Genetic association of rs7754840 and rs7756992 polymorphisms in the CDKAL1 gene and gestational diabetes mellitus in selected Filipino pregnant women
Maria Ruth B. Pineda-Cortel, Karlo Baybayan, Peter Louie Bello, Latiffa Lois Camenforte, Stefany Jane Ching,Kathleen Conti, Jeremiah Jose Ignacio, Jiovanni Diaz, Adrian Villavieja, Jefferyl Kae Pandac, Emilyn U. Alejandro
1Department of Medical Technology, Faculty of Pharmacy, University of Santo Tomas, Manila, Philippines
2The Graduate School, University of Santo Tomas, Manila, Philippines
3Institute of Clinical Laboratory Sciences, Silliman University, Dumaguete City, Philippines
4Research Center for the Natural and Applied Sciences, University of Santo Tomas, Manila, Philippines
5Department of Integrative Biology and Physiology, University of Minnesota, USA
ABSTRACT
KEYWORDS: Gestational diabetes mellitus; GDM; Single nucleotide polymorphism; SNP; Genetic association; rs7756992;rs7754840; CDKAL1; Filipino pregnant women
1. Introduction
Gestational diabetes mellitus (GDM), defined as a metabolic disorder characterized by insulin resistance and hyperglycemia of varying severity that is first detected during pregnancy, is one of the most common pregnancy complications and is associated with both long-term and short-term complications on both the mother and the offspring[1]. Its prevalence varies among population and may range from 1% to 30%[2]. The likelihood of developing GDM varies among populations, with Hispanic and Asian women having greater risk, and Filipino women having a prevalence rate of 29.3%[3].
GDM has been strongly linked to type 2 diabetes mellitus (T2DM),which is characterized by insulin resistance in peripheral tissues and impaired insulin production in pancreaticβ-cells[1]. Like T2DM patients, GDM women appear to develop glucose intolerance during pregnancy in part due to insulin resistance, and subsequently they develop hyperglycemia when theβ-cells fail to produce sufficient insulin to control glucose homeostasis[3]. A known long-term complication associated with GDM is that women with GDM have increased risk to develop T2DM later in her life[4]. This increased susceptibility to develop T2DM later in life is observed in different populations[5]. Similarly, it has been shown that women with family history of T2DM are at higher risk to have GDM during pregnancy[6]. These studies showing the association between T2DM and GDM suggest the potential similarity in the genetic factors affecting the pathogenesis of these two conditions.
Although the pathophysiology of GDM is not yet fully understood,several risk factors have been implicated to contribute in the development of the disorder, such as maternal obesity, maternal age, family history of T2DM, previous adverse pregnancy outcomes, belonging to a high-risk ethnic group, genetic factors affecting insulin secretion and glucose uptake, among others[7].Although genetic factors play a major factor in the development of hyperglycemia and insulin resistance, this can also be exacerbated by the presence of a low-level inflammatory response[8], resulting in proinflammatory and cytokine-mediatedβ-cell destruction[9].
Consequently, development of GDM can cause adverse effects on both mother and offspring. As mentioned previously, pregnant women who have developed GDM have increased risk of developing T2DM. GDM is also associated with increased incidence of hypertension during pregnancy and postpartum[10], as excess blood glucose can lead to endothelial cell and vascular damage[11].Moreover, because of an excess of glucose in the placenta, fetal cell overgrowth[12] resulting in fetal macrosomia[13] has been observed among GDM offspring. There is also increased incidence of neonatal respiratory distress syndrome in the offspring of GDM mothers, due to fetal lung underdevelopment associated with preterm labor[14]and inhibition of surfactant protein A production by increased insulin levels[15]. Also, the low-level systemic inflammation present in GDM mothers also induces excessive inflammatory response on the neonate, resulting in adverse outcomes following neonatal infections[16]. Aside from other risk factors like anatomical[17],endocrine/hormonal[18], and immunological abnormalities[19] as well as pathogenic infections[20], GDM can also be associated with increased risk of pregnancy loss[21].
