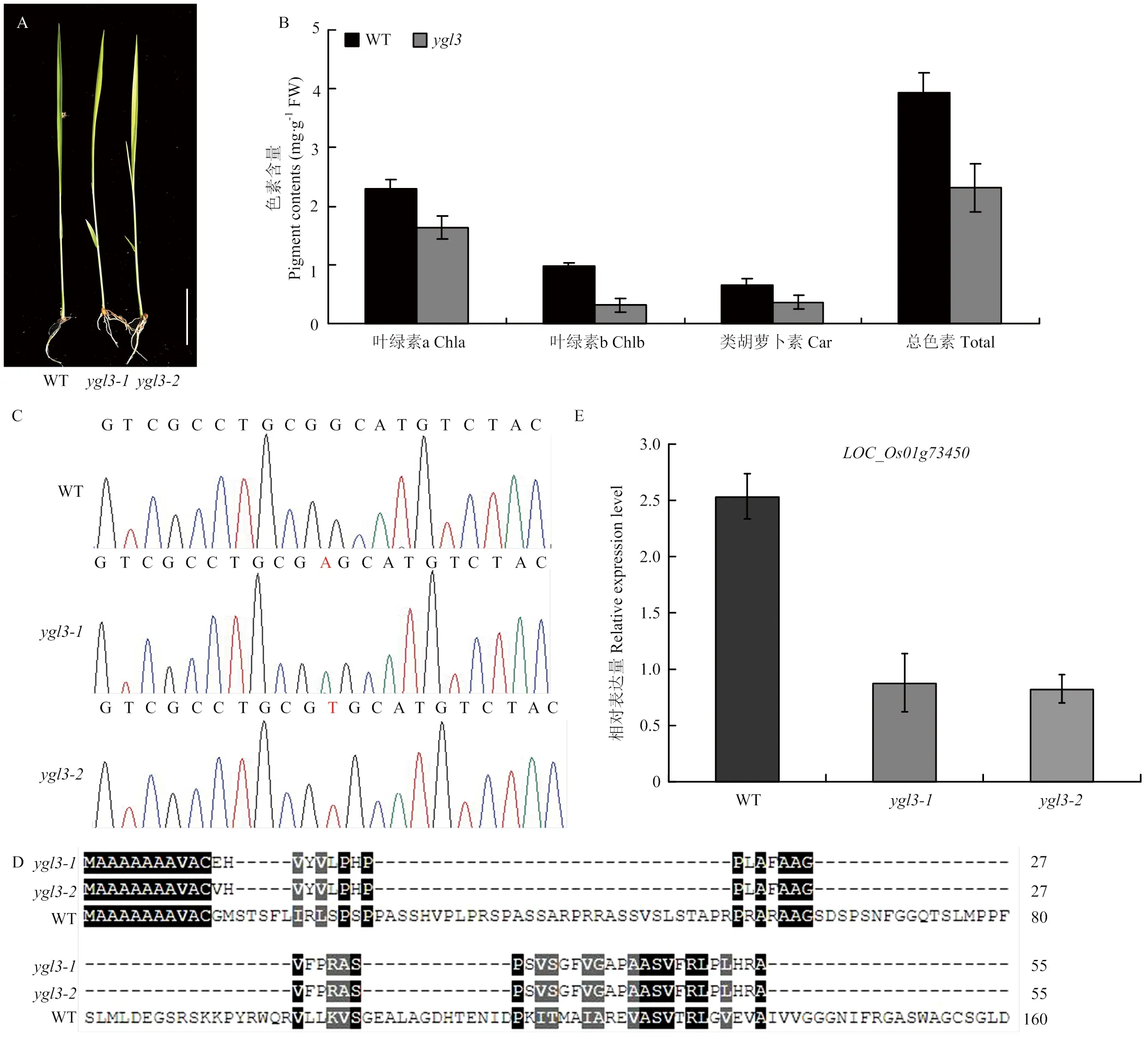
A:苗期野生型和突变体ygl3的表型;B:苗期野生型和突变体ygl3光合色素含量测定;C:CRISPR-Cas9敲除位点的鉴定;D:野生型和突变体氨基酸序列比对;E:LOC_Os01g73450表达分析
2.2 叶绿体形态结构观察
为探究野生型和突变体叶绿体发育的差异,选取苗期突变体和野生型的叶片进行透射电镜观察。结果表明,野生型叶绿体结构正常,规则地贴壁分布,叶绿体内基粒结构正常,类囊体片层结构有序排列(图2-A和图2-B),而突变体叶绿体形态异常,类囊体片层显著减少,无序排列(图2-C和图2-D)。以上结果说明,与野生型相比,突变体的叶绿体发育异常。
2.3 YGL3的组织表达分析
为研究的表达模式,在Rice eFP Browser(http://bar.utoronto.ca/efprice/cgi-bin/efpWeb.cgi)网站上对的表达模式进行预测,结果显示,在各个组织中都有表达,呈组成型表达,叶片中表达水平最高。为了验证预测结果,选取中花11植株的根、叶鞘、叶、穗以及种子提取RNA,反转录成cDNA用于qRT-PCR分析。所得结果和预测结果基本一致,证实呈组成型表达(图3)。
2.4 YGL3及其同源蛋白氨基酸序列的比对
编码一个含有351个氨基酸定位于质体的尿嘧啶核苷酸激酶。在水稻基因组中尿嘧啶核苷酸激酶基因只有1个拷贝。通过对水稻、玉米、高粱、拟南芥、渐尖二型花以及野生稻等物种中尿嘧啶核苷酸激酶进行氨基酸序列进行比对,发现水稻YGL3与玉米、高粱的尿嘧啶核苷酸激酶氨基酸序列有约60%的相似性(图4),尿嘧啶核苷酸激酶在结构域上较为保守,N端差异较大。
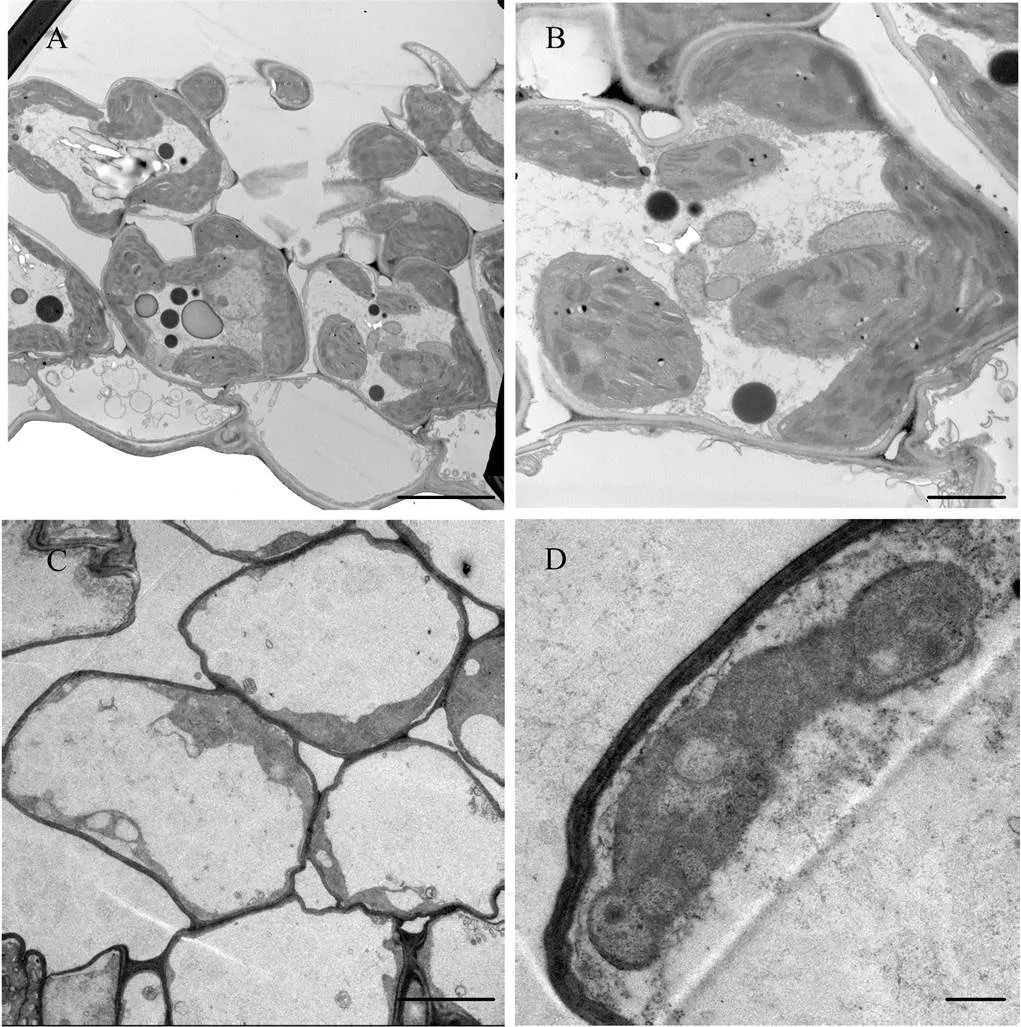
A、B:野生型叶绿体结构;C、D:突变体ygl3叶绿体结构。Bar=5 μm(A、C)和Bar=2 μm(B、D)

图3 YGL3的组织表达分析
2.5 叶绿素合成基因表达分析
