水牛Novel-miR-57对Bcap-37和BMECs细胞DOK4基因的调控作用
蔡小艳 李雅辉 鲍正潘 陈秋萍 李胜 周宇 邓凯 石德顺 刘庆友


摘要:【目的】篩选Novel-miR-57调控靶基因,并明确其对靶基因的调控作用及生物功能,为揭示水牛乳腺上皮细胞(BMECs)的分化机理提供科学依据。【方法】利用MiRscan预测Novel-miR-57二级结构;以自编软件Ensembl(v80)注释的水牛mRNA截取3'-非翻译区(3'-UTR)作为预测数据库,采用Miranda(v3.3a)对Novel-miR-57进行靶基因预测;运用实时荧光定量PCR筛选重点靶基因。以化学合成的Novel-miR-57模拟物Mimics和抑制剂Inhibitor,分别转染人类乳腺癌细胞(Bcap-37)及BMECs细胞,以验证Novel-miR-57与靶基因的表达相关性。【结果】Novel-miR-57前体序列形成7个茎环结构,成熟序列位于第1、2和3个茎环结构间,其结合自由能为-53.70 kcal/mol。以结合自由能低于-20.00 kcal/mol为标准,最终筛选出34个可能的靶基因,共与42条KEGG信号通路存在关联,其富集的信号通路主要有代谢通路(ID:bta01100)、PI3K-Akt(信号通路(ID:bta04151)、MAPK信号通路(ID:bta04010)和细胞因子—细胞因子受体相互作用(ID:bta04060)等;经实时荧光定量PCR检测分析发现DLX3、CANCNG3、DOK4、NFKBID、C17orf53、RTN1和FBXO10等7个靶基因在非泌乳期的相对表达量极显著高于泌乳期(P<0.01),二者间相差100.0倍以上,且与Novel-miR-57的相对表达量呈负相关。7个靶基因中仅DOK4基因与Novel-miR-57的表达具相关性,以200 nmol/L Inhibitor转染B-cap37细胞能显著提高DOK4基因表达(P<0.05,下同),添加100 nmol/L Mimics则显著抑制DOK4基因表达。以100 nmol/L Mimics转染BMECs细胞,Novel-miR-57和DOK4基因的相对表达量显著提高;而以200 nmol/L Inhibitor转染BMECs细胞,Novel-miR-57和DOK4基因的表达均受到显著抑制。【结论】Novel-miR-57含有7个茎环结构,且其成熟序列位于第1~3个茎环上。Novel-miR-57过表达可下调Bcap-37细胞DOK4基因表达或上调BMECs细胞DOK4基因表达,即Novel-miR-57对靶基因的调控作用因乳腺细胞生理状态不同而存在差异。
关键词: 水牛;Novel-miR-57;靶基因;BMECs细胞;Bcap-37细胞;调控作用
中图分类号: S823.83 文献标志码: A 文章编号:2095-1191(2021)02-0269-11
Abstract:【Objective】In order to provide scientific basis for revealing the differentiation mechanism of buffalo mammary epithelial cells(BMECs), the regulatory target gene of Novel-miR-57 was screened to clarify its regulatory function and biological function on target genes. 【Method】MiRscan was used to predict the secondary structure of Novel-miR-57. The target gene of Novel-miR-57 was predicted by Miranda(v3.3a) using buffalo mRNA truncated 3-untranslated region (3'-UTR) annotated by Ensembl(v80) as prediction database. Key target genes were screened by real-time fluorescence quantitative PCR. To verify the correlation between Novel-miR-57 and target gene expression, chemically synthesized mimics and inhibitor were transfected into human breast cancer cells(Bcap-37) and BMECs cells, respectively. 【Result】The precursor sequence of Novel-miR-57 formed seven stem-loops, and the mature sequence was located between the first, second and third stem-loops, and its binding free energy was -53.70 kcal/mol. With the binding free energy lower than -20.00 kcal/mol as the standard, 34 possible target genes were finally screened out, which were associated with 42 KEGG signaling pathways. The enriched signaling pathways mainly included metabolic pathway(ID:bta01100), PI3K-Akt(ID:bta04151), MAPK signaling pathway(ID:bta04010) and cytokine-cytokine receptor interaction (ID:bta04060). The real-time fluorescence quantitative PCR showed that the relative expression of seven target genes,DLX3, CANCNG3, DOK4, NFKBID, C17orf53, RTN1 and FBXO10, were significantly higher in non-lactation period than in lactation pe-riod(P<0.01), and the difference between them was more than 100.0 times, which were negatively correlated with the relative expression of Novel-miR-57. Only the expression of DOK4 gene was correlated with the expression of Novel-miR-57 among the seven target genes. Transfection of B-cap37 cells with 200 nmol/L inhibitor could significantly increase the expression of DOK4 gene(P<0.05, the same below), while addition of 100 nmol/L mimics could significantly inhibit the expression of DOK4 gene. The relative expression of Novel-miR-57 and DOK4 gene was significantly increased when BMECs cells were transfected with 100 nmol/L mimics. The expression of Novel-miR-57 and DOK4 gene were significantly inhibited when BMECs cells were transfected with 200 nmol/L inhibitor. 【Conclusion】Novel-miR-57 contains se-ven stem-loops, and its mature sequence locates on the first to the third stem rings. Overexpression of Novel-miR-57 can down-regulate DOK4 gene expression in Bcap-37 cells or up-regulate DOK4 gene expression in BMECs cells, that is, Novel-miR-57 has different regulating effects on target genes due to different physiological states of breast cells.