The association of GDM with genetic factors and its linkage with candidate genes have been studied in the past decades[22]. Studies that focused on investigating the genetic susceptibilities of GDM and T2DM revealed that these conditions share common genetic background, with analogous degree of effect sizes on the same risk alleles[23]. Similarly, pathogenesis between the two conditions are also comparable, that is, both are associated with obesity and insulin resistance[24].With interest on the association of genetic variations, particularly that of CDKAL1, with GDM during pregnancy, we performed the present study. We aimed to determine the association between the two intronic variants of CDKAL1 gene, namely rs7756992 (A/G) and rs7754840 (C/G), and that of the risk of developing GDM among a Filipino population. Moreover, we aimed to assess whether alteration in lipid profile and glycosylated hemoglobin (HbA1c) values were observed between GDM and non-GDM patients.
2. Material and methods
2.1. Participants
In this cross-sectional study, pregnant women were recruited from partner hospitals and maternity clinics within Metro Manila,Philippines from July 2018 to January 2019. These institutions were the following: Dr. Jose Fabella Memorial Hospital, Santa Ana Hospital, Ospital ng Maynila, Ospital ng Sampaloc, and Blooming Babies Maternity Clinic. All participants gave informed consent to participate in the study and ethical approval was granted by the local ethics committee. A total of 200 pregnant women were included in the study, with the sample size determined by using GPower ver.3.1.9.7 (Universitat Dusseldorf), 101 of which had developed GDM and 99 of which were normoglycemic.
The selection and recruitment of the control and GDM mothers were based on the study’s inclusion and exclusion criteria. A patient was eligible to participate if she was of full Filipino descent,between 18 to 45 years old, within 21 to 28 weeks of pregnancy upon recruitment based on the patient’s ultrasound result, and was willing to participate in the study. However, pregnant women who had a history of other pre-existing health conditions such as body organ and/or metabolic disorders, and sexually transmitted infections such as hepatitis B, syphilis, and human immunodeficiency virus,were excluded from participating in the study.
Pregnant women who were qualified to participate were interviewed by trained interviewers using a structured questionnaire. Participant’s height, pre-oral glucose tolerance test (OGTT), weight (their weight during their first checkup), and their weight during OGTT (their current weight as of the time the OGTT test was performed) were requested from the patients through their patient information forms.Thereafter, the participants were screened for GDM by the one-step OGTT with 75 g of glucose load and their GDM status was assessed by using the criteria set by the International Association of Diabetes and Pregnancy Study Groups[25]. The OGTT test was performed upon the physician’s request, during the second trimester, between their 21st to 28th week of gestation. In brief, diagnosis of GDM was made when a patient’s OGTT result met one or more of the following criteria: fasting blood sugar of 5.11 mmol/L; first hour blood glucose of ≥10.0 mmol/L; and, second hour blood glucose of>8.5 mmol/L.
2.2. Blood collection and sample preparation
Blood samples were collected from the antecubital fossa region of the arm of the patients after they had undergone a standard fasting procedure. Samples for fasting blood sugar and lipid profile were collected first and were placed in gold-top evacuated tubes with serum separator. KEDTA-filled tubes were also filled for HbA1c testing and DNA extraction purposes. Thereafter, the participants were required to drink a 75-gram glucose load and blood samples were collected one and two hours later for their first hour and second hour blood glucose samples, respectively.
The serum separator gel tubes were spun at 4 000 ×g for 20 min to separate serum from red blood cells. The same procedure was also followed for lavender-top tubes for the separation of buffy coat samples after HbA1c testing was completed. After centrifugation,the buffy coat was carefully aspirated and placed in sterile microcentrifuge tubes which were then labelled and stored at 4 ℃until used for the isolation of DNA.
2.3. Biochemical measurements
Blood glucose measurements (fasting blood sugar, first hour and second hour blood glucose) employed an enzymatic colorimetric assay procedure according to the manufacturer’s protocol (Human,Germany). Lipid profile encompassed a number of parameters which included total cholesterol, triglyceride, high-density lipoprotein(HDL), and low-density lipoprotein (LDL). Total cholesterol was measured spectrophotometrically by using the Cholesterol Liquicolor Kit (Human, Germany). For triglyceride determination,an enzymatic colorimetric assay was employed according to the instructions provided by the manufacturer (Human, Germany).HDL concentrations were determined by initially precipitating other lipoproteins, and then measuring HDL by using the Cholesterol Liquicolor kit (Human, Germany). The Friedewald formula was used to determine LDL instead of directly measuring its concentration from the sample. Lastly, whole blood from lavender-top tubes were used as samples in measuring for HbA1c using the NycoCard™HbA1c (Alere Technologies AS, Oslo, Norway) boronate affinity assay procedure. All measurements and assays were performed according to standard laboratory procedures.