与野生型相比,突变体的色素含量显著降低,暗示的突变影响叶绿素合成途径。因此,通过对野生型和突变体中叶绿素合成基因进行qRT-PCR分析。与野生型相比,突变体中叶绿素合成基因(编码谷氨酰-tRNA合成酶)、(编码尿卟啉原氧化脱羧酶)和(编码尿卟啉原脱羧酶)的转录水平显著降低(图5)。叶绿素合成基因、、、的表达水平在突变体中上调或无差异。因此,叶绿素合成途径相关基因表达异常可能是导致突变体叶绿素含量降低的原因之一。
2.6 YGL3与MORF8在酵母中互作
为进一步阐明YGL3的分子功能,构建pGBKT7- YGL3融合表达载体,利用水稻叶片酵母cDNA文库,进行酵母双杂交筛选YGL3的互作蛋白。鉴定获得4个与YGL3互作的克隆,均为RNA编辑因子MORF8的片段(图6-A)。随后,构建pGADT7-MORF8融合表达载体,进行YGL3与MORF8的互作验证。与阴性对照相比,YGL3-BD与MORF8-AD组合可以在缺陷培养基(SD-Leu/-Trp/-His/-Ade)上生长,表明在酵母体内YGL3与MORF8存在互作(图6-B)。
3 讨论
叶绿体是绿色植物特有的细胞器,正常的叶绿体对植物生长发育至关重要[24]。叶绿素生物合成和叶绿体发育的调控网络极其复杂,需要鉴定更多的基因完善叶绿体发育的调控路径。目前,已克隆的水稻叶色相关基因超过120个。在已克隆的基因中,有直接编码叶绿素合成与降解的基因如[25]、[26]和[27],有调控叶绿体发育的基因如[28]、[29]和[30]。本研究鉴定叶色突变体在苗期植株叶片为黄化,与野生型相比,突变体叶片叶绿素a、叶绿素b、总色素和类胡萝卜素含量均显著降低。叶绿体透射电镜观察结果表明,与野生型相比,突变体叶绿体形态异常,类囊体片层结构排列无序。水稻黄叶突变体叶绿体基粒片层显著降低[9];黄绿叶突变体的叶绿体形态不规则,有较多空的囊泡状结构[31]。本研究中叶绿体形态异常,类囊体片层减少,分布不规则,推测突变体黄叶的表型可能由叶绿体发育异常和光合色素含量下降引起。

NP_188498.1:拟南芥;NP_001137013.1:玉米;OEL23965.1:渐尖二型花;XP_002456998.1:高粱;XP_015693434.1:野生稻;YGL3:水稻。完全或部分保守的氨基酸分别是蓝色和粉色标示