Key words: buffalo; Novel-miR-57; target gene; BMECs cell; Bcap-37 cell; regulating effects
Foundation item: National Natural Science Foundation of China(31960680); Ningxia Key Research and Development Project(2018BEB04031)
0 引言
【研究意义】microRNAs(miRs)是一类约22 bp的非编码小分子RNA,大量存在于哺乳动物体内(李新云等,2017)。miRs通过与靶mRNA分子的3'-非翻译区(3'-UTR)不完全结合以抑制或降解mRNA翻译(Stark et al.,2005),进而调控细胞的发育、分化及增殖等重要生命活动过程(王塑天等,2013;Jiao et al.,2019)。miRs還参与乳腺发育,包括维持乳腺上皮前体细胞、促使乳腺上皮管道长出及促进乳腺上皮细胞增殖分化等(Wang et al.,2018)。因此,研究miRs对乳腺细胞基因的调控作用及其功能,可为揭示乳腺泌乳机制和提高泌乳性能奠定基础。【前人研究进展】目前,有关miRs的研究主要集中在功能调控和作用机理等方面,如miR-103通过靶向PANK3(Pantothenate kinase 3)调控水牛乳脂代谢(蔡小艳,2016);miR-200b通过负调控靶基因Pten(Phosphatase and tensin homologue)表达而正调控奶牛乳腺泌乳(边艳杰等,2018);miR-183通过靶向MST1(Mammalian sterile 20-like kinase 1)基因来调节山羊奶中脂肪的消化吸收、合成及分解(Chen et al.,2018);miR-3880通过与奶山羊乳腺上皮细胞OGR1(Ovarian cancer G protein-coupled receptor 1)基因3'-UTR结合而调控其mRNA和蛋白的表达(侯金星等,2019);miR-4312通过靶向PDCD4(Programmed cell death protein 4)以促进细胞增殖和抑制细胞凋亡(苑红,2019);miR-221通过靶向STAT5a(Signal transducer and activator of transcription 5A)和IRS1(Recombinant insulin receptor substrate 1)基因调控牛乳腺上皮细胞增殖(Jiao et al.,2019);miR-92a具有调控奶山羊乳腺发育和泌乳性能的潜能(包黎娟等,2020);miR-28-3p能促进三阴性乳腺癌细胞系MDA-MB-468(人乳腺癌细胞)的增殖、侵袭和抑制其凋亡(李杰等,2020);miR-380-5p能抑制乳腺癌细胞增殖和促进乳腺癌细胞凋亡(孙云菊等,2020)。此外,有少数学者针对测序新发现miRs的靶基因及其功能进行研究。崔晓钢等(2015)基于物种间序列同源比对新发现44条牛候选miRs,选取其中4条进行实时荧光定量PCR验证,并预测对应的靶基因,但尚未进一步研究其调控功能。郭丽霞(2017)基于高通量技术从5头黄牛和5头水牛的大脑组织中测序获得Novel-miR-25和Novel-miR-13,并证实二者均能调控大脑功能及神经元的分化和发育。臧树成等(2018)研究发现,处于冷应激状态下大鼠肝脏中的Novel-miR-133-5p异常上调,可能对机体的生长发育、免疫调节及遗传等有重要影响。练雨(2019)对经脂多糖(LPS)处理的猪子宫内膜上皮进行测序,结果发现Novel-mir-106-5p能抑制NF-tB激活,降低NF-tB磷酸化水平。Li等(2020)从96 条日本牙鲆miRs中筛选出pol-miR-novel-171,并证实其靶向负调控FAM49B(Family with sequence similarity 49,member B)的3'-UTR。张月(2020)研究表明,奶山羊乳腺中的Novel-miR-3880可通过PI3K/AKT/mTOR/S6Kl和Bcl-2/Bax通路调节乳腺上皮细胞细胞生长、乳脂合成和乳酪蛋白分泌,进而促进乳腺发育。【本研究切入点】尽管目前已有较多关于miRs调控牛和水牛乳腺基因表达的研究,但针对水牛乳腺组织新发现miRs的调控基因研究鲜见报道。本课题组前期基于Solexa高通量测序和实时荧光定量PCR,在创建的miRs文库中发现并鉴定出5个新的水牛miRs表达模式(蔡小艳等,2017a,2017b),其中Novel-miR-57在水牛泌乳期和非泌乳期的表达量差异极显著,是5个新发现水牛miRs中差异最明显的miRs(Cai et al.,2017),因此深入研究Novel-miR-57对揭示水牛乳腺细胞的泌乳作用机制具有重要意义。【拟解决的关键问题】通过MiRscan预测Novel-miR-57二级结构,以化学合成的Novel-miR-57模拟物Mimics和抑制剂Inhibitor分别转染人类乳腺癌细胞(Bcap-37)及水牛乳腺上皮细胞(BMECs),筛选其调控靶基因,并明确Novel-miR-57对靶基因的调控作用及生物功能,以期为揭示BMECs细胞的分化机理提供科学依据。
1 材料与方法
1. 1 试验材料
