Early genetic diagnosis of clarithromycin resistance in Helicobacter pylori
Xiao-Hua Li, Yong-Yi Huang, Lin-Ming Lu, Li-Juan Zhao, Xian-Ke Luo, Ru-Jia Li, Yuan-Yuan Dai, Chun Qin,Yan-Qiang Huang, Hao Chen
Abstract
Key Words: Helicobacter pylori; Clarithromycin-resistance; Diagnostic gene; Early genetic diagnosis; Helicobacter pylori strains
INTRODUCTION
Helicobacter pylori(H. pylori) is recognized as an important human pathogen that colonizes the gastric mucus, resulting in superficial gastritis, atrophic gastritis, and gastric cancer[1 -3 ]. Present treatments forH. pyloriinfection include proton pump inhibitors, bismuth in combination with amoxicillin, metronidazole and clarithromycin[4 ,5 ]. The rate of drug resistance is increasing because of the wide use of antibiotics and high resistance rates to clarithromycin, metronidazole, and levofloxacin are associated with the failure ofH. pylorieradication[6 -8 ]. The World Health Organization designated clarithromycin-resistantH. pyloria high priority bacterium for antibiotic research and development[9 ].
At present, the mechanism of antibiotic resistance ofH. pyloriis not completely understood[10 ,11 ]. It is widely accepted that the resistance to these antimicrobials is related to mutations inH. pylorigenes, and clarithromycin-resistant strains present three point mutations in the region of domain V of 23 S ribosomal RNA (rRNA):A2142 G, A2142 C, and A2143 G[12 ,13 ]. In addition to the mutations, the efflux pump cluster is also involved in the development of resistance to clarithromycin[14 ,15 ].However, there may be gene mutation sites that are not yet known, and the mechanism of drug resistance warrants further study.
We isolated and culturedH. pylorifrom the population in Bama County, which is a township known for the longevity of its residents in Guangxi, and randomly selected three strains of multiple drug-resistantH. pyloriwith resistance to clarithromycin.Complete genome sequences were analyzed to study the genomic characteristics of the strains and to elucidate the underlying mechanism of drug resistance inH. pylori.
MATERIALS AND METHODS
Isolation and culture of H. pylori
This study had received a strict medical ethics review from Youjiang Medical University for Nationalities. Written informed consent was obtained from each patient.Gastric mucosa tissue samples were collected from the People's Hospital of Bama Yao Autonomous County in patients’ gastric body and pylorus with gastritis or gastric ulcers. Isolation and culture ofH. pyloriwere performed at the Research Center for the Prevention and Treatment of Drug Resistant Microbial Infection, Youjiang Medical University for Nationalities. Patients investigated had not taken any antibiotics for at least 4 wk before examination. The isolation and identification ofH. pyloriwere performed as previously described[16 ,17 ]. The bacteria were cultured on Columbia agar plates containing 5 % fresh defibrinated sheep blood. The microaerophilic conditions included 5 % O2 , 10 % CO2 , and 85 % N2 at 37 °C for 3 to 5 days. Suspicious colonies were confirmed by Gram staining, urease, oxidase, and catalase activity testing, and urease gene polymerase chain reaction (PCR).
Antibiotic susceptibility testing
The antibiotic resistance ofH. pyloriwas measured by dilution methods with reference to the protocols of the Clinical and Laboratory Standards Institute (Wayne, PA, United States)[18 ]. Briefly, the density of H. pylori was adjusted to be 1 × 106 CFU/mL and incubated at 37 °C for 3 to 5 d under microaerophilic conditions. After incubation, the plates were visually examined and the minimal inhibitory concentration (MIC) was determined to be the lowest concentration that resulted in no turbidity. Metronidazole(Aladdin, d1707126 ), amoxicillin (Xiansheng pharmaceutical, Co., Ltd, China),levofloxacin (Shandong Lukang Pharmaceutical Group Saite Co., Ltd, China), and clarithromycin (Yangzi River Pharmaceutical Group Co., Ltd, China) were also used.
Complete genome sequencing and analysis
Drug-resistant strains were selected and sent to the Shenzhen Huada Gene Co., Ltd(China) for complete genome analysis. After the DNA samples were delivered, the quality of the samples was tested and then used to construct a BSlibrary. The purified genomic DNA samples including genomic DNA, bacterial artificial chromosomes, or long-length PCR products were sheared into smaller fragments by CovarisS/E210 or using a Bioruptor. The overhangs resulting from fragmentation were converted into blunt ends using T4 DNA polymerase, Klenow fragment, and T4 polynucleotide kinase. After adding an ‘A’ base to the 3 ' end of the blunt phosphorylated DNA fragments, adapters were ligated to the ends of the DNA fragments. The desired fragments were purified though gel-electrophoresis, selectively enriched, and amplified by PCR. The index tag was introduced into the adapter at the PCR stage as appropriate and a library quality test was conducted. Finally, the qualified BSlibrary was used for sequencing. Genomic component and gene function analyses were performed, including gene prediction, tRNA, sRNA, and gene annotation, and prediction of open reading frames by GO.
Drug-resistant gene detection
Drug-resistant genes were predicted based on the results of the complete genome sequence analysis and selected for detection by reverse transcription PCR (RT-PCR).The reaction for cDNA synthesis was held at 25 °C for 10 min, 42 °C for 60 min, and then 99 °C for 5 min. The reaction consisted of 32 cycles with each cycle composed of 1 min at 95 °C, 4 min at 56 °C, and 7 min at 70 °C. After a final extension of 15 min at 72 °C, the RT-PCR products were visualized by electrophoresis on 1 % agarose gel and 15 % acrylamide gel with a 200 -bp ladder size marker.
