Genetic Diversity and Population Structure of the Oriental Garden Lizard,Calotes versicolor Daudin,1802 (Squamata:Agamidae) along the Mekong River in Thailand and Lao PDR
Chairat TANTRAWATPAN ,Weera THONGNETR ,Warayutt PILAP ,Warong SUKSAVATE ,Takeshi AGATSUMA ,Wittaya TAWONG ,Trevor N.PETNEY and Weerachai SAIJUNTHA*
1 Division of Cell Biology,Department of Preclinical Sciences,Faculty of Medicine and Center of Excellence in Stem Cell Research,Thammasat University,Pathumthani 12120,Thailand
2 Walai Rukhavej Botanical Research Institute,Biodiversity and Conservation Research Unit,Mahasarakham University,Maha Sarakham 44150,Thailand
3 Department of Forest Biology,Faculty of Forestry,Kasetsart University,Bangkok 10900,Thailand
4 Division of Environmental Health Sciences,Kochi Medical School,Kochi University,Oko,Nankoku 783-8505,Japan
5 Department of Agricultural Science,Faculty of Agriculture,Natural Resources and Environment,Naresuan University,Phitsanulok 65000,Thailand
6 Departments of Zoology and Paleontology and Evolution,State Museum of Natural History Karlsruhe,Karlsruhe 76133,Germany
Abstract Calotes versicolor Daudin,1802,is geographically widespread along the Mekong River basin.The Mekong River is play important role as a significant natural barrier to several terrestrial animals living on different sides.This study aims to analyze the genetic diversity and population structure of C.versicolor populations collected from different sides of Mekong River using mitochondrial cytochrome c oxidase subunit 1 (CO1) sequences.We obtained sequences of 200 individuals from 18 sampling localities from left and right sides of the Mekong River in Lao PDR and Thailand respectively.Overall,91 haplotypes were detected,which reflect high levels of genetic diversity in this species at the study areas.Haplotype network and phylogenetic analyses revealed that there were six major lineages (lineage C -lineage H) of C.versicolor populations within the Mekong River,whereas lineages A and B have previously been found from China and Vietnam.The genetic distance among C.versicolor was significantly related to spatial distance,however,the Mekong River had no significant effect on genetic distance.Our findings,together with previous studies,suggests that C.versicolor in Asia is a species complex with other cryptic lineages being likely but there is a need for further exploration.Thus,comprehensive genetic,biological and ecological studies of C.versicolor should be conducted throughout its entire distribution range.
Keywords Agamid,CO1,haplotype,lizard,phylogeny,reptile
1.Introduction
The Oriental garden lizard,Calotes versicolor
Daudin,1802,is one of a few geographically widespread tropical lizards (Radder,2006).Calotes versicolor
is also known as the“changeable crested lizard”,due to its wide variation in coloration and ability to change colors significantly during the breeding season(Jiet al.
,2002).Recent field surveys confirm its distribution stretching from Oman in the West,across Southern and South-East Asia to the East,the Maldives,Réunion,Mauritius,the Seychelles,and more recently it has been introduced to Florida in the United States of America (Radder,2006).This lizard is naturally found in open forest and shrub but has adapted tremendously well to urban environments and can be found in agricultural areas,parks,empty lots and gardens where it is usually seen off the ground in low vegetation (Radder,2006).It helps to control insect populations,occasionally feeding on small lizards,mollusks,crustaceans,baby birds and rodents,or seeds (Sudasinghe and Somaweera,2015).It is also a prey item of snakes and birds (Matyot,2004),as well as being used for cooking in some localities in Southeast Asia (unpublished data).In Thailand,the local people,especially in the northeastern region (I-san) often cook and eat several lizards in the generaLeiolepis
,Varanus
andCalotes
.It is a famous dish for I-san people called“Koi Kapom”,which is ‘spicy lizard salad’(unpublished data).It is hunted massively in its breeding season (around March-April).Therefore,populations ofC.versicolor
may be dramatically reduced in the near future.It is important to gather information relating to its habitat,biology,and ecology,including genetic diversity,in order to facilitate the conservation of this species.A comprehensive genetic investigation ofC.versicolor
was previously conducted in Hainan Island and adjacent mainland China,including a locality in northern Vietnam and found high genetic variation with two major lineages,lineage A and B (Huanget al.