水牛乳腺组织采自广西南宁市屠宰场,一般于凌晨2:00—4:00时采集;泌乳期样本为可明显观察到有白色乳液一直流出的乳腺组织,非泌乳期样本是无法从表面上看出有白色乳液流出,且用手按压后也无乳液流出的乳腺组织。2种乳腺组织样本各3个重复,以生理盐水清洗干净后,立即使用消毒眼科手术剪在所有样本的组织内部取样1.0~2.0 g,装入2 mL的EP管中,然后立即将其放进液氮罐,带回实验室置于-80 ℃冰箱保存。BMECs细胞为亚热带农业生物资源保护与利用国家重点实验室分离获得;Bcap-37细胞由亚热带农业生物资源保护与利用国家重点实验室保存提供。DMEM培养基、FBS和Lipo2000转染试剂购自Life Technology公司;一步快速反转试剂盒购自TaKaRa公司;荧光定量SYBR Green Master和Mix罗氏转染试剂购自瑞士Roche公司。仪器设备主要有CO2培养箱(Thermo公司)、低温离心机(Beckman公司)及荧光定量PCR仪等(蔡小艳等,2016)。
1. 2 Novel-miR-57二级结构预测
登录MiRscan(http://genes.mit.edu/mirscan/),输入Novel-miR-57基因序列,单击Submit即可获得结果(刘长征和余佳,2012;王伟等,2019)。
1. 3 引物设计与合成
采用Primer 3.0进行PCR扩增引物设计,并委托生工生物工程(上海)股份有限公司合成。Novel-miR-57的茎环反转录引物为5'-GTCGTATCCAGTG CAGGGTCCGAGGTATTCGCACTGGATACGACTCGGTC-3',U6的逆转录引物为5'-CGCTTCACGA ATTTGCGTGTCAT-3';Novel-miR-57的实时荧光定量PCR上游引物为5'-GGAAATACCGGCACGAGA C-3',下游引物为5'-GTGCAGGGTCCGAGGT-3';内参基因U6的实时荧光定量PCR上游引物为5'-CTC GCTTCGGCAGCACA-3',下游引物为5'-AACGCT TCACGAATTTGCGT-3'。Novel-miR-57预测靶基因的实时荧光定量PCR扩增引物见表1。
1. 4 实时荧光定量PCR
参照蔡小艳等(2016)的方法,将0.1 g乳腺组织置于研钵中,倒入液氮迅速研磨,加入TRIzol试剂进行总RNA提取;提取获得的总RNA采用紫外分光光度计测定其浓度,并以1.0%琼脂糖凝胶电泳进行鉴定。使用AMV试剂盒(TaKaRa)反转合成cDNA:反应液配制均在冰上操作,依照AMV试剂盒说明加入各种试剂进行反转录。将反转录合成的cDNA稀释至100 ng/mL,实时荧光定量PCR反应体系20.0 μL: cDNA模板1.0 μL,SYBR GreenMaster 10.0 μL,上、下游引物(10 μmol/L)各0.3 μL,无RNA酶水8.4 μL。扩增程序:95 ℃预变性10 min;95 ℃ 15 s,60~55 ℃ 1 min,进行40个循环,收集荧光。每个样品设3个重复,以U6基因为Novel-miR-57的内参基因。
1. 5 靶基因预测
因水牛全基因组尚未公布,故以Ensembl(v80)注释的水牛mRNA截取3'-UTR作为预测数据库,采用Miranda(v3.3a)对Novel-miR-57进行靶基因预测,筛选出结合自由能小于-20 kcal/mol的3'-UTR,即获得有关Novel-miR-57靶基因总数。
1. 6 化学合成Novel-miR-57的Mimics和Inhibitor转染Bcap-37细胞和BMECs细胞Novel-miR-57 Pri序列为:CACTCAGAGATAA GAGGCTGGGTTCGACTGGGAGAGGATGCAAGTTTCAGGCTAAATACCGGCACGAGACCGATAGTCAACAAGTACCATAAGGGAAAGTTGAAAAGAACTTTGAGATGGTACGTGTGCATGCTAGAGGAACCGTTGCTGTATTAATGAAAGGGCACCTGGCACGTGGAAACACCCCTAAGATGTTCATT。
根据其成熟序列化学合成Novel-miR-57的Mi-mics(正义链:5'-AAAUACCGGCACGAGACCGA-3',反义链:5'-UCGGUCUCGUGCCGGUAUUU-3')和Inhibitor(5'-UCGGUCUCGUGCCGGUAUUU-3'),以及Mimics对照(MNC)序列(正义链:5'-UUUGUA CUACACAAAAGUACUG-3',反义链:5'-CAGUAC UUUUGUGUAGUACAAA-3')和Inhibitor对照(INC)序列(5'-CAGUACUUUUGUGUAGUACAAA-3')。参照Lipofectamine 2000(Thermo Fisher Scientific)试剂说明转染Bcap-37细胞和BMECs细胞。于直径60 mm的Bcap-37细胞培养皿中分别加入50、75和100 nmol/L Mimics及200 nmol/L Inhibitor,并设空白对照组(NC);根据Bcap-37细胞的转染结果,分别以100 nmol/L Mimics和200 nmol/L Inhibitor转染BMECs细胞,采用实时荧光定量PCR检测Novel-miR-57和靶基因的表达情况。
1. 7 统计分析
实时荧光定量PCR检测结果采用2-△△Ct法进行换算,然后以SPSS 19.0进行差异显著性分析。
2 结果与分析
2. 1 Novel-miR-57二级结构预测结果
前体茎环结构是miRNA的主要特征之一,且miRNA的二级结构会影响其加工及成熟miRNA进入不同AGO(Argonaute)蛋白质,因此本研究通过MiRscan对Novel-miR-57的二级结构进行预测分析,结果如图1所示。Novel-miR-57前体序列(黑色)形成7个不一样的小茎环,其成熟序列位于第1、2和3个茎环间的核苷酸序列(红色)上。Novel-miR-57的结合自由能为-53.70 kcal/mol。