Knockout of mutant genes
Hp1181 and hp1184 knockout mutants were constructed by insertion of the KAN resistance cassette. Double-knockout mutants were made by natural transformation of the KAN resistance cassette with pBSII KS (as presented by Bi HK, Laboratory of Nanjing Medical University, China) containing an internal fragment interrupted with a cat cassette from pAV35 , with selection for both KAN- and CHL-resistant colonies.Insertion of the KAN and cat resistance cassette at the desired locations in theH. pyloriputative efflux genes was validated by PCR.

Table 1 Drug resistance characteristics of three drug-resistant strains (minimal inhibitory concentration: μg/mL)
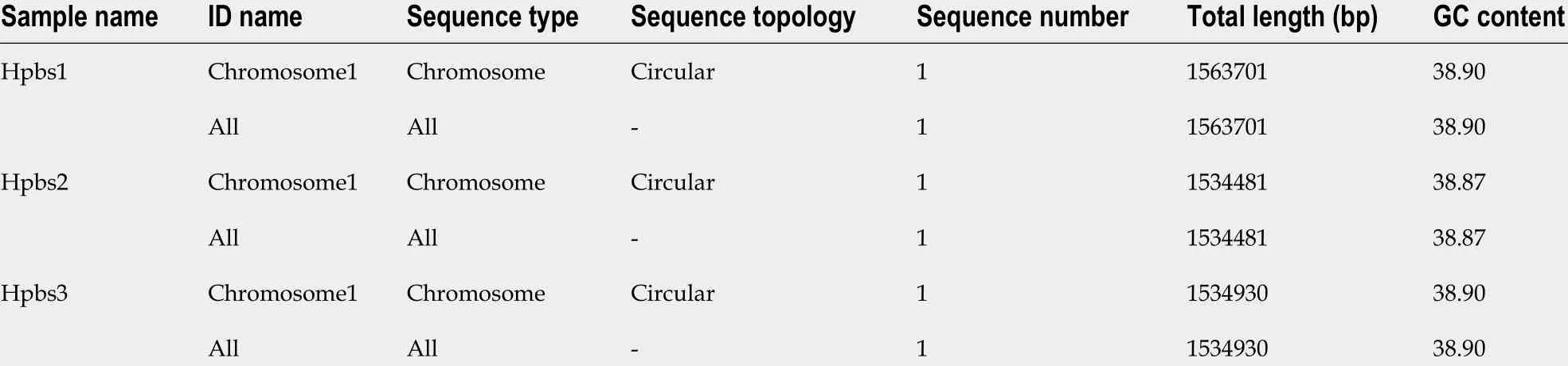
Table 2 Sequence information of three drug-resistant strains
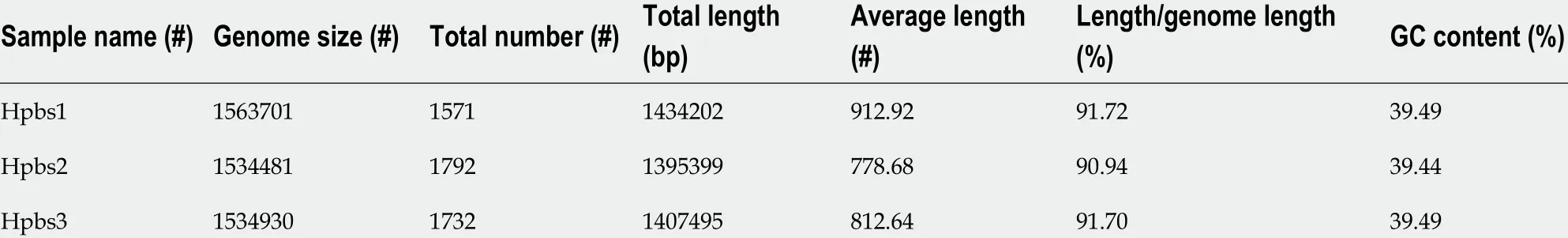
Table 3 Gene information of three drug-resistant strains
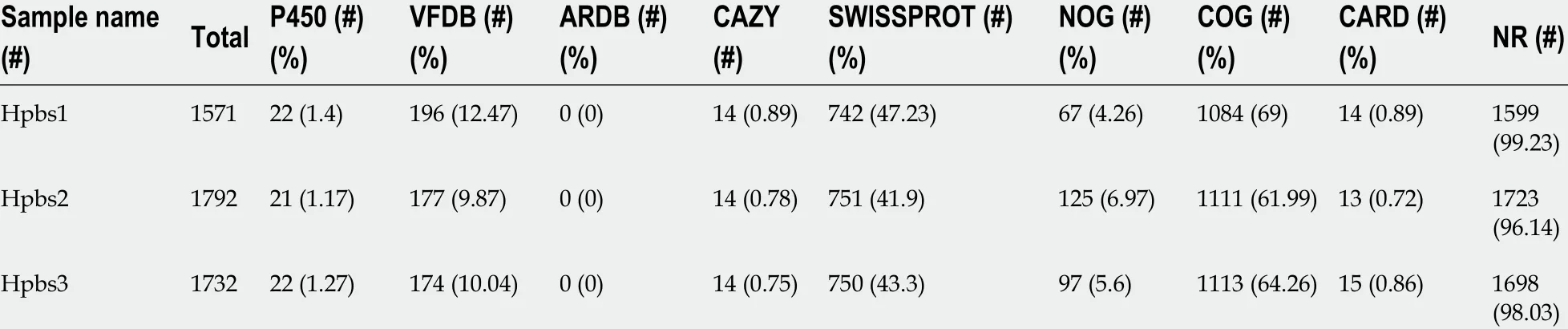
Table 4 Gene annotation statistics A
Induction of drug resistance
The MIC of clarithromycin to hp26695 was detected. Drug resistance was induced by 1 /4 MIC. The culture medium was changed every 2 d and MIC was detected every 4 d. The concentration of induced drug was changed with MIC.