,2013).But there has been no study investigating the genetic diversity withinC.versicolor
in Southeast Asia.There are,however,morphometric studies comparingC.versicolor
in the Lower Mekong Basin in Thailand and Lao PDR (Thongnetret al.
,2015),and southern and northern Thailand (Prakobkarnet al.
,2016),which show some significant differences in characteristics between different populations.This suggests thatC.versicolor
inThailand may comprise a number of cryptic lineages within a species complex.The Mekong River flows 4800 km from Tibet through China,Myanmar,Lao PDR,Thailand,Cambodia,to its delta in Vietnam,and then into the South China Sea,draining an area of 795 000 km(Mekong River Commission,2011).The river forms and extensive sections of the border between Thailand from Lao PDR,except in the northern region.The Mekong River is also known as a significant natural barrier to gene flow for some freshwater and terrestrial animals living on different sides of it (Saijunthaet al.
,2019;Tantrawatpanet al.
,2020).This study aims to test whether the Mekong River plays an important role as a natural barrier to gene flow among populations of the lizard.Thus,C.versicolor
populations living on the left and right sides of the Mekong River in Lao PDR and Thailand were collected and examined to address these goals.The genetic diversity and genetic structure ofC.versicolor
populations were also examined.In order to make meaningful comparison of our work with the previous study of Huanget al