2. 2 Novel-miR-57靶基因预测结果
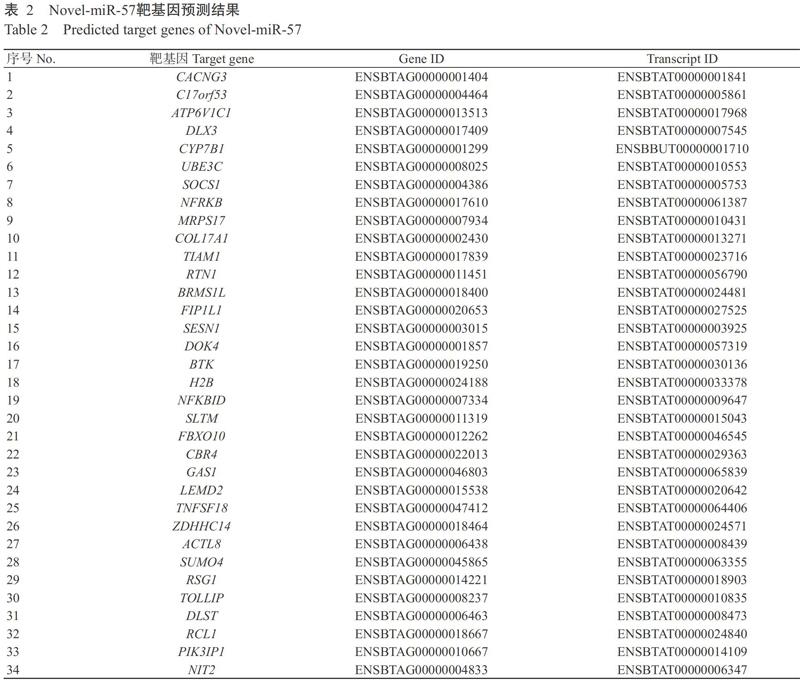
miRNA主要通过结合与调控靶基因实现其生理功能,故使用Miranda(v3.3a)预测Novel-miR-57的靶基因,挑选出结合自由能低于-20.00 kcal/mol的所有靶基因,最终筛选出34个可能的靶基因(表2),经基因资料搜索及信号通路分析,然后筛选部分代表靶基因进行实时荧光定量PCR检测分析。
2. 3 Novel-miR-57靶基因的GO功能注釋及KEGG信号通路富集分析结果
使用GO数据库比对分析Novel-miR-57靶基因,最终获得729个相关类别(Term),其中有89个类别的P<0.05,有2个类别的P<0.01,分别是水解酶活性和线性酰胺中碳—氮键(非缩氨酸)(GO:0016811)及细胞内信号转导负调控(GO:1902532)。将预测筛选获得的34个靶基因输入KEGG数据库,进行系统的KEGG信号通路富集分析,结果发现这些靶基因与42条KEGG信号通路存在关联,其富集的信号通路主要有代谢通路(Metabolic pathways,ID:bta01100)、PI3K-Akt(磷脂酰肌醇三激酶—蛋白激酶B或PKB)信号通路(ID:bta04151)、MAPK(Mitogen-activated protein kinase)信号通路(ID:bta04010)和细胞因子—细胞因子受体相互作用(Cytokine-cytokine receptor interaction,ID:bta04060)等。
2. 4 靶基因的实时荧光定量PCR检测分析结果
针对34个预测靶基因进行实时荧光定量PCR检测分析,结果发现在所有乳腺组织中有5个靶基因的相对表达量较低。对比各靶基因在泌乳期和非泌乳期的相对表达量,发现有11个靶基因(图2)在泌乳期的相对表达量相当或显著高于非泌乳期(P<0.05,下同),有10个靶基因(图3)在非泌乳期的相对表达量相当或显著高于泌乳期;而DLX3(Distal-less homeobox 3)、CACNG3(Calcium voltage-gated channel auxiliary subunit gamma 3)、DOK4(Downstream of tyrosine kinase/docking protein)、NFKBID(NFKB inhibitor delt)、C17Orf53(Chromosome 17 open reading frame 53)、RTN1(Isoform RTN1-C)和FBXO10(F-box protein 10)等7个靶基因(图4)在非泌乳期的相对表达量极显著高于泌乳期(P<0.01),二者间相差100.0倍以上,且与Novel-miR-57的相对表达量呈负相关,故将其列为靶向基因的重点筛选对象。
2. 5 靶基因在Bcap-37细胞上的验证结果
实时荧光定量PCR检测结果(图5)表明,仅DOK4基因与Novel-miR-57的表达具相关性,添加Novel-miR-57的Inhibitor能显著提高DOK4基因表达,加入100 nmol/L Mimics则显著抑制DOK4基因表达。
2. 6 转染Mimics和Inhibitor对BMECs细胞Novel-miR-57表达的影响
由图6可知,以100 nmol/L Mimics转染BMECs细胞,Novel-miR-57的相当表达量显著高于其对照组(MNC);而添加200 nmol/L Inhibitor后Novel-miR-57的相对表达量显著低于其对照组(INC),说明Novel-miR-57转染成功,达到预期效果。
2. 7 转染Mimics和Inhibitor对BMECs细胞DOK4基因表达的影响
由图7可知,以100 nmol/L Mimics转染BMECs细胞,DOK4基因的相对表达量显著提高;而以200 nmol/L Inhibitor转染BMECs细胞,DOK4基因的表达受到显著抑制。综合转染Mimics和Inhibitor对BMECs细胞Novel-miR-57表达的影响可知,Novel-miR-57在BMECs细胞中对DOK4基因表达呈正调控作用。
3 讨论
miRs对乳腺组织起重要的调控作用,可影响乳腺发育,包括维持乳腺上皮前体细胞、促使乳腺上皮管道长出及促进乳腺上皮细胞增殖分化等(Wang et al.,2018;Zheng et al.,2019),其相关研究主要集中在已知miRs的靶向基因挖掘及其功能探析等方面,如miR-27a通过靶向PPAR-γ(Peroxisome proliferato-ractivated receptor gamma)调控甘油三酰合成(Tang et al.,2017),miR-146通过靶向TRAF6(TNF receptor associated factor 6)和HMG(High mobility group)调控乳腺上皮细胞炎症因子(Wang et al.,2017),miR-454通过靶向PPAR-γ调控牛乳腺上皮细胞甘油三酯合成(Zhang et al.,2018)。崔晓钢等(2015)虽然从新发现的44条牛候选miRs中选取4条进行实时荧光定量PCR验证,并预测对应的靶基因,但尚未深入研究靶基因的功能。Novel-miR-57是本课题组从水牛体内新发现的miRs,在水牛泌乳期和非泌乳期的表达量差异极显著,在其功能研究过程中尚存在诸多未知问题和难题。一是Novel-miR-57二级结构尚属于预测阶段,无法确定其二级结构是否会影响miRs加工及成熟miRs进入不同的AGO蛋白质(张伟等,2020);二是在预测Novel-miR-57靶基因时,由于水牛全基因组尚未公布,因此只能使用自编软件Ensembl(v80)注释的水牛mRNA截取3'-UTR作为预测数据库,而导致靶基因预测数量相对较少;三是验证Novel-miR-57靶基因时由于缺乏相关的参考资料,只能将已筛选出的34个靶基因全部进行实时荧光定量PCR检测分析,最后在细胞水平仅发现DOK4基因与Novel-miR-57存在表达相关性,而以往的研究发现单个miRs可调控多个靶基因表达(吴宁昭,2017;袁茂等,2019),因此對于Novel-miR-57的靶基因及功能作用尚有待进一步研究。