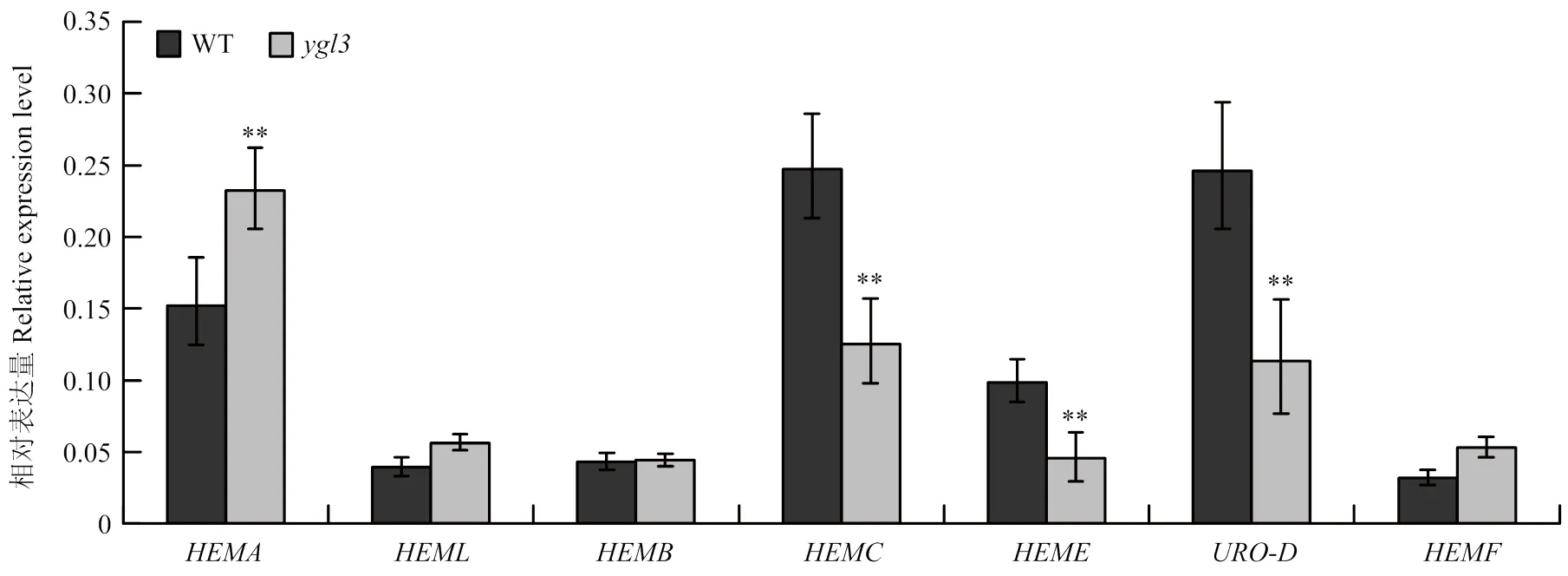
HEMA:编码谷氨酰-tRNA合成酶;HEME:编码尿卟啉原脱羧酶;HEML:编码谷氨酰-1-半醛转氨酶;HEMB:编码胆色素原合酶;HEMC:编码羟甲基后胆色素原合酶;HEMF:编码粪卟啉Ⅲ氧化酶;URO-D:编码尿卟啉原氧化脱羧酶。**:在0.01水平差异极显著
目前,水稻研究进入后基因组学时代,越来越多的等位基因被克隆,等位基因的发现有助于进一步阐明该基因的分子功能。因此,收集鉴定更多的水稻叶色突变体,将会推动对叶色调控分子路径的认识。本研究中的黄叶基因是水稻第1染色体核基因编码的尿嘧啶核苷酸激酶OsUMPK。Zhu等[32]通过图位克隆鉴定到的等位基因,突变体在整个生育期均表现出黄绿叶。此外,Chen等[33]通过甲基磺酸乙酯诱变蜀恢527鉴定到的等位突变体,研究中分离鉴定的发生单碱基替换,产生新的截断转录本。与野生型相比,突变体叶片黄绿色,成熟期突变体矮化。本研究是通过CRISPR-Cas9技术编辑中花11获得,突变位点在第一外显子上,与、的突变位点不同,3个等位突变体叶片表现为黄化,叶片色素含量降低,但下降的幅度均不一致,可能是遗传背景导致的。

DDO:基本营养缺陷培养基SD/-Leu/-Trp;QDO:选择营养缺陷培养基SD-Leu/-Trp/-His/-Ade
在拟南芥中存在一个同源基因,其T-DNA插入突变体表现出矮化、叶色异常的表型,PUMPKIN参与质体基因-UCC、-UAC、、以及的内含子剪切[34]。目前,水稻已经报道了17个PPR蛋白、1个MORF蛋白以及3个剪切因子参与质体基因转录后调控。编码一个PLS-DYW亚家族的PPR蛋白,其突变导致植株白化致死,参与质体基因转录本编辑和内含子剪切[35]。PLS-DYW亚家族的PPR蛋白DUA1调控低温下与的RNA编辑[36]。编码一个P家族PPR蛋白,参与转录本编辑和4个质体基因(、、和)内含子剪切[37]。编码一个叶绿体内含子剪切促进因子,影响多个质体基因内含子剪切[38]。本研究利用酵母双杂交系统筛选到一个YGL3的互作蛋白MORF8。MORF家族蛋白在拟南芥已经被证实参与质体基因转录后调控,如RNA编辑、RNA剪切[39-40]。然而,在水稻中,只鉴定出一个MORF蛋白WSP1的功能,突变体表现出叶片白条纹和白穗的表型,参与多个质体基因(、、和)RNA编辑以及内含子的剪切[41]。的突变是否参与质体基因转录后调控,将是下一步研究的重点。