.,(2013),the same DNA region,namely mitochondrial cytochrome c oxidase subunit 1 (CO1
),was chosen as a marker.This maternally inherited marker has proven to be useful in population genetic studies,including the definition of cryptic species of theCalotes
(Huanget al
.,2013;Saijunthaet al
.,2017;Saijunthaet al
.,2020).2.Materials and Methods
2.1.Specimen collection and DNA extraction
A total of 200 specimens ofC.versicolor
were sampled from 18 different localities along the lower Mekong River in Thailand and Lao PDR (Figure 1 and Table 1).AdultC.versicolor
were caught using the fishing pole method (Saijunthaet al.
,2017).Their tails were excised around 5 mm from the end,soaked in 80%alcohol and kept at room temperature until DNA extraction.After collection of the apical portion,the lizard’s tails were cleaned with 70% alcohol and the animals released back into their natural habitat.Total genomic DNA was extracted using E.Z.N.A.Tissue DNA kit (Omega bio-tek,USA) following the manufacture’s protocol.All DNA samples were kept at -20 °C until used.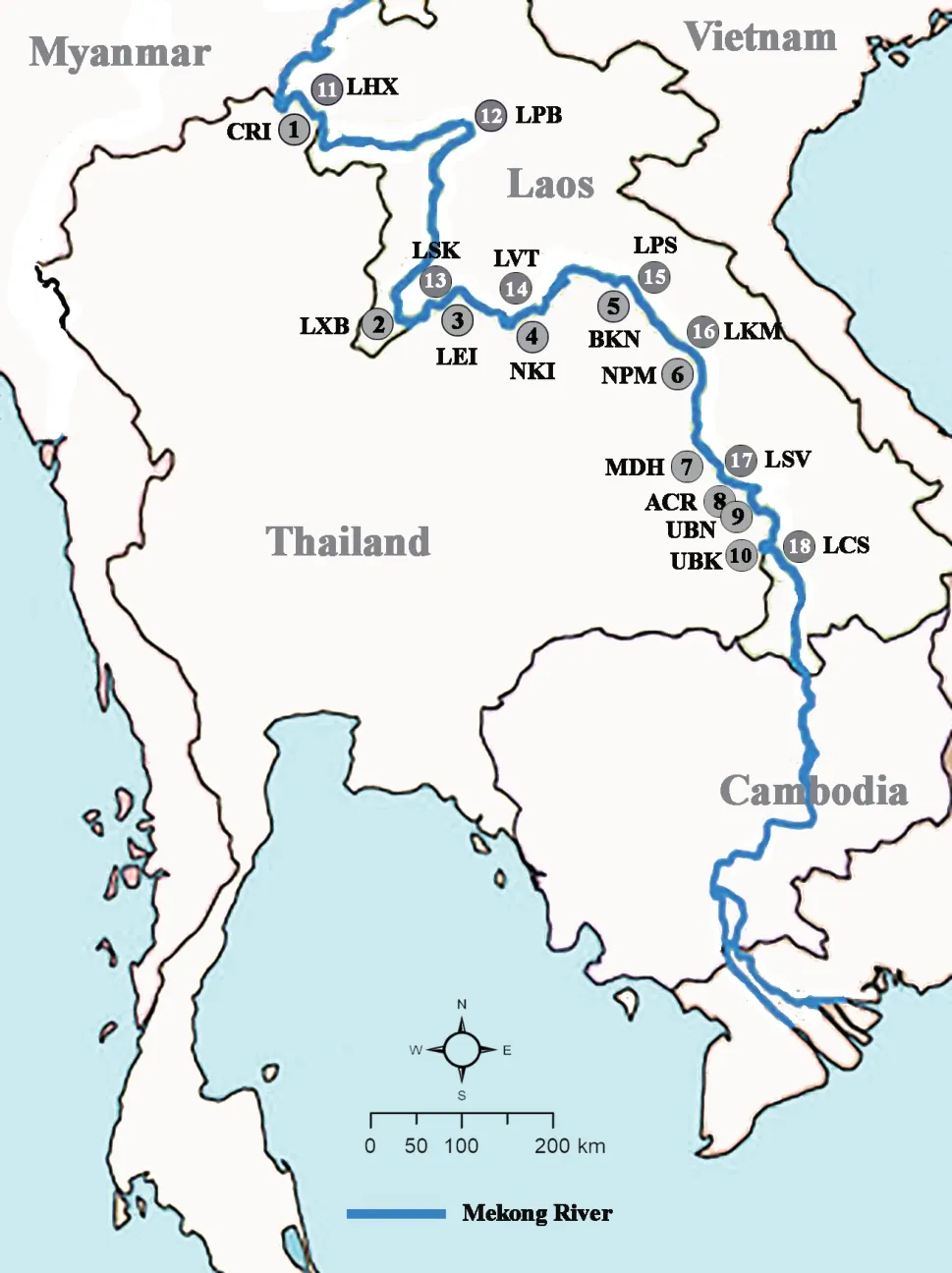
Figure 1 A map of the 18 collection localities of Calotes versicolor aside the Mekong River in Thailand and Lao PDR.
2.2.Polymerase Chain Reaction and DNA sequencing
A partial region of the mitochondrial cytochromec
oxidase subunit 1 (CO1
) gene was amplified with the primers L5037 (5’-GAG TAG ACC CAG GAA CCR AAG TTC-3’) and H6448(5’-GTA TAC CGG CTA ATC CAA GCA TGT-3’) (Huanget al.