本研究结果表明,仅DOK4基因与Novel-miR-57的表达具相关性,且Novel-miR-57在BMECs细胞中对DOK4基因表达呈正调控作用,即Novel-miR-57的靶基因是DOK4基因。DOK4为酪氨酸激酶下游分子(Win et al.,2018),是细胞膜中的适配器及支架蛋白的组成成分,由15406个氨基酸残基组成,定位于反义链上,同时是胰岛素受体底物(Cai et al.,2003;Hooker et al.,2012),涉及多个信号通路,包括GDNF-家族配体和受体互作等。已有学者在灵长类动物和牛的乳腺细胞中预测到DOK4b基因,被视为酪氨酸激酶发送信号的抑制剂(Zhang et al.,2020)。此外,DOK4的mRNA和蛋白大量存在于多种上皮细胞中,对上皮细胞的分化有一定影响,由此推测DOK4基因受Novel-miR-57表达调控,进而影响上皮细胞分化。DOK4还可能与下游效应分子一起作用,然后与Ret(RET proto-oncogene)的1062Tyr结合,且Shc(Generic shell script compiler)及其他效应器分子也会竞争性与Ret的1062Tyr结合,并根据细胞环境不同而改变对Elk/Elk-1的激活作用。在Caco-2细胞系中,DOK4基因的作用是防止Ret介导的Elk-1信号通路被激活,但该作用在293T细胞中较弱(Grimm et al.,2001;Itoh et al.,2005;Guittard et al.,2018),具体作用机制还需进一步探究。
大部分动物的miRs与靶基因结合后会下调基因表达(汪劼,2016),但在近年来的相关报道中发现动物miRs呈正调控或去抑制作用,即在少数动物中miRs能促进靶基因上调表达。Vasudevan等(2007)研究发现,miRs既可降低靶基因表达,又能促使靶基因执行活化功能。当细胞处于休眠期时,miRs能上调基因表达;当细胞处于循环/增殖期时,miRs则抑制基因表达(Vasudevan et al.,2008),且这种作用与ARE(AU rich element)(富含腺嘌呤/尿嘧啶原件)相關。ARE是一种活化miRs翻译的信号,能与AGO、FXRP(Fragile X retardation-1 protein)等miRISC复合物结合,而影响miRs翻译及上调基因表达。von Roretz和Gallouzi(2008)研究表明,ARE对miRs介导的mRNA衰减调控有一定影响。此外,miRs的表达具有细胞特异性,当miR-24在抑制胃癌的BCL2L11细胞中表达时,能促进细胞生长,减少细胞凋亡(Zhang et al.,2016);而在下调肝癌的Bcl-2细胞内,miR-24表现为促进细胞凋亡(杨鹏等,2019)。因此,Novel-miR-57是否通过类似机制对DOK4基因进行调控有待进一步探究。Guittard等(2018)研究表明,通过引入外源性siRNA干扰,可同时抑制Caco-2细胞中的DOK4基因和DOK4b基因表达,但对α-管蛋白无任何影响。本研究也发现,内源性Novel-miR-57可下调DOK4基因表达,为后期揭示Novel-miR-57在其他细胞系中的功能作用提供了参考依据。
4 结论
Novel-miR-57含有7个茎环结构,且其成熟序列位于第1~3个茎环上。Novel-miR-57过表达可下调Bcap-37细胞DOK4基因表达或上调BMECs细胞DOK4基因表达,即Novel-miR-57对靶基因的调控作用还因细胞生理状态不同而存在差异。
参考文献:
包黎娟,刘育含,马毅,安小鹏,张月,张梦,王建刚,堵斌,李广,曹斌云. 2020. miR-92a对奶山羊乳腺上皮细胞增殖及凋亡的调控分析[J]. 畜牧兽医学报,51(1):137-149. doi:10.11843/j.issn.0366-6964.2020.01.016. [Bao L J,Liu Y H,Ma Y,An X P,Zhang Y,Zhang M,Wang J G,Du B,Li G,Cao B Y. 2020. The regulation of mir-92a on proliferation and apoptosis of dairy goats mammary epithelial cells[J]. Acta Veterinaria et Zootechnica Sinica,51(1):137-149.]
边艳杰,韩金汾,段江燕. 2018. miR-200b对奶牛乳腺上皮细胞泌乳功能的影响[J]. 河南农业科学,47(7):124-131. doi:10.15933/j.cnki.1004-3268.2018.07.020. [Bian Y J,Han J F,Duan J Y. 2018. Effect of miR-200b on lactation ability of bovinemammary epithelial cells[J]. Journal of Henan Agricultural Sciences,47(7):124-131.]