2.4. DNA extraction and single nucleotide polymorphism(SNP) genotyping
Genomic DNA were isolated from buffy coat samples by using the Invitrogen™ PureLinkGenomic DNA Extraction Kit (Thermo Fisher Scientific, USA) which involved four major steps: lysis,binding, washing, and eluting of genomic DNA samples. The concentration and purity of the isolated DNA were determined by using the nanodrop technique and absorbance was read in a FLUOstarOmega Plate Reader (BMG LABTECH, Germany) prior to storage at -20 ℃ until use.
Amplification of the target gene polymorphisms was performed through real time quantitative polymerase chain reaction (RT-qPCR) by using the Rotor-Gene-Q PCR (Qiagen,Germany) with the following thermal cycling conditions: hold for 10 min at 92 ℃; and 30 s at 92 ℃ and 90 s at 60 ℃ which were repeated for 45 cycles. Genotyping was performed by allelic discrimination assay. In order to discriminate rs7754840 and rs7756992 gene polymorphism, TaqManPre-Designed SNP Genotyping Assays were used (Assay ID C_29246232_10, Context Sequence GGGGAAGAAGTA GTAATGTTGGAAA [C/G]GTTGACTTGATAGAGGATTTTGTAA; Assay ID C_2504058_20,context sequence ATATTCCCCCCTGTATTTTAGTTTT[A/G]GATCTACAGTTATGTAGCAATGAGC, respectively), including the specific primers and fluorescently-labelled VIC and FAM probes to detect the genetic alleles.
2.5. Statistical analysis
A combination of Microsoft Excel 2016 and IBM SPSS ver. 21 was used in the study. Median and interquartile range were used to describe the various physical and biochemical characteristics between pregnant women with and without GDM, while Mann-Whitney U-test was used to compare the measured values between the two groups. Chi-square test was used to determine the distribution of the CDKAL1 gene SNPs (rs7756992 (A/G) and rs7754840 (C/G)) between GDM and non-GDM groups. Prior to performing genetic association analysis, the distribution of both rs7756992 and rs7754840 were tested for conformity to the Hardy-Weinberg equilibrium. The Hardy-Weinberg equilibrium is a principle wherein genotype frequencies are expected to remain constant between generations, assuming there are no outside factors[26]. Measurement and assessment of the association between the presence of rs7756992 (A/G) and rs7754840 (C/G) genotypes and allotypes and patient OGTT results were done by using pointbiserial correlation. Lastly, multiple logistic regression model was used to investigate the effect of different genotypes and genetic models on GDM development.
2.6. Ethical considerations
The study protocol was approved by the University of Santo Tomas Graduate School Ethics Review Committee and was determined humane and ethically correct under the protocol number: E-2016-02-R3. All participants provided informed consent before participation.
3. Results
3.1. Physical and biochemical data
The patients’ physical characteristics are shown in Table 1. Patient weight was significantly higher in the GDM group compared to the non-GDM group (P=0.01). The GDM patients also showed significantly higher body mass index compared to the non-GDM patients (P<0.5). For the patients’ biochemical characteristics, the GDM patients had significantly higher glucose parameters such as fasting blood sugar, OGTT 1st and OGTT 2nd hour (all P<0.5)(Table 2).
3.2. Genotype distribution and association with GDM
For the rs7754840 polymorphism, the CG and GG genotypes were non-significantly higher in GDM patients than non-GDM women(CG χ=1.27, P=0.26; GG χ=0.02, P=0.90). The homozygous CC genotype were found to be higher in non-GDM women but the difference was not statistically significant (P=0.18). Distribution of both the C and G alleles for rs7754840 were not significant.For rs7756992, both AG and AA genotype frequencies were nonsignificantly higher in GDM patients (AG χ=0.51, P=0.48; AA χ=1.14, P=0.29). GG genotype frequency was significantly higher in non-GDM women (χ=4.97, P=0.03). Distribution of the A allele in rs7756992 were significant at P value < 0.05 (χ=9.86, P=0.02),while G allele showed no significance (Table 3).