,2013).Standard polymerase chain reactions (PCR) were performed in a total of 25 μL,including approximately 100 ng of template DNA,1 μL of each primer (each 10 pmol/μL),2.5 μL of 10xEx-Taq
buffer (Mgplus),2 μL dNTPs (each 2.5 mmol/L),0.125 μL ofEx-Taq
DNA polymerase (5 U/μL),and deionized water.PCR was conducted with the following conditions:an initial denaturing step at 95 °C for 4 min;35 cycles of denaturing at 94 °C for 35 s,annealing at 65 °C for 45 s,and extending at 72 °C for 90 s;and a final extending step of 72°C for 8 min.PCR products were electrophoresed in 1% agarose gels,visualized with GelRedNucleic Acid Gel Stain (Biotium,Inc.,Hayward,CA).The amplified band was cut and purified by using E.Z.N.A.Gel Extraction kit (Omega bio-tek,USA).The purified PCR products were cycle-sequenced at Eurofins Genomics Company,Japan.All new sequences were deposited in GenBank under accession numbers MT438484 to MT438683.2.3.Sequence and haplotype analyses
All sequences generated in this study were checked and edited using the software program ABI sequence scanner v1.0.Multiple sequence alignment was performed using a BioEdit version 7.2.6 (Hall,1999).Molecular variation indices and population structure patterns based on global AMOVA were calculated in Arlequin ver 3.5.1.3 (Excoffier and Lischer 2010).Genetic distance (P
-distance) was calculated using MEGA X (Kumaret al.
,2018).Nei’s genetic distance (Nei,1978),isolation-bydistance (IBD),and isolation-by-barrier (IBB) were calculated using a randomized Mantel test (Mantel,1976) of ade4 package(Bougeard and Dray,2018) in the R program (R Core Team,2013).Haplotype data was generated using the DnaSp v5 program (Librado and Rozas 2009).A maximum parsimony haplotype network was constructed in the Network 5.0.0.0 program based on a median-joining network (Bandeltet al.
,1999).2.4.Phylogenetic analyses
We performed a suite of analyses to infer the phylogenetic relationship ofC.versicolor
from 18 different localities along the Mekong River in Thailand and Lao PDR,as well as the 11 sequences ofC.versicolor
representing lineage A and the 24 sequences representing lineage B from China and Vietnam that were available in GenBank.For Bayesian Inference (BI) and Maximum Likelihood (ML)analysis,the best-fitting substitution model for theCO1