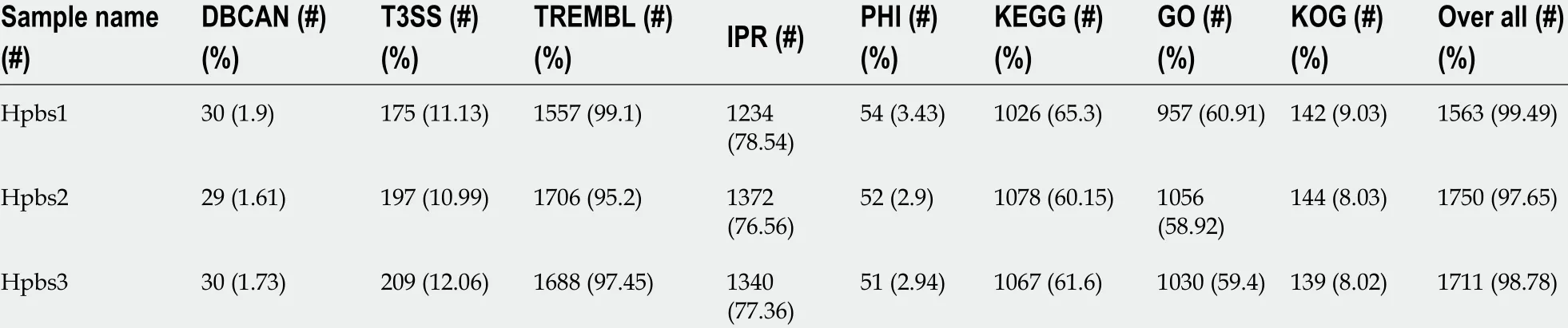
Table 5 Gene annotation statistics B
RESULTS
Bacterial resistance
Three drug-resistant strains were isolated and identified by Gram staining, urease,oxidase, catalase activity testing, and urease gene PCR. The drug resistance information of these strains is summarized in Table 1 .
Bacterial sequence information
Based on the valid data from the previous sequencing platform, the CleanData could be assembled for each sample and the optimal assembly results were obtained after multiple adjustments. The assembly sequence was analyzed by correcting single base,circular judgment, and plasmid comparison. The results of the genome assembly statistics of each sample are displayed in Table 2 . These three strains have been uploaded to the NCBI Biosample database: Hpbs1 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN10461767 ). Hpbs2 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN10663081 ), and Hpbs3 (https://www.ncbi.nlm.nih.gov/biosample/?term=SAMN10663175 ).
Gene information
Gene prediction was applied to determine gene composition. The statistics are shown in Table 3 .
Circular genome analysis
GC skew analysis was carried out using (G-C)/(G+C) calculations based on genomic sequences of strains. The results of gene distribution, ncRNA distribution, and gene annotation are demonstrated in Figure 1 . Hpbs1 had 835 genes, 26 tRNAs, 6 rRNAs,and 2 sRNAs in the positive chain. It also had 736 genes, 10 tRNAs, 0 rRNAs, and 5 sRNAs in the negative chain and 157 repeats without positive or negative chain. There are 943 genes, 26 tRNAs, 6 rRNAs, 3 sRNAs, 849 genes, 10 tRNAs, 0 rRNA, 3 sRNAs,and 153 repeats in Hpbs2 ; there are 869 genes, 26 tRNAs, 6 rRNAs, 3 sRNAs, 863 genes, 10 tRNAs, 0 rRNA, 3 sRNAs, and 155 repeats in Hpbs3 .
Gene annotation
Functional annotation was accomplished by analysis of protein sequences. We aligned genes with databases to obtain their corresponding annotations. To demonstrate the biological meaning, the highest quality alignment result was chosen as a gene annotation. Functional annotation was completed by blast resistance genes with different databases. In this project we have finished annotations using 17 databases,including P450 , VFDB, ARDB, CAZY, SWISSPROT, NOG, COG, CARD, NR, DBCAN,T3 SS, TREMBL, IPR, PHI, KEGG, GO, and KOG. The annotation results are shown in Tables 4 and 5 .
Analysis of drug-resistant gene database
The drug resistance gene numbers of three strains were different in the CARD(Comprehensive Antibiotic Resistance Database), which are 14 , 13 , and 15 genes,respectively. However, after sorting, it was found that some genes were repetitive. The specific numbers and characteristics of genes are presented in the Tables 6 and 7 .NP_207975 .1 and NP_207972 .1 were efflux pump genes of 26695 strain, i.e., hp1181 andhp1184 genes. Their drug resistance was verified by RT-PCR, as illustrated in Figure 2 .After knocking out the drug-resistant genes, drug sensitivity was significantly improved, as shown in Figure 3 .