4 结论
黄绿叶突变体表型由基因突变导致。突变体苗期叶绿素a、叶绿素b、总色素和类胡萝卜素含量显著减少,叶绿体发育异常,类囊体片层减少。在水稻中呈组成型表达,且叶片中表达最高。YGL3与RNA编辑因子MORF8在酵母中存在互作。是已报道的的等位基因,因编码框发生单碱基插入导致蛋白翻译提前终止而引起黄叶表型。
[1] Eberhard S, Finazzi G, Wollman F A. The dynamics of photosynthesis.Annual Review of Genetics, 2008, 42: 463-515.
[2] Pogson B J, Albrecht V. Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiology, 2011, 155(4): 1545-1551.
[3] Awan M A, Konzak C F, Rutger J N, Nilan R A. Mutagenic effects of sodium azide in riceCrop Science, 1980, 20(5): 663-668.
[4] Fambrini M, Castagna A, Dalla Vecchia F, Degl'Innocenti E, Ranieri P, Guidi L, RascioN, PugliesiC.Characterization of a pigment-deficient mutant of sunflower (L.) with abnormal chloroplast biogenesis, reduced PSII activity and low endogenous level of abscisic acid. Plant science, 2004, 167(1): 79-89.
[5] Yoo S C, Cho S H, Sugimoto H, Li J, Kusumi K, Koh H J, Iba K, Paek N C. Riceandencoding the large and small subunits of ribonucleotide reductase are required for chloroplast biogenesis during early leaf development. Plant Physiology, 2009, 150(1): 388-401.
[6] Dong H, Fei G L, Wu C Y, Wu F Q, Sun Y Y, Chen M J, Ren Y L, Zhou K N, Cheng Z J, Wang J L, Jiang L, Zhang X, Guo X P, Lei C L, Su N, Wang H Y, Wan J M. A ricemutant reveals new insights into the role and sssembly of plastid caseinolytic protease in higher plants. Plant Physiology, 2013, 162(4): 1867-1880.
[7] Beale S I. Green genes gleaned. Trends in Plant Science, 2005, 10(7): 309-312.
[8] Zeng Z Q, Lin T Z, Zhao J Y, Zheng T H, Xu L F, Wang Y H, Liu L L, Jiang L, Chen S H, Wan J M.gene, encoding glutamyl-tRNA reductase (GluTR) is essential for chlorophyll biosynthesis in rice (). Journal of Integrative Agriculture, 2020, 19(3): 612-623.