data set was the general time reversibility with the gamma distribution model (GTR+G) selected by MrModeltest ver 2.2 (Nylander,2008).We constructed a ML tree using MEGA X (Kumaret al.
,2018) with nodal support estimated using 1000 bootstrap re-samples.BI was performed using MrBayes version 3.1.2(Ronquist and Huelsenbeck,2003).The number of generations used in BI was 20 000 000,sampling every 100 generations.The first 10 000 sampled trees were discarded as burn-in until the average standard deviation values of the run dipped below 0.01 and the potential scale reduction factor (PSRF) value approached 1.0.The consensus tree and posterior probability values were calculated using the remaining 100 000 trees.3.Results
3.1.Sequences analyses
Comparison between the 1254 bp ofCO
1 gene of 200 samples from 18 different localities along both sides of the Mekong River revealed high genetic variation with191 variable sites consisting of 15 singleton variable sites with two variants and 176 parsimony informative sites including 156,19 and one informative sites with two,three and four variants,respectively.Nucleotide and haplotype diversity within populations ranged between 0.000 to 1.000 ± 0.096 and 0.0000 to 0.0286 ± 0.0042,respectively (Table 2).The number of haplotypes(N) within each population ranged between 1 and 14 haplotypes and a total of 91 haplotypes were found and classified.Five haplotypes were shared between different populations,whereas the others were uniquely found in a particular locality (Table 2).Genetic (p
) distance ranged between 0.0003 and 0.0492 (Table 3).The genetic distance between populations within lineages ranged between 0.0003 and 0.0153,whereas genetic distance between populations of different lineages ranged between 0.0313 and 0.0492 (Table 3).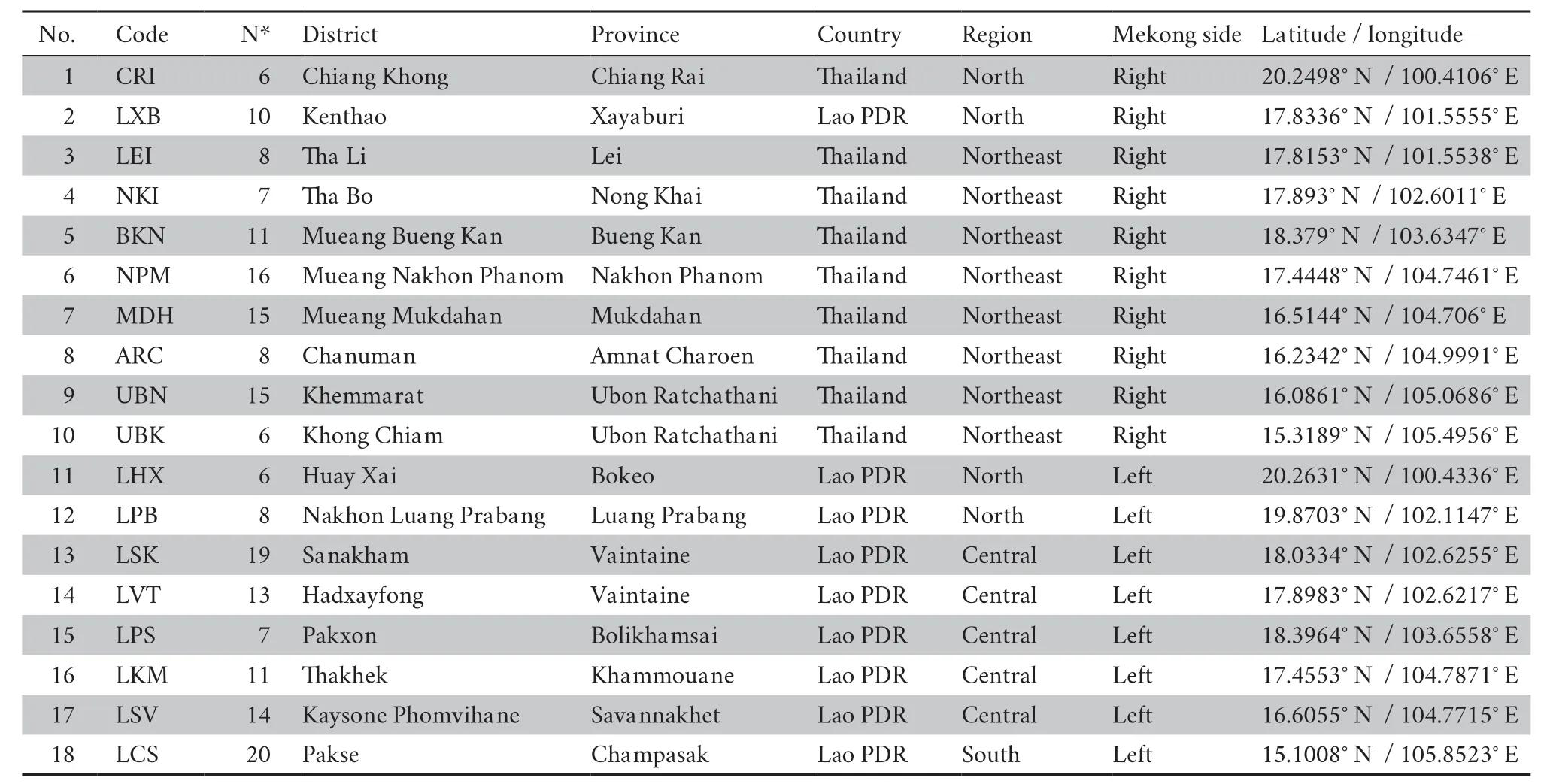
Table 1 Details of sampling localities of Calotes versicolorcollected in this study.
3.2.Haplotype analyses
Based on haplotype analyses,the 91 haplotypes generated in this study were grouped into six lineages based on mutational steps ≥ 20,i.e.lineage C -lineage H.Linage A and lineage B,which were previously classified by Huanget al.