蔡小艳,鲍正攀,古景开,邓凯,张晓溪,任艳萍,沈朋雷,石德顺,刘庆友. 2017a. 水牛泌乳期与非泌乳期乳腺组织miRNAs表达谱鉴定及差异表达分析[J]. 畜牧兽医学报,48(9):1635-1647. doi:10.11843/j.issn.0366-6964. 2017. 09.008. [Cai X Y,Bao Z P,Gu J K,Deng K,Zhang X X,Ren Y P,Shen P L,Shi D S,Liu Q Y. 2017a. Identify and analyze the expression profile and mechanism of bu-ffalo mammary gland miRNAs in the lactation and non-lactation periods[J]. Acta Veterinaria et Zootechnica Sinica,48(9):1635-1647.]
蔡小艳,李胜,陈秋萍,王萍,邓凯,刘庆友,石德顺. 2016. 水牛bbu-miR-103-1在泌乳期与非泌乳期表达模式及靶向基因的初步研究[J]. 畜牧兽医学报,47(11):2191-2201. doi:10.11843/j.issn.0366-6964.2016.11.006. [Cai X Y,Li S,Chen Q P,Wang P,Deng K,Liu Q Y,Shi D S. 2016. Expression pattern and target regulation gene of bbu-miR-103-1 from lactation and non-lactation periods in B. bubalis[J]. Acta Veterinaria et Zootechnica Sinica,47(11):2191-2201.]
蔡小艳,王鹏程,邓凯,王萍,冯万有,张晓溪,任艳萍,刘庆友,石德顺. 2017b. 水牛泌乳期与非泌乳期乳腺组织差异miRNAs的表达模式鉴定及分析[J]. 中国畜牧兽医,44(11):3220-3228. doi:10.16431/j.cnki.1671-7236.2017. 11.016. [Cai X Y,Wang P C,Deng K,Wang P,Feng W Y,Zhang X X,Ren Y P,Liu Q Y,Shi D S. 2017b. Identification and analysis of differential expression miRNAs in the lactation and non-lactation of buffalo mammary gland[J]. China Animal Husbandry & Veterinary Medicine,44(11):3220-3228.]
蔡小艳. 2016. 水牛泌乳期和非泌乳期miRNAs表达谱分析及miR-103和Novel-miR-57的靶向基因研究[D]. 南宁:广西大学. doi:10.7666/d.Y3277300. [Cai X Y. 2016. Research on miRNA expression profiles in lactation and non-lactation and the target genes of miR-103 and Novel-miR-57 in buffalo[D]. Nanning:Guangxi University.]
崔曉钢,杨少华,谢岩,张胜利,张勤,孙东晓. 2015. 应用生物信息学预测牛新microRNA及验证[J]. 畜牧兽医学报,46(8):1317-1324. doi:10.11843/j.issn.0366-6964.2015.08. 006. [Cui X G,Yang S H,Xie Y,Zhang S L,Zhang Q,Sun D X. 2015. Computational and experimental identification of novel microRNAs in bovine by bioinforma-tics[J]. Acta Veterinaria et Zootechnica Sinica,46(8):1317-1324.]
郭丽霞. 2017. 高通量测序鉴定黄牛和水牛大脑组织中的microRNAs[D]. 昆明:云南大学. [Guo L X. 2017. Identification microRNAs in brain of cattle and buffalo by high
throughput sequencing[D]. Kunming:Yunnan University.]
侯金星,安小鹏,杜晓岩,曹斌云,沈文正,李云甫. 2019. MiR-3880对奶山羊乳腺上皮细胞OGR1基因的靶向调控作用[J]. 家畜生态学报,40(5):13-17. doi:10.3969/j.issn.1673-1182.2019.05.003. [Hou J X,An X P,Du X Y,Cao B Y,Shen W Z,Li Y F. 2019. MiR-3880 regulates the expression of OGR1 gene in mammary epithelial cells of dairy goats[J]. Acta Ecologiae Animalis Domastici,40(5):13-17.]
李杰,刘天旭,吕微,张评梅,段玉青,王郁,刘丽华. 2020. miR-28-3p通过抑制BIN1表达促进三阴性乳腺癌MDA-MB-468细胞的恶性生物学行为[J]. 中国肿瘤生物治疗杂志,27(1):55-61. doi:10.3872/j.issn.1007-385x.2020. 01.009. [Li J,Liu T X,Lü W,Zhang P M,Duan Y Q,Wang Y,Liu L H. 2020. miR-28-3p promotes the malignant biological behaviors of triple negative breast cancer MDA-MB-468 cells via inhibiting BIN1[J]. Chinese Journal of Cancer Biotherapy,27(1):55-61.]
李新云,付亮亮,程会军,赵书红. 2017. microRNA调控哺乳动物骨骼肌发育[J]. 遗传,39(11):1046-1053. doi:10. 16288/j.yczz.17-112. [Li X Y,Fu L L,Cheng H J,Zhao S H. 2017. Advances on microRNA in regulating mammalian skeletal muscle development[J]. Hereditas(Beijing),39(11):1046-1053.]
练雨. 2019. Ssc-novel-mir-106-5p在LPS诱导的母猪子宫内膜上皮细胞炎症反应中的作用[D]. 重庆:西南大学. [Lian Y. 2019. Effects of ssc-nove1-miR-106-5p on LPS-induced inflammatory response in porcine endometrial epi-thelial cells[D]. Chongqing:Southwest University.]
刘长征,余佳. 2012. microRNA鉴定与功能分析技术[M]. 北京:化学工业出版社. [Liu C Z,Yu J. 2012. microRNA identification and function analysis technology[M]. Beijing:Chemical Industry Press.]
孙云菊,张星,陆幼波,董建兰. 2020. miRNA-380-5p在乳腺癌细胞增殖和凋亡中的作用机制[J]. 中国妇幼保健,35(11):2097-2100. doi:10.19829/j.zgfybj.issn.1001-4411. 2020.11.044. [Sun Y J,Zhang X,Lu Y B,Dong J L. 2020. Mechanism of miRNA-380-5p in proliferation and apoptosis of breast cancer cells[J]. Maternal and Child Health Care of China,35(11):2097-2100.]