[9] Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, Wang C, Zhai H Q, Wan J M. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiology, 2007, 145(1): 29-40.
[10] Kong W Y, Yu X W, Chen H Y, Liu L L, Xiao Y J, Wang Y L, Chaolong Wang C L,Yun Lin Y,Yu Y, Wang C M, Jiang L, Zhai H Q, Zhao Z G, Wan J M. The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice. Plant Molecular Biology, 2016, 92(1): 177-191.
[11] 杨颜榕, 黄纤纤, 赵亚男, 汤佳玉, 刘喜. 水稻叶色基因克隆与分子机制研究进展. 植物遗传资源学报, 2020, 21(4): 794-803.
Yang Y Y, Huang Q Q, Zhao Y N, Tang J Y, Liu X. Advances on gene isolation and molecular mechanism of rice leaf color genes. Journal of Plant Genetic Resources, 2020, 21(4): 794-803. (in Chinese)
[12] 赵绍路, 刘凯, 宛柏杰, 朱静雯, 刘艳艳, 唐红生, 严国红, 孙明法. 水稻叶色突变研究进展. 大麦与谷类科学, 2018, 35(6): 1-6.
Zhao S L, Liu K, Wan B J, Zhu J W, Liu Y Y, Tang H S, Yan G H, Sun M F. Advances on mutation of rice leaf color.Barley and Cereal Sciences, 2018, 35(6):1-6. (in Chinese)
[13] Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L. A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nature Communications, 2011, 2: 477.
[14] Tan J, Tan Z, Wu F, Sheng P, Heng Y, Wang X, Ren Y, Wang J, Guo X, Zhang X, Cheng Z, Jiang L, Liu X, Wang H, Wan J M. A novel chloroplast-localized pentatricopeptide repeat protein involved in splicing affects chloroplast development and abiotic stress response in rice. Molecular Plant, 2014, 7(8): 1329-1349.
[15] Lin D Z, Zheng K L, Liu Z H, Li Z K, Teng S, Xu J L, Dong Y J. Riceencoding a component of the TAC complex is required for chloroplast development under cold stress. Plant Genome, 2018, 11(1): 160065
[16] Adam Z, Rudella A, van Wijk K J. Recent advances in the study of Clp, FtsH and other proteases located in chloroplasts. Current Opinion in Plant Biology, 2006, 9(3): 234-240.
[17] Sjögren L L, Clarke A K. Assembly of the chloroplast ATP-dependent Clp protease inis regulated by the ClpT accessory proteins. The Plant Cell, 2011, 23(1): 322-332.
[18] Chen Y L, Chen L J, Chu C C, Huang P K, Wen J R, Li H M. TIC236 links the outer and inner membrane translocons of the chloroplast. Nature, 2018, 564(7734): 125-129.
[19] He Y, Shi Y, Zhang X, Xu X, Wang H, Li L, Zhang Z, Shang H, Wang Z, Wu J L. The OsABCI7 transporter interacts with OsHCF222 to stabilize the thylakoid membrane in rice. Plant Physiology, 2020, 184(1): 283-299.
[20] Nakamura H, Muramatsu M, Hakata M, Ueno O, Nagamura Y, Hirochika H, Takano M, Ichikawa H. Ectopic overexpression of the transcription factorinduces chloroplast development in non-green rice cells. Plant Cell Physiology, 2009, 50(11): 1933-1949.
[21] Zhao C, Xu J, Chen Y, Mao C, Zhang S, Bai Y, Jiang D, Wu P. Molecular cloning and characterization of, a rice chromatin-remodeling factor required for early chloroplast development in adaxial mesophyll. Planta, 2012, 236(4): 1165-1176.
[22] Porra R J, Thompson W A, Kriedemann P E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica Et Biophysica Acta-Bioenergetics, 1989, 975(3): 384-394.
[23] 刘喜, 周春雷, 任雅琨, 杨春艳, 何旎清, 柳周, 江玲, 万建民. 水稻叶色白化转绿突变体的遗传分析与基因定位. 南京农业大学学报, 2015, 38(5): 712-719.