(2013),contained the haplotypes from China and Vietnam (Figure 2).Lineage C contained the three haplotypes from UBK in northeastern Thailand and 14 haplotypes from LCS in southern Lao PDR.Lineage D consisted of eight and four haplotypes from LSV and LKM in central Lao PDR,respectively.Lineage E consisted of the six haplotypes from CRI in north Thailand,as well as the two and one haplotypes from LHX and LSK in northern and central Lao PDR,respectively.Lineage E also included the three haplotypes from LEI in northeastern Thailand.Linage F contained ten and four haplotypes of UBN and ACR in northeastern Thailand,and also three and one haplotypes from MDH and BKN in northeastern Thailand,respectively.Lineage G contained all haplotypes of NKI in northeastern Thailand,as well as LVT,LXB,and LPB in Lao PDR,including three haplotypes from LEI.Lineage H contained three,three and six haplotypes from MDH,BKN and NPM in northeastern Thailand,and included two haplotypes from LPS in central Lao PDR (Figure 2).3.3.Genetic structure analyses
The genetic structure of the populations was examined by AMOVA (Table 4).This revealed thatC.versicolor
populations were genetically substructured and correlated with the six major lineages (lineage C-lineage H) classification (F
=0.0639,P
< 0.05),similar to the classification from haplotype network analysis.The isolationby-distance (IBD) test also showed that genetic distance was significantly correlated withR
=0.3894 (P
< 0.001).However,it was not significantly related to the groups divided by the Mekong River (F
=0.0182,P
> 0.05).The isolation-by-barrier(IBB) test also show that the Mekong River had no significant effect on genetic distance ofC.versicolor