Prior to conducting further statistical analysis, the genotypic distributions were first tested for conformity to Hardy-Weinberg equilibrium, with the frequencies expected to be exactly or as close as p, 2pq, and q[26]. A distribution of P<0.05 demonstrates deviation from Hardy-Weinberg equilibrium, and conventionalpractice is to exclude it from further analysis.
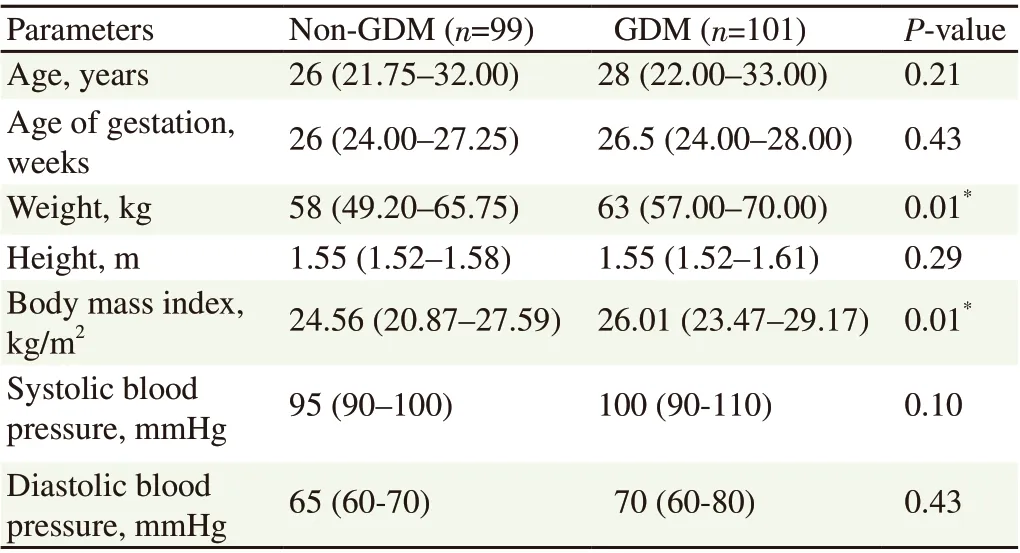
Table 1. Comparison of the physical characteristics of GDM and non-GDM pregnant women.

Table 2. Comparison of the biochemical characteristics of GDM and non-GDM pregnant women.
Hardy-Weinberg equilibrium analysis of SNPs of the CDKAL1 gene shows that the rs7754840 polymorphism has a P value of 0.17 for GDM, and a P value of 0.99 for non-GDM, suggesting that the genotype distribution of SNP satisfied Hardy-Weinberg equilibrium.Conversely, the distributions of rs7756992 polymorphism genotypes did not agree with expected values (P<0.05), and was thus excluded from further analysis (Table 4).
Point-biserial correlation was employed to determine the relationship between the blood glucose parameters and the genotypes and alleles of the rs7754840 polymorphism of the CDKAL1 gene.For the rs7754840 (C/G) SNP, no significant correlation was observed between presence of its genotypes and alleles with OGTT parameters (P>0.05) (Table 5).
Table 6 summarizes the variants of the rs7754840 (C/G) SNP under additive, dominant, and recessive models. Multiple logistic regression analysis was used to demonstrate the effect of the different genotypes and genetic models to GDM, while the odds ratio determined the strength of the effect between the genotypic characteristics and the GDM status of the participants. The genotype distribution of rs7754840 (C/G) contrasted between GDM and non-GDM groups but gave non-significant odds ratios in additive model (OR 1.43; 95%CI 0.82-2.50), dominant model (OR 1.21; 95% CI 0.57-2.59), and recessive model (OR 1.63; 95% CI 0.86-3.09) (all P>0.05).