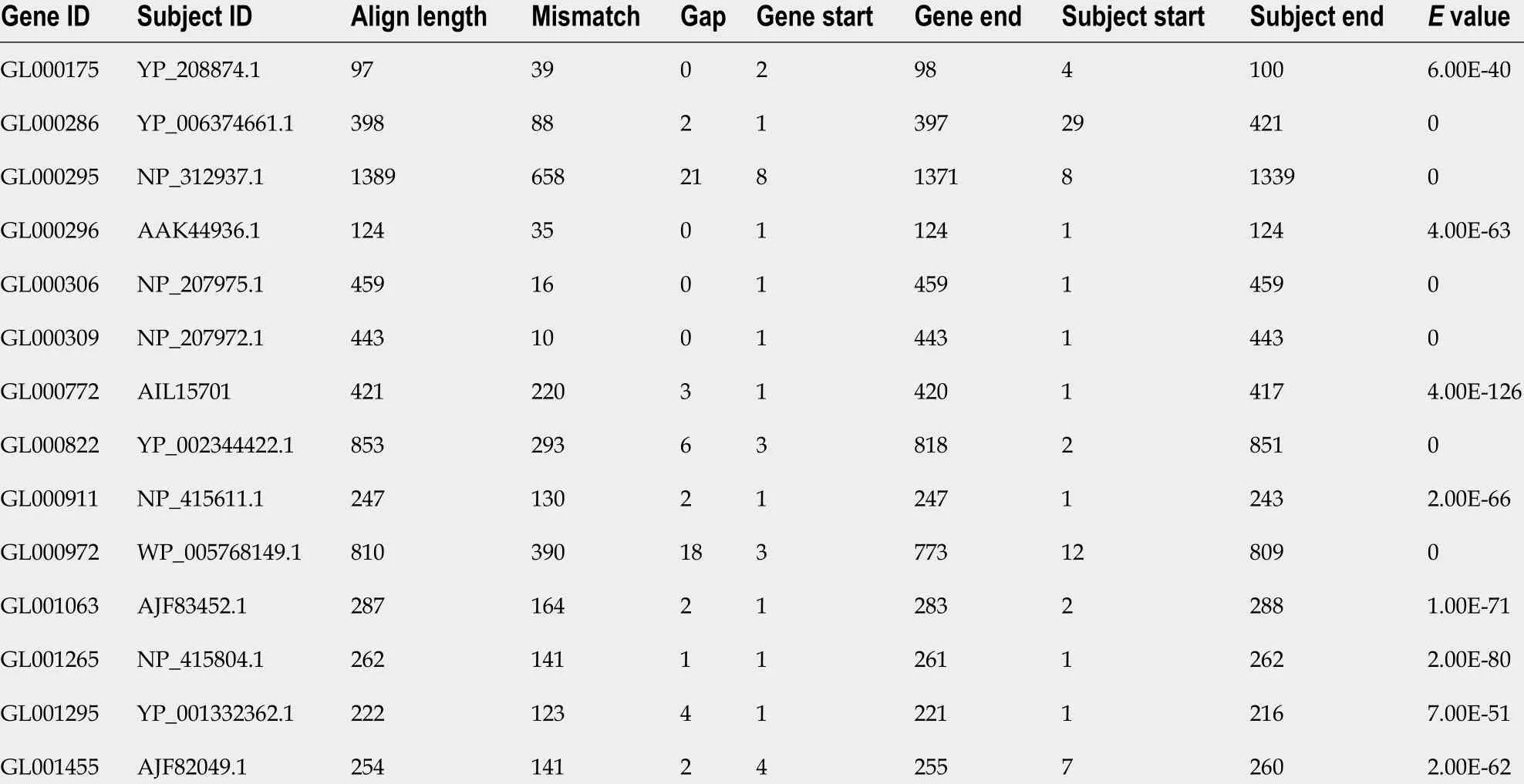
Table 6 Analysis of gene resistance in CARD
Identification of 2 3 S rRNA gene mutations
As three strains were resistant to clarithromycin, so we analyzed and identified the sites of clarithromycin-resistant mutations. We found that three strains had mutations in A2142 G, A2143 G, G2144 T, and some had mutations in other sites, as shown in Table 8 .
Gene mutation induced in drug-resistant strains
After induction with clarithromycin, hp26695 drug resistance was enhanced on the 12 th day, reached the highest level on day 16 , and increased to 8 μg/mL on the 24thday. The expression ofhp1181 and hp1184 was also increased with increasing clarithromycin resistance, especiallyhp1184 , as shown in Figure 4 . Only A2142 G and A 2143 G mutations were detected in 23 S RNA, with no other mutation sites being found,as shown in Table 9 . These data indicated that these two genes may be involved early in the regulation of clarithromycin resistance.
DISCUSSION
The treatment ofH. pyloriinfection remains reliant on bismuth tetralogy at present.H.pyloriis eradicated clinically using common antibiotics including clarithromycin,amoxicillin, metronidazole, tetracycline and levofloxacin. However, in recent years,the growing rate of antibiotic resistance has resulted in the failure ofH. pylorieradication[19 ,20 ]. The most serious resistance has developed to drugs including metronidazole, clarithromycin, and levofloxacin star. The common mechanisms of bacterial resistance involve the production of inactivated enzymes, change in the target position of antibacterial drugs, change in the permeability of bacterial outer membrane, effects on the active outflow system, and formation of bacterial biofilm and cross resistance[21 -23 ]. There are some differences in the mechanisms of drug resistance of each kind of bacteria; however, the same kind of bacteria still have different resistances to the same antibiotic in different areas[24 ]. The mechanism of drug resistance ofH. pyloriremains unclear and needs further study.