withR
=0.0113 (P
>0.05) (Figure 3).3.4.Phylogenetic analyses
The phylogenetic analyses ofC.
versicolor
based onCO1
sequences usingCalotes amma alticristatus
as out-group based on ML and BI methods were reciprocally demonstrated that eight well-supported lineages were observed(Figure 4).Lineage A and B contained the 37 sequences from China and Vietnam available in GenBank.Our samples were classified into six lineages,namely lineage C to H in concordance with the haplotype network analysis.Lineage C and D generated in the current study were closely clustered with lineage A and B reported by a previous study.Lineage E and F were subdivided into three and two sub-lineages,namely sub-lineage E1 -E3 and F1 -F2,respectively.Sublineage E1 and E2 contained the sequences from LEI and CRI,respectively,whereas sub-lineage E3 consisted of the sequences from LEI,LHX and LSK (Figure 4).Sub-lineage F1 contained the sequences from UBN and ACR,whereas sub-lineage F2 consisted of the sequences from MDH and BKN (Table 3 and Figure 4).Lineage E and G were found on both sides of the Mekong River,i.e.
the northernmost (CRI) to northeast (NKI)of Thailand and the northernmost (LHX) to central (LVT) Lao PDR.Lineage F and H were found on the Thailand side from BKN to UBN,whereas the LPS in Lao PDR belonged to lineage H.Lineage D was found in the Lao PDR at LKM and LSV,whereas lineage C was found on both sides,UBK in Thailand and LCS in Lao PDR (Figure 4).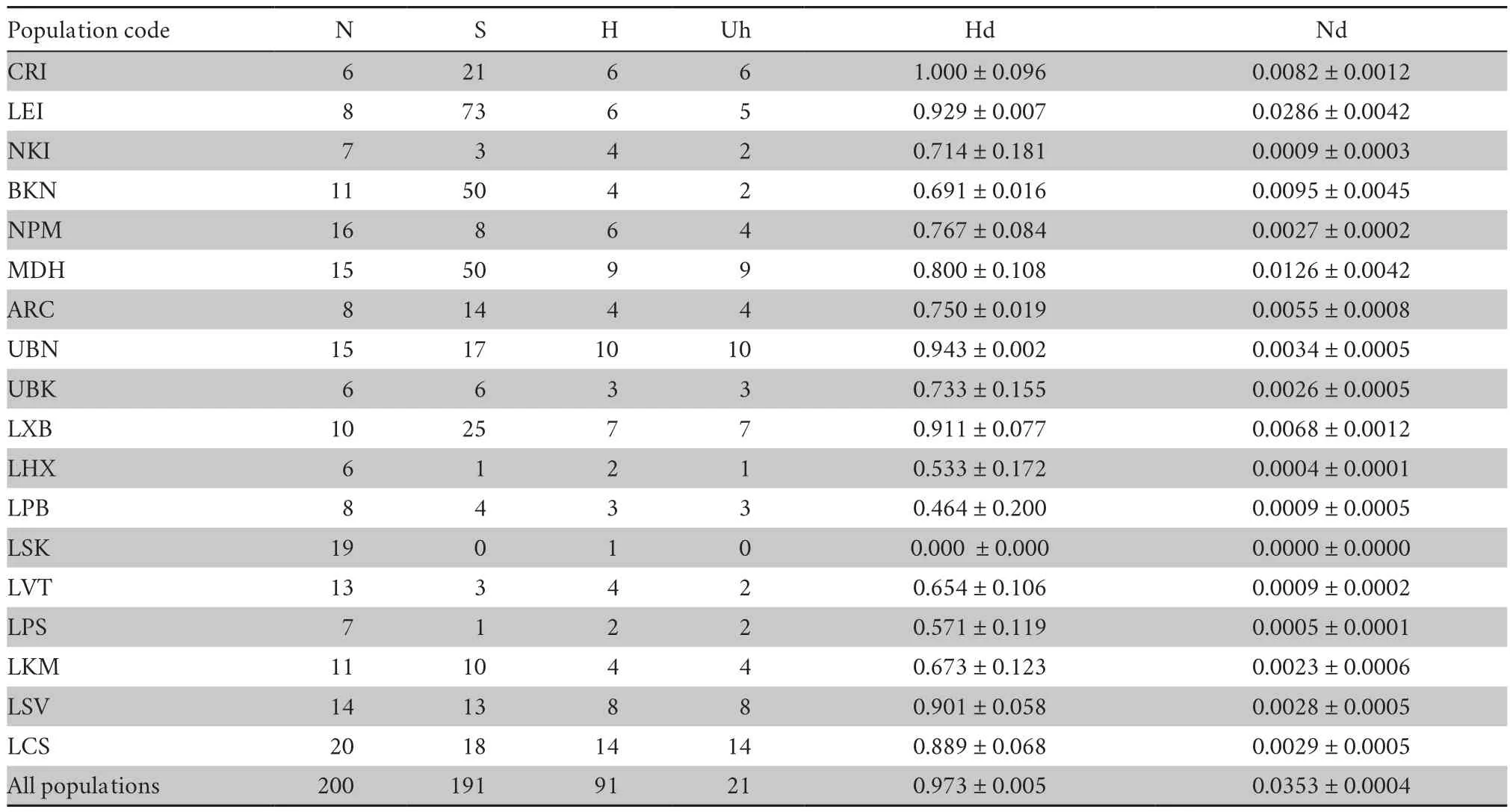
Table 2 Summary statistics of molecular variation in 18 populations/localities of Calotes versicolor in Thailand and Lao PDR.