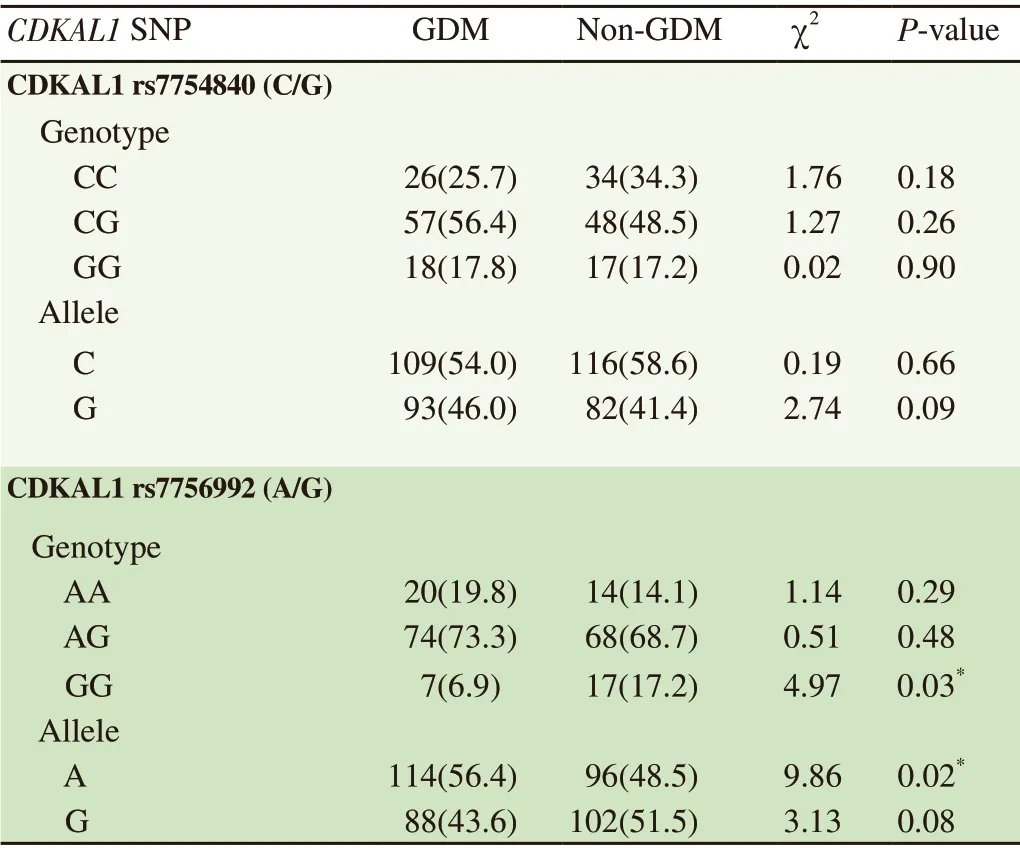
Table 3. CDKAL1 rs7754840 (C/G) and rs7756992 (A/G) genotype and allele distribution between GDM and non-GDM groups [n(%)].

Table 4. Hardy-Weinberg equilibrium analysis of genotypic distribution of rs7754840 (C/G) and rs7756992 (A/G).
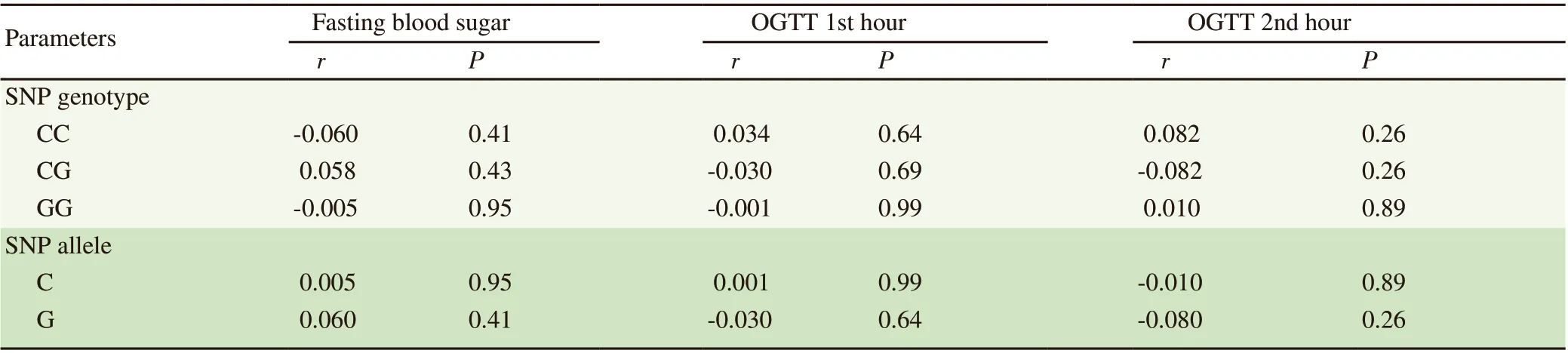
Table 5. Point-biserial correlation of CDKAL1 gene rs7754840 (C/G) genotypes and alleles with OGTT parameters.