We selected drug-resistant strains using metronidazole, clarithromycin, and levofloxacin for genome sequencing analysis. We found that there were no significant differences in the number of drug-resistant genes in the CARD database. This may be because two kinds of antibiotic resistance can develop and the drug-resistant genes inH. pyloriare mainlyhp1181 and hp1184 . Hp1181 encodes a putative NDA translocase that is related to the major facilitator superfamily and is an integral membrane protein;hp1184 encodes another translocase that belongs to the MATE family, resulting in the aforementioned susceptibility. These can contribute to resistanceviaa multidrug-resistant efflux protein, active-efflux of antibiotics, and other efflux pump genes, such asHefA. After knockout of these two genes, the MICs of the drugs were significantly decreased and the sensitivity was increased. It is noteworthy that in addition to these two genes, theGE2270Agene ofEnterococcusandMurAgene ofEscherichia colialso show a correlation. It is likely that the drug-resistant plasmids of other strains invadeH. pylorithrough transformation or other mechanisms. Bacteria other thanH. pyloriinthe gastric mucosa of patients can indirectly confirm this view. The main reason for this may be long-term acid resistant treatment, gastric erosion, or intestinal bacterial reflux. This will lead to drug resistance becoming more difficult to prevent and control. In addition, all three strains have clarithromycin resistance. The mechanism of resistance to clarithromycin is mainly reflected in the mutations A2142 G, A2143 G, and G2144 T. In addition, it is common that there are several mutations in the same strain.

Table 7 Characteristics of drug-resistant genes in CARD
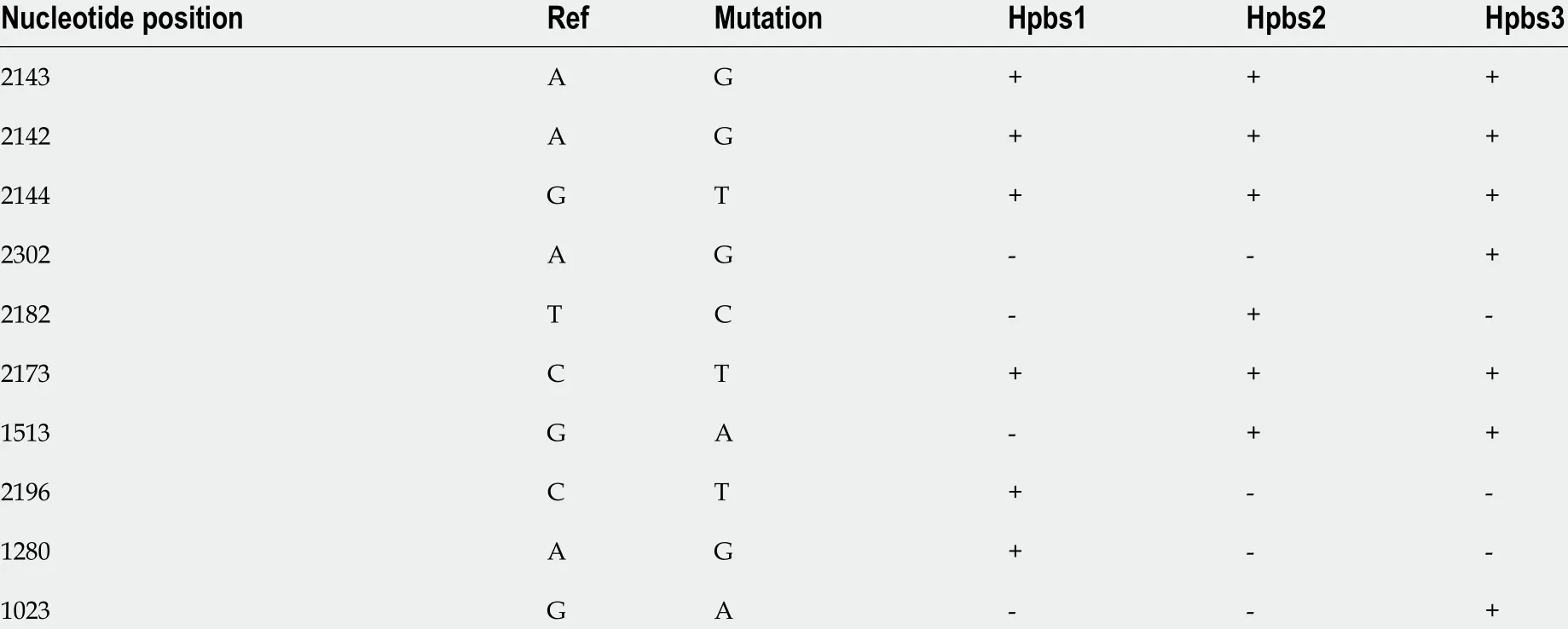
Table 8 Mutations in the 23 S rRNA genes of Helicobacter pylori strains

Table 923 S rRNA mutations of Helicobacter pylori strains
Hp1181 and hp1184 are related to multidrug resistance and to clarithromycin resistance, which has been previously reported in the literature[25 ,26 ]. The RNA expression ofhp1181 and hp1184 were increased with the emergence of clarithromycin resistance, withhp1184 showing the fastest increase. Therefore, these genes are also involved in the regulation of drug resistance and may be one of the mechanisms ofH.pyloriresistance to clarithromycin. Compared with the clinical isolates, 23 S RNA mutation sites ofH. pyloriwere less frequent in artificially induced strains that had only A2142 G and A2143 G mutations. These may be attributed to the single factor of artificial induction that is not as complex as human stomach environment. More importantly,hp1181 and hp1184 mutations may be the earliest and most persistent response to clarithromycin resistance, and they may be the main target genes for the diagnosis, prevention, and treatment of clarithromycin resistance.