Liu X, Zhou C L, Ren Y K, Yang C Y, He N Q, Liu Z, Jiang L, Wan J M. Genetic analysis and gene mapping of virescent albino leaf mutantin rice. Journal of Nanjing Agricultural University, 2015, 38(5): 712-719. (in Chinese)
[24] Stern D B, Hanson M R, Barkan A. Genetics and genomics of chloroplast biogenesis: maize as a model system. Trends in Plant Science, 2004, 9(6): 293-301.
[25] Jung K H, Hur J, Ryu C H, Choi Y, Chung Y Y, Miyao A, Hirochika H, An G. Characterization of a rice chlorophyll- deficient mutant using the T-DNA gene-trap system. Plant Cell Physiology, 2003, 44(5): 463-472.
[26] Yang Y L, Xu J, Huang L C, Leng Y J, Dai L P, Rao Y C, Chen L, Wang Y Q, Tu Z J, Hu J, Ren D Y, Zhang G H, Zhu L, Guo L B, Qian Q, Zeng D L., encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. Journal of Experimental Botany, 2016, 67(5): 1297-1310.
[27] Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, Tanaka A. Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. The Plant Cell, 2007, 19(4): 1362-1375.
[28] Kusumi K, Sakata C, Nakamura T, Kawasaki S, Yoshimura A, Iba K. A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. The Plant Journal, 2011, 68(6): 1039-1050.
[29] Huang W, Zhang Y, Shen L, Fang Q, Liu Q, Gong C, Zhang C, Zhou Y, Mao C, Zhu Y, Zhang J, Chen H, Zhang Y, Lin Y, Bock R, Zhou F. Accumulation of the RNA polymerase subunitdepends on RNA editing by OsPPR16 and affects chloroplast development during early leaf development in rice. New Phytologist, 2020, 228(4): 1401-1416.
[30] Kusumi K, Yara A, Mitsui N, Tozawa Y, Iba K. Characterization of a rice nuclear-encoded plastid RNA polymerase gene. Plant Cell Physiology, 2004, 45(9): 1194-1201.
[31] Deng X J, Zhang H Q, Wang Y, He F, Liu J L, Xiao X, Shu Z F, Li W, Wang G H, Wang G L. Mapped clone and functional analysis of leaf-color geneL. ssp.). PLoS One, 2014, 9(6): e99564.
[32] Zhu X, Guo S, Wang Z, Du Q, Xing Y, Zhang T, Shen W, Sang X, Ling Y, He G. Map-based cloning and functional analysis of, which controls leaf colour in rice (). BMC Plant Biology, 2016, 16(1): 134.
[33] Chen F, Dong G, Ma X, Wang F, Zhang Y, Xiong E, Wu J, Wang H, Qian Q, Wu L, Yu Y. UMP kinase activity is involved in proper chloroplast development in rice. Photosynthesis Research, 2018,137(1): 53-67.
[34] Schmid LM, Ohler L, Möhlmann T, Brachmann A, Muiño JM, Leister D, Meurer J, Manavski N. PUMPKIN, the sole plastid UMP kinase, associates with group II introns and alters their metabolism. Plant Physiology, 2019, 179(1): 248-264.
[35] Tang J P, Zhang W W, Wen K, Chen G M, Sun J, Tian Y L, Tang W J, Yu J, An H Z, Wu T T, Kong F, Terzaghi W, Wang C M, Wan J M. OsPPR6, a pentatricopeptide repeat protein involved in editing and splicing chloroplast RNA, is required for chloroplast biogenesis in rice. Plant Molecular Biology, 2017, 95(4/5): 345-357.
[36] Cui X A, Wang Y W, Wu J X, Han X, Gu X F, Lu T G, Zhang Z G. The RNA editing factoris crucial to chloroplast development at low temperature in rice. New Phytologist, 2019, 221(2): 834-849.
[37] Wang Y, Ren Y L, Zhou K N, Liu L L, Wang J L, Xu Y, Zhang H, Zhang L, Feng Z M, Wang L W, Ma W W, Wang Y L, Guo X P, Zhang X, Lei C L, Cheng Z J, Wan J M.encodes a novel p-Type PPR protein required for chloroplast biogenesis during early leaf development. Frontiers in Plant Science, 2017, 8: 1116.