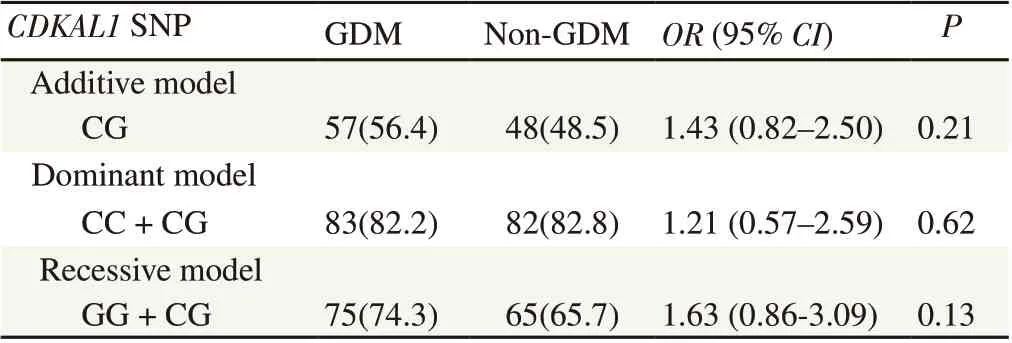
Table 6. Logistic regression analysis of genotype distribution and genetic models of rs7754840 (C/G) with GDM status [n(%)].
4. Discussion
In this study, we focused on the CDKAL1 gene, which encodes for cyclin-dependent kinase 5 regulatory associated protein 1-like.The CDKAL1 gene is commonly described in most studies as a T2DM and/or GDM susceptibility gene and yet the mechanism linking this variant with diabetes is unclear[27]. CDKAL1 gene is located in chromosome 6p22.3 falling within an intronic region of the 700 kb CDKAL1 locus and encodes for the CDKAL1 protein.CDKAL1 protein is a methylthiotransferase that acts as a tRNA modification enzyme that enhances translation of numerous transcripts in pancreatic beta cells, including that of proinsulin[28].More specifically, it modifies tRNA(UUU), a codon important for the conversion of proinsulin to insulin. In a study involving knock out mice (CDKAL1 -/-), genetic ablation of CDKAL1 in beta cells causes glucose intolerance in vivo by affecting the first-phase insulin exocytosis[29].
Previous studies have shown that defects in CDKAL1 protein or variations in CDKAL1 gene may affect insulin secretion[27,30]. For instance, in the study of metabolically well offspring of T2DM patients, the presence of CDKAL1 is related to impaired first-phase insulin release, which provides a good explanation why CDKAL1 gene variations or polymorphisms have been widely linked with risk and development of T2DM[31]. It has been shown in the study that CDKAL1 controls first-phase insulin exocytosis by facilitating adenosine triphosphate (ATP) generation, glucose-induced potassium ATP channel responsiveness, and calcium channel activity, and any defect in these processes increases the risk of developing diabetes[30].
In this study, we delved into the association of GDM and the polymorphism in two genetic loci of the CDKAL1 gene. The results of the present study revealed that the presence of rs7754840 gene variants were not significantly associated with risk of developing GDM among Filipino pregnant women. Varying results on the association of the rs7754840 gene polymorphism with GDM are expected due to variability within study populations among several studies. Our results, insofar as CDKAL1 rs7754840 is concerned,concur with those from independent studies on South Indian[32],Japanese[33], and Egyptian[34] populations.