The genetic characteristics of multidrug-resistant strains in this area were preliminarily identified: The relationship betweenhp1181 or hp1184 and clarithromycin resistance was ascertained through genome sequencing analysis and gene function identification of drug-resistantH. pylorifrom Bama County, Guangxi Province. Our study further provided an improved experimental basis for the prevention and treatment of drug resistance ofH. pylori.
CONCLUSION
Hp1181 and hp1184 mutations may be the earliest and most persistent response to clarithromycin resistance, and they may be the main target genes for the diagnosis,prevention, and treatment of clarithromycin resistance.
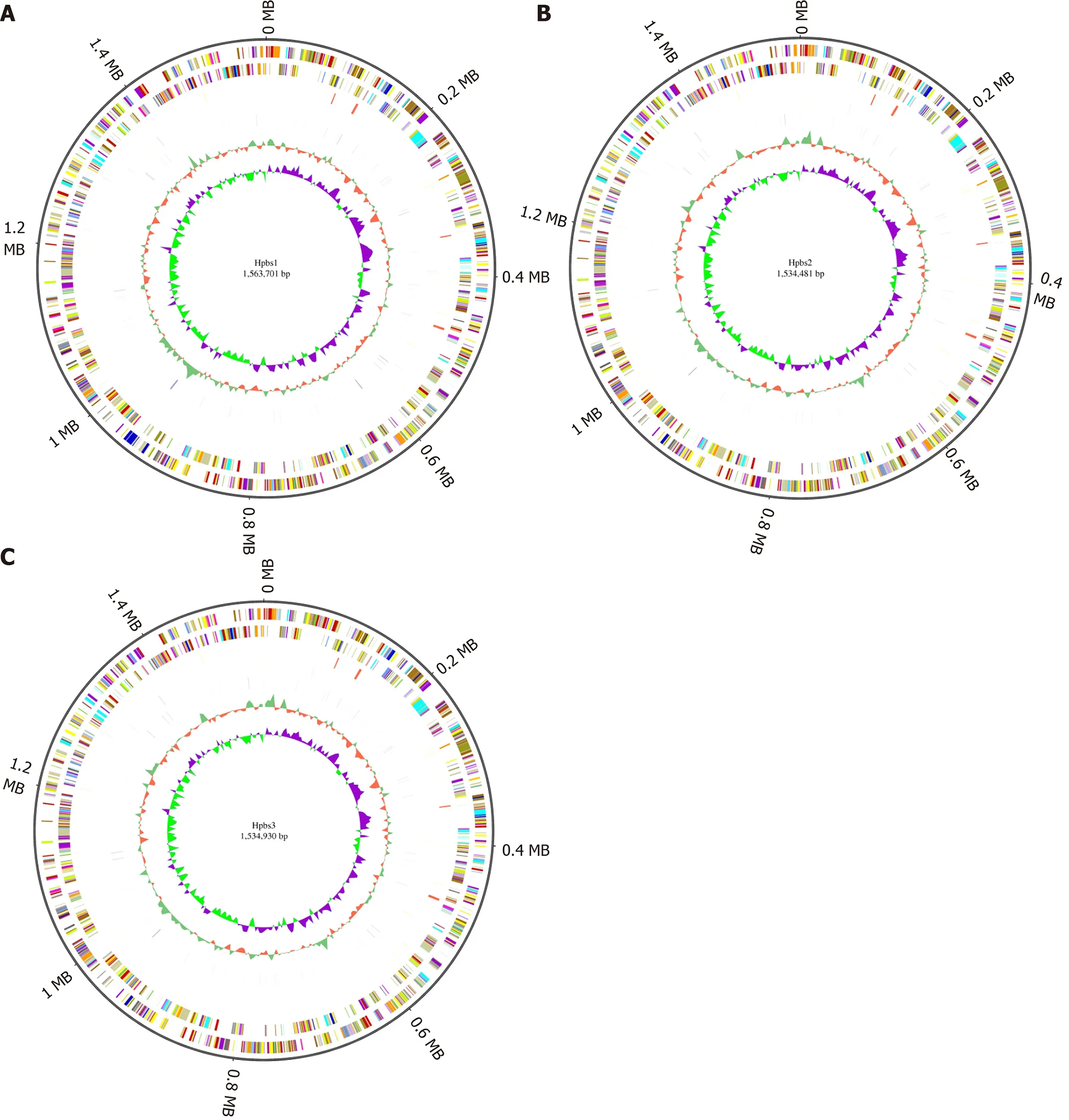
Figure 1 Circular genome analysis of three drug-resistant strains. A: Hpbs1 ; B: Hpbs2 ; C: Hpbs3 .