汪劼. 2016. 闖入动物王国的植物miRNA[J]. 生命的化学,36(3):404-408. doi:10.13488/j.smhx.20160323. [Wang J. 2016. Plant miRNA breaking into animal kingdom[J]. Chemistry of Life,36(3):404-408.]
王塑天,李敏霞,娄娟,左二伟,陆阳清,卢克焕,杨小淦. 2013. miRNA-145、Oct4和Sox2基因在小鼠早期胚胎发育分化中的表达情况[J]. 南方农业学报,44(12):2075-2079. doi:10.3969/j:issn.2095-1191.2013.12.2075. [Wang S T,Li M X,Lou J,Zuo E W,Lu Y Q,Lu K H,Yang X G. 2013. Expression of miRNA-145,Oct4 and Sox2 in early embryonic development and differentiation in mouse[J]. Journal of Southern Agriculture,44(12):2075-2079.]
王伟,黄晓宇,闫尊强,马晓文,王鹏飞,谢开会,雒瑞瑞,高小莉,马艳萍,滚双宝. 2019. 猪miR-339-3p靶基因预测分析及部分靶基因验证[J]. 农业生物技术学报,27(5):885-896. doi:10.3969/j.issn.1674-7968.2019.05.012. [Wang W,Huang X Y,Yan Z Q,Ma X W,Wang P F,Xie K H,Luo R R,Gao X L,Ma Y P,Gun S B. 2019. Target gene prediction analysis and partial target gene verification of pig(Sus scrofa) miR-339-3p[J]. Journal of Agricultural Biotechnology,27(5):885-896.]
吴宁昭. 2017. miRNA-1和miRNA-133在鸭骨骼肌发育中的表达及功能初步研究[D]. 扬州:扬州大学. [Wu N Z. 2017. The expression of miRNA-1 and miRNA-133 and its function on duck skeletal cell proliferation and diffe-rentiation[D]. Yangzhou:Yangzhou University.]
杨鹏,滕红丽,罗雪兰,李丹,颜鸳渊,欧和生,覃裕旺. 2019. miR-24对肝癌MHCC97H细胞增殖、凋亡及迁移能力的影响[J]. 广西医科大学学报,36(9):1397-1402.doi:10.16190/j.cnki.45-1211/r.2019.09.002. [Yang P,Teng H L,Luo X L,Li D,Yan Y Y,Ou H S,Qin Y W. 2019. Effect of miR-24 on cell proliferation,apoptosis and migration of liver cancer MHCC97H cellline[J]. Journal of Guangxi Medical University,36(9):1397-1402.]
袁茂,江明锋,徐亚欧,林亚秋,蒋小松,杨朝武,余春林,李志雄. 2019. 藏鸡不同发育阶段腿部肌肉组织转录组及microRNA联合分析[J]. 畜牧兽医学报,50(12):2400-2412. doi:10.11843/j.issn.0366-6964.2019.12.004. [Yuan M,Jiang M F,Xu Y O,Lin Y Q,Jiang X S,Yang C W,Yu C L,Li Z X. 2019. Analysis of transcriptome and microRNA in leg muscle of Tibetan chicken at different developmental stages[J]. Acta Veterinaria et Zootechnica Sinica,50(12):2400-2412.]
苑红. 2019. miR-4312促进乳腺上皮细胞增殖并抑制其凋亡[D]. 呼和浩特:内蒙古大学. doi:10.27224/d.cnki.gnmdu.2019.000076. [Yuan H. 2019. miRNA-4312 promotes poliferation and inhibits apoptosis in mammary epithelial cell[D]. Huhhot:Inner Mongolia University.]
臧树成,郭文晋,甄莉,连帅,王立鹏,袁建彬,李文杰,杨焕民. 2018. 急性冷应激大鼠肝脏中novel-miR-133-5p生物学功能预测[J]. 中国兽医学报,38(8):1585-1591. doi:10.16303/j.cnki.1005-4545.2018.08.21.[Zang S C,Guo W J,Zhen L,Lian S,Wang L P,Yuan J B,Li W J,Yang H M. 2018. Biological function prediction of novel-miR-133-5p in the liver of acute cold stress rat[J]. Chinese Journal of Veterinary Science,38(8):1585-1591.]
張伟,王世银,石国庆,邓双义,刘晓娜,杨力伟. 2020. 巴什拜羊miR-486多态性及其表达规律研究[J]. 农业生物技术学报,28(1):92-100. doi:10.3969/j.issn.1674-7968.2020. 01.009. [Zhang W,Wang S Y,Shi G Q,Deng S Y,Liu X N,Yang L W. 2020. Polymorphism of miR-486 and its expression pattern in Bashby sheep(Ovis aries)[J]. Journal of Agricultural Biotechnology,28(1):92-100.]
张月. 2020. Novel-miR-3880对奶山羊乳腺上皮细胞功能和乳腺发育的调控作用研究[D]. 杨凌:西北农林科技大学. doi:10.27409/d.cnki.gxbnu.2020.000123. [Zhang Y. 2020. The regulatory effects of novel-miR-3880 on mammary epithelial cell function and mammary gland deve-lopment in dairy goats[D]. Yangling:Northwest A & F University.]
Cai D S,Dhe-Paganon S,Melendez P A,Lee J,Shoelson S E. 2003. Two new substrates in insulin signaling,IRS5/DOK4 and IRS6/DOK5[J]. The Journal of Biological Chemistry,278(28):25323-25330. doi:10.1074/jbc.M21 2430200.
Cai X Y,Li S,Chen Q P,Wang P,Deng K,Liu Q Y,Shi D S. 2017. Expression pattern and target gene of bbu-miR-103-1 in buffalo(Bubalus bubalis) at lactation and non-lactation periods[J]. Animal Husbandry and Feed Science,9(3):157-164. doi:10.19578/j.cnki.ahfs.2017.03. 007.