In a number of genome-wide association studies, CDKAL1 is recognized as a strong candidate gene for the development of T2DM in various study populations[35-37] and polymorphisms of this gene had been investigated by previous researchers in patients with T2DM[27,37]. Due to the similarities in pathophysiology and development of T2DM and GDM, association between CDKAL1 gene expression and GDM development is continuously being studied. Previous studies showed significant associations of CDKAL1 gene polymorphisms namely, rs7754840 and rs7756992, with GDM development. Among Malaysian women with GDM, significant odds ratios were observed in the presence of rs7754840 and rs7756992(OR 2.19, 95% CI 1.30-3.69 and OR 2.04, 95% CI 1.62-2.58,respectively; P<0.05), implying an increased risk to develop GDM when these SNPs are present[36]. Similarly, in the study of Rosta et al, a significant risk association was found between CDKAL1 rs7754840 and GDM development (OR 1.51, P=0.016)[38]. Although various studies have identified significant associations with these SNPs and GDM risk, some studies have also shown insignificant results. A study done by Noury et al showed insignificant association between rs7754840 and GDM[34].
Numerous other variants of the CDKAL1 gene were also identified and studied with regards to their association with GDM. Tarnowski et al found out that among Polish pregnant women, CDKAL1 rs10946398 CC genotype is associated with the need for insulin therapy among the participants, but cannot be considered as a predictor for GDM risk[39]. Meanwhile, SNPs including rs4712527(A/G), rs9350276 (A/G), rs7748720 (A/G), and rs6938256 (A/G)were found to be correlated with a reduced risk of developing GDM in a Chinese population of pregnant women. In the same study population, rs9295478 (G/A), rs6935599 (A/G), and rs7747752 (C/G) were associated with an increased predisposition to developing GDM[40]. Another study by Ju et al on seven common variants of CDKAL1 gene (rs4712523, rs10946398, rs7756992, rs7766070,rs9368222, rs6931514, and rs9465871) showed no significant associations with gestational glycemic traits[41]. Variations in the results of association studies between CDKAL1 gene polymorphisms and GDM may be attributed to population-based variability of genetic variations.
Genotypic and allelic distribution of rs7756992 among the selected Filipino pregnant women did not conform to the Hardy-Weinberg equilibrium, and further analysis on the SNP was no longer conducted. Various factors could have led to its departure from Hardy-Weinberg equilibrium, such as mutation, natural selection, non-random mating, genetic drift and gene flow[26], and identification of the possible reason for deviation was not included in the scope of this study. Analyzing a small sample size could have an effect on our results and could differ as the sample size grows.Therefore, investigating a larger number of Filipino pregnant women could strengthen the results of this study. Indirect factors that could also contribute to development of GDM, like socioeconomic status,education, degree of access to healthcare, should also be considered in future studies.
In conclusion, the CDKAL1 gene rs7754840 polymorphism has no association with GDM risk. Nonetheless, the result is important as this study is able to show that genetic variability may or may not affect the risk to develop certain conditions, that is, depending on population and/or ethnicity of population under study.
Conflict of interest statement
The authors have declared no conflicts of interest.
Funding
This work was supported by the Department of Science and Technology-Philippine Council for Health Research and Development (Grant No. 18-0200). Emilyn U. Alejandro was supported by the Balik Scientist Program of the Department of Science and Technology and the National Institutes of Health.
Authors’ contributions
Maria Ruth Pineda-Cortel is the research head and has contributed to experimentation, writing and editing the journal article. Karlo Baybayan, Peter Louie Bello, Latiffa Lois Camenforte, Stefany Jane Ching, Kathleen Conti, Jeremiah Jose Ignacio, Adrian Villavieja,and Jefferyl Kae Pandac were responsible for patient recruitment and experimentation. Jiovanni Diaz, Adrian Villavieja, and Jefferyl Kae Pandac contributed to the writing of the journal article. Adrian Villavieja and Emilyn U. Alejandro edited the contents of the journal article. Asian Pacific Journal of Reproduction2021年4期
Asian Pacific Journal of Reproduction2021年4期
- Asian Pacific Journal of Reproduction的其它文章
- Descriptive histomorphological evaluation of the testis and caudal epididymis following treatment with rooibos (Aspalathus linearis), honeybush (Cyclopia intermedia) and sutherlandia (Lessertia frutescens) in healthy and streptozotocininduced diabetic rats
- Oral supplementation of selenium improves post-thaw sperm quality in Saanen bucks
- Taxifolin attenuates ischemia-reperfusion induced oxidative ovarian damage in rats
- Proposed age-stratified reference intervals of FSH derived from normozoospermic men
- Etiopathogenesis of reproductive tract infections and the emerging role of bitter taste receptors: A scoping review