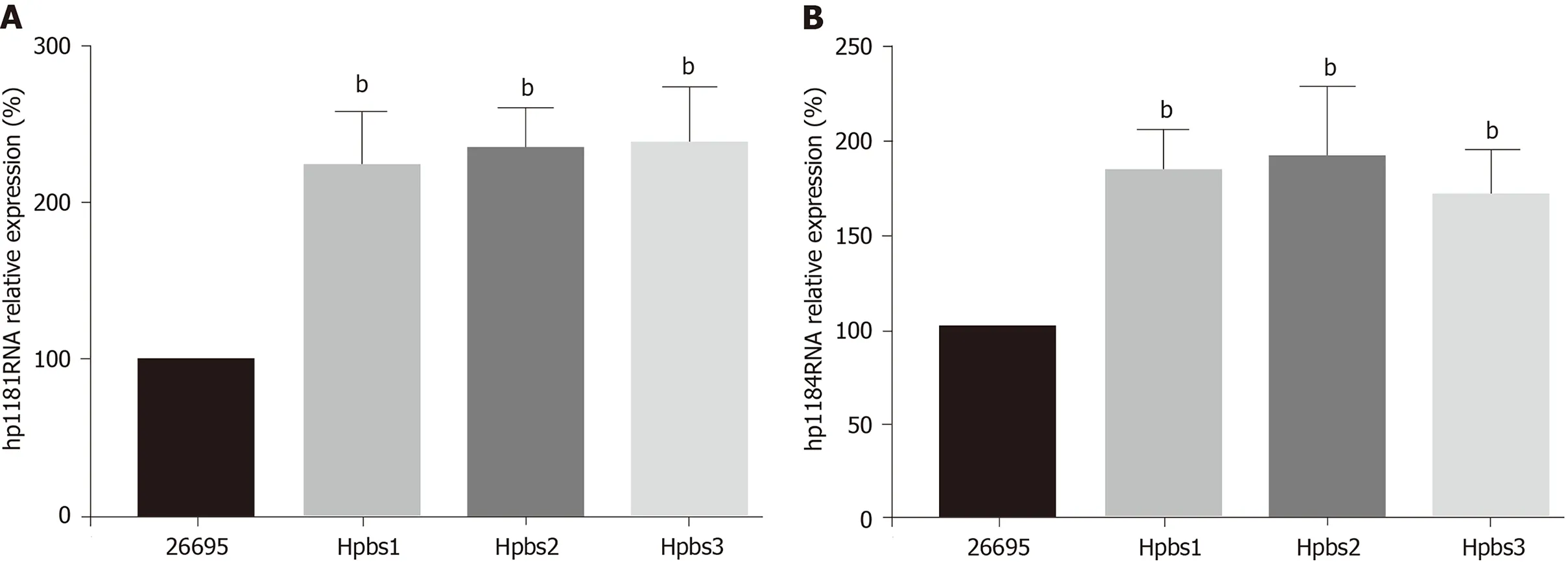
Figure 2 Hp1181 and hp1184 gene expression in drug-resistant strains. A: Hp1181 ; B: Hp1184 . bP < 0 .01 .
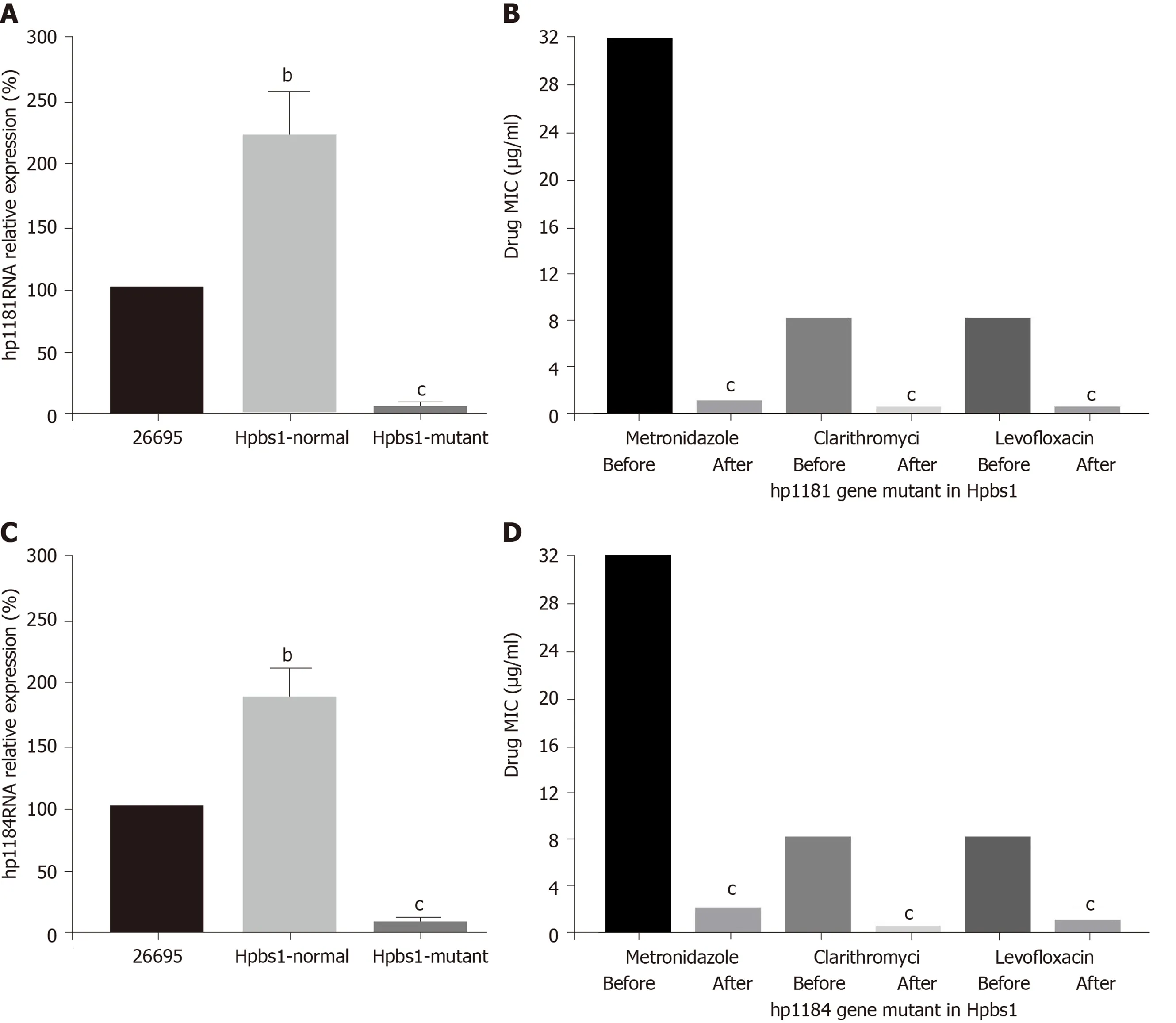
Figure 3 Drug sensitivity is improved after knockout of the drug-resistant genes. A: Hp1181 knockout; B: Minimal inhibitory concentration (MIC) afterhp1 181 knockout; C: Hp1184 knockout; D: MIC after hp1184 knockout. MIC: Minimal inhibitory concentration. bP < 0 .01 ; cP < 0 .001 .