Chen Z,Shi H P,Sun S,Luo J,Zhang W,Hou Y,Loor JJ. 2018. MiR-183 regulates milk fat metabolism via MST1 in goat mammary epithelial cells[J]. Gene,646:12-19. doi:10.1016/j.gene.2017.12.052.
Grimm J,Sachs M,Britsch S,Di Cesare S,Schwarz-Romond T,Alitalo K,Birchmeier W. 2001. Novel p62dok family members,dok-4 and dok-5,are substrates of the c-Ret receptor tyrosine kinase and mediate neuronal differentiation[J]. The Journal of Cell Biology,154(2):345-354.doi:10.1083/jcb.200102032.
Guittard G,Pontarotti P,Granjeaud S,Rodrigues M,Abi-Rached L,Nunès J A. 2018. Evolutionary and expression analyses reveal a pattern of ancient duplications and functional specializations in the diversification of the Downstream of Kinase(DOK) genes[J]. Developmental and Comparative Immunology,84:193-198. doi:10.1016/j.dci. 2018.02.011.
Hooker E,Baldwin C,Lemay S. 2012. New insights into Dok-4 PTB domain structure and function[J]. Biochemical and Biophysical Research Communications, 427(1):67-72. doi:10.1016/j.bbrc.2012.08.148.
Itoh S,Lemay S,Osawa M,Che W Y,Duan Y T,Tompkins A,Brookes P S,Sheu S S,Abe J I. 2005. Mitochondrial Dok-4 recruits Src kinase and regulates NF-κB activation in endothelial cells[J]. The Journal of Biological Chemistry,280(28):26383-26396. doi:10.1074/jbc.M410262200.
Jiao B L,Zhang X L,Wang S H,Wang L X,Luo Z X,Zhao H B,Khatib H,Wang X. 2019. MicroRNA-221 regulates proliferation of bovine mammary gland epithelial cells by targeting the STAT5a and IRS1 genes[J]. Journal of Dairy Science,102(1):426-435. doi:10.3168/jds.2018-15108.
Li W R,Guan X L,Jiang S,Sun L. 2020. The novel fish miRNA pol-miR-novel_171 and its target gene FAM49B play a critical role in apoptosis and bacterial infection[J]. Developmental and Comparative Immunology,106:103616.doi:10.1016/j.dci.2020.103616.
Stark A,Brennecke J,Bushati N,Russell R B,Cohen S M. 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3'UTR evolution[J]. Cell,123(6):1133-1146. doi:10.1016/j.cell.2005. 11.023.
Tang K Q,Wang Y N,Zan L S,Yang W C. 2017. miR-27a controls triacylglycerol synthesis in bovine mammary epithelial cells by targeting peroxisome proliferator-activated receptor gamma[J]. Journal of Dairy Science,100(5):4102-4112. doi:10.3168/jds.2016-12264.
Vasudevan S,Tong Y C,Steitz J A. 2007. Switching from repression to activation:MicroRNAs can up-regulate translation[J]. Science,318(5858):1931-1934.doi:10.1126/science.1149460.
Vasudevan S,Tong Y C,Steitz J A. 2008. Cell cycle control of microRNA-mediated translation regulation[J]. Cell Cycle,7(11):1545-1549. doi:10.4161/cc.7.11.6018.
von Roretz C,Gallouzi I E. 2008. Decoding ARE-mediated decay:Is microRNA part of the equation?[J]. The Journal of Cell Biology,181(2):189-194. doi:10.1083/jcb. 200712054.
Wang X P,Luoreng Z M,Zan L S,Li F,Li N. 2017. Bovine miR-146a regulates inflammatory cytokines of bovine mammary epithelial cells via targeting the TRAF6 gene[J]. Journal of Dairy Science,100(9):7648-7658. doi:10. 3168/jds.2017-12630.
Wang X H,Zhang L,Jin J,Xia A T,Wang C M,Cui Y J,Qu B,Li Q Z,Sheng C Y. 2018. Comparative transcriptome analysis to investigate the potential role of miRNAs in milk protein/fat quality[J]. Scientific Reports,8(1):6250. doi:10.1038/s41598-018-24727-y.
Win S,Than T A,Kaplowitz N. 2018. The regulation of JNK signaling pathways in cell death through the interplay with mitochondrial SAB and upstream post-translational effects[J]. International Journal of Molecular Sciences,19(11):3657. doi:10.3390/ijms19113657.
Zhang H Y,Duan J J,Qu Y J,Deng T,Liu R,Zhang L,Bai M,Li J L,Ning T,Ge S H,Wang X,Wang Z Z,Fan Q,Li H L,Ying G G,Huang D Z,Ba Y. 2016. Onco-mir-24 regulates cell growth and apoptosis by targeting BCL2l11 in gastric cancer[J]. Protein and Cell,7(2):141-151. doi:10.1007/s13238-015-0234-5.
Zhang J,Padarti A,Jiang X T,Abou-Fadel J. 2020. Redefining PTB domain into independently functional dual cores[J]. Biochemical and Biophysical Research Communications,524(3):595-607. doi:10.1016/j.bbrc.2020.01.114.
Zhang M Q,Gao J L,Liao X D,Huang T H,Zhang M N,Wang M Q,Tian Y,Bai J,Zhou C H. 2018. miR-454 re-gulates triglyceride synthesis in bovine mammary epithe-lial cells by targeting PPAR-γ[J]. Gene,691:1-7. doi:10. 1016/j.gene.2018.12.048.
Zheng C Y,Zou X,Zhao B C,Zhang M L,Lin H J,Luo C H,Xu Z M,Shao L Y,Fu S X. 2019. miRNA-185 regulates retained fetal membranes of cattle by targeting STIM1[J]. Theriogenology,126:166-171. doi:10.1016/j.theriogenology.2018.11.030.
(責任编辑 兰宗宝)