[38] Liu C H, Zhu H T, Xing Y, Tan J J, Chen X H, Zhang J J, Peng H F, Xie Q J, Zhang Z M.is involved in the splicing of chloroplast group I and II introns in rice. Journal of Experimental Botany, 2016, 67(18): 5339-5347.
[39] Glass F, Hartel B, Zehrmann A, Verbitskiy D, Takenaka M. MEF13 requires MORF3 and MORF8 for RNA editing at eight targets in mitochondrial mRNAs in. Molecular Plant, 2015, 8(10): 1466-1477.
[40] Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Hartel B, Brennicke A. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proceedings of the National Academy of Sciences of the United States of America, 2012, 109(13): 5104-5109.
[41] Zhang Z, Cui X, Wang Y, Wu J, Gu X, Lu T. The RNA editing factoris essential for chloroplast development in rice. Molecular Plant, 2017, 10(1): 86-98.
Identification and Gene Functional Analysis of Yellow Green Leaf Mutant
XU ZiYi, CHENG Xing, SHEN Qi, ZHAO YaNan, TANG JiaYu, LIU Xi
Huaiyin Normal University/Jiangsu Key Laboratory for Eco-agriculture Biotechnology Around Hongze Lake/Jiangsu Collaborative Innovation Center of Regional Modern Agriculture and Environment Protection, Huaian 223300, Jiangsu
【】To enrich and deepen people’s understanding of the molecular mechanism of plant leaf color, the phenotype identification and gene cloning of the yellow green leaf mutant() were carried out to clarify the molecular function ofand lay the foundation for elucidating the molecular mechanism ofregulating rice leaf color.【】Two stable genetic allelic yellow green leaf mutants,and, were isolated from the CRISPR-Cas9 knockout mutant library of Zhonghua 11. The phenotype of the mutant was identified, and the chlorophyll contents of the wild-type andwere determined. The chloroplast structure of the wild-type andwas observed by transmission electron microscope. qRT-PCR was used to analyze the tissue expression of, and BioXM2.6 software was used for sequence alignment of YGL3 and its homologs. Yeast two hybrid was used to screen the interacting proteins of YGL3.【】Compared with the wild type, the leaves ofwere yellowing, and the contents of chlorophyll, carotenoid and total photosynthetic pigment at seedling stage inwere significantly decreased. Transmission electron microscopy showed that the chloroplast morphology ofwas abnormal, and the thylakoid lamellar structure was less, whereas the chloroplast morphology of the wild type was normal and the thylakoid lamellar structure was orderly arranged. CRISPR-Cas9 knock-out site identification showed that thegene had a single base insertion, which resulted in the early termination of protein translation. The gene encoding 351 amino acids was mutated into a truncated protein with 55 amino acids. Compared with the wild type, the expression level ofwas significantly down-regulated in the mutants. qRT-PCR showed thatwas expressed in roots, panicles, seeds, leaf sheaths and leaves.was highly expressed in leaves.encodes a plastid localized UMP kinase. The YGL3 protein was conserved in,and. YGL3 shared the high sequence homology (59.4% amino acid identity) to. qRT-PCR showed that chlorophyll synthesis genes, including,and,were significantly down-regulated in, whereas the expression levels of,andwere no significant difference between the wild type and. Yeast two hybrid screen showed that YGL3 interacted with RNA editing factor MORF8.【】The phenotype of the yellow leaf mutantresulted from themutation.was an allele of the yellow green leave genewas highly expressed in leaves, and YGL3 interacted with MORF8 in yeasts.
rice (L.); yellow green leaf;; CRISPR-Cas9; chloroplast development

10.3864/j.issn.0578-1752.2021.15.001
2020-12-14;
2021-02-10
江苏省高等学校自然科学研究面上项目(19KJB180009)、淮阴师范学院大学生创新创业计划(202019009XJ)
许子怡,E-mail:2631140968@qq.com。通信作者刘喜,E-mail:1240623244@qq.com
(责任编辑 李莉)