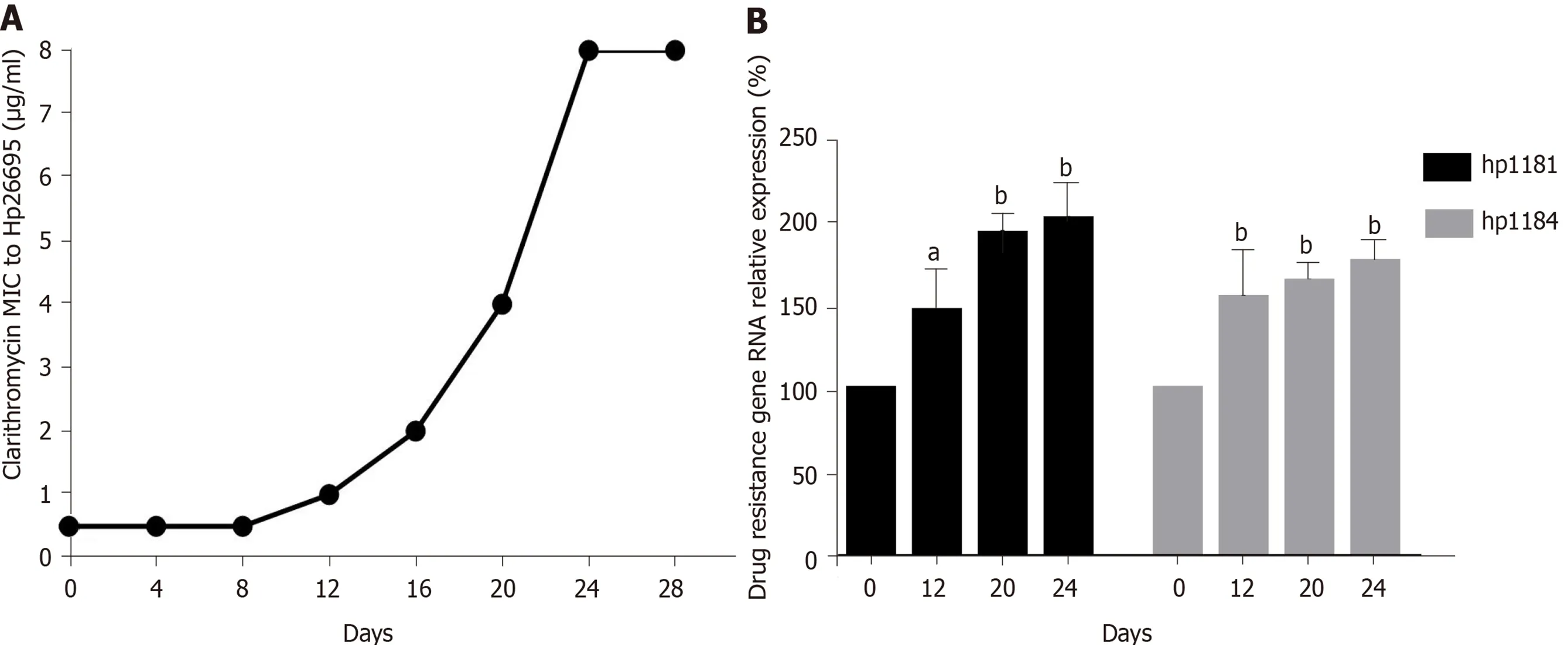
Figure 4 Induction of resistance to clarithromycin and expression of drug-resistant genes in Helicobacter pylori. A: Induction of clarithromycin resistance; B: Expression of drug-resistant genes. aP < 0 .05 ; bP < 0 .01 .
ARTICLE HIGHLIGHTS

ACKNOWLEDGEMENTS
The authors thank Huang YN and Huang MY working in Guangxi Bama Yao Autonomous County People Hospital who helped to collect gastric mucosal samples from clinical patients.
 World Journal of Gastroenterology2021年24期
World Journal of Gastroenterology2021年24期
- World Journal of Gastroenterology的其它文章
- COVID-19 and its effects on the digestive system
- Weight loss interventions in living donor liver transplantation as a tool in expanding the donor pool: A systematic review and meta-analysis
- Disorders of the brain-gut interaction and eating disorders
- Stem cell injection for complex anal fistula in Crohn’s disease: A single-center experience
- Altered profiles of fecal bile acids correlate with gut microbiota and inflammatory responses in patients with ulcerative colitis
- Chronic intestinal failure and short bowel syndrome in Crohn’s disease