4.Discussion
To our knowledge,this is the first report of comprehensive genetic variation and genetic structure analyses ofC.versicolor
in the Lower Mekong Basin in Southeast Asia.We found high levels of genetic variation in theC.versicolor
populations along the Mekong River from Thailand and Lao PDR,as indicated by the 91 haplotypes ofCO1
.The significant genetic distance among populations indicates that theC.versicolor
populations examined in this study can be subdivided into at least six lineages.Each linage may contain several cryptic groups,such as lineages E and F that can also be subdivided into sub-lineages.This evidence that there are other cryptic lineages needs to be explored in the future.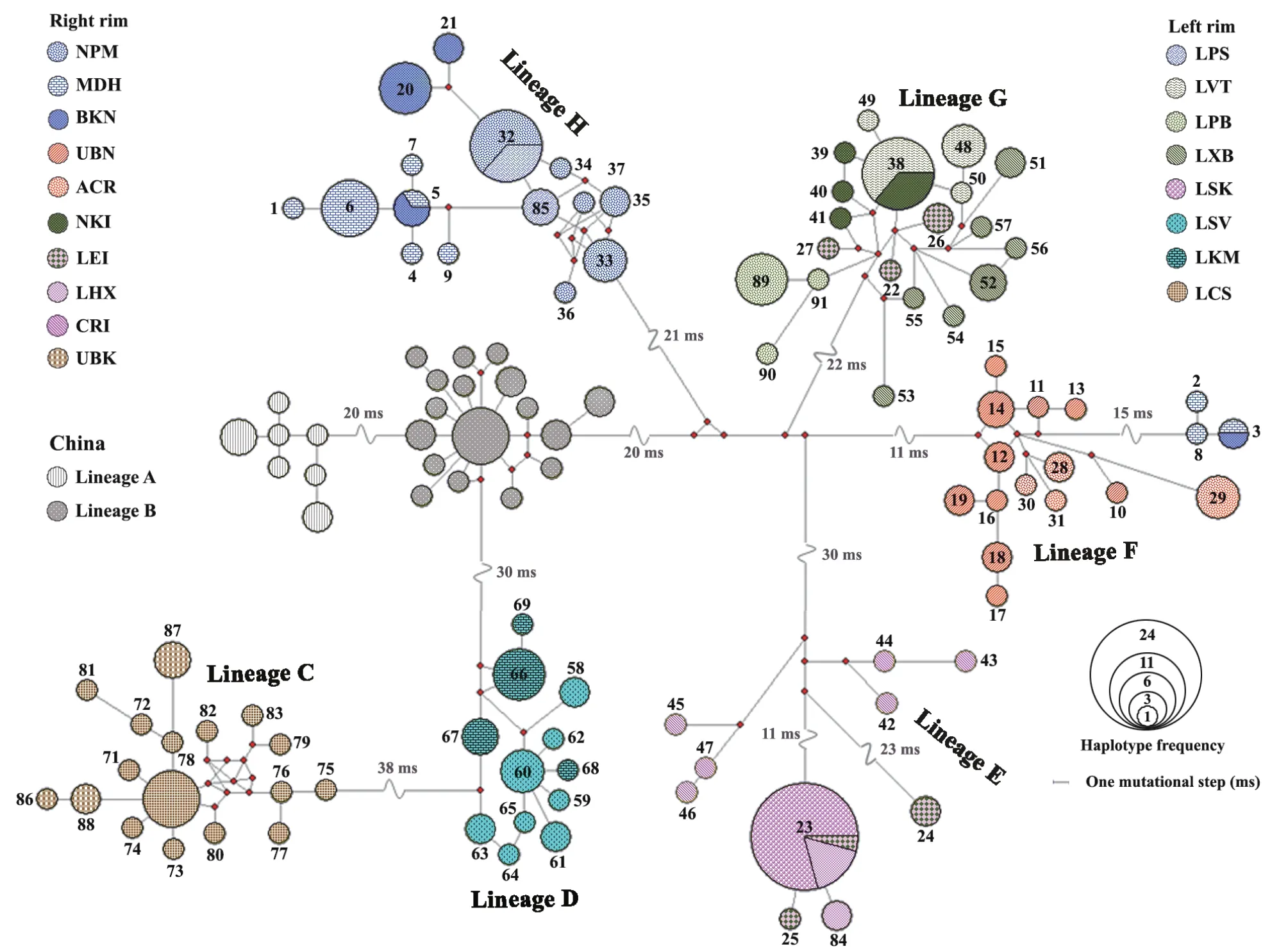
Figure 2 Minimum spanning haplotype network of Calotes versicolorgenerated based on partial CO1 sequences corresponds to their geographical localities separated into 18 different localities along the Lower Mekong River in Thailand and Lao PDR,including 34 sequences of C.versicolor from China retrieved from GenBank.The area of the circles represents the proportion of specimen number found in each haplotype.
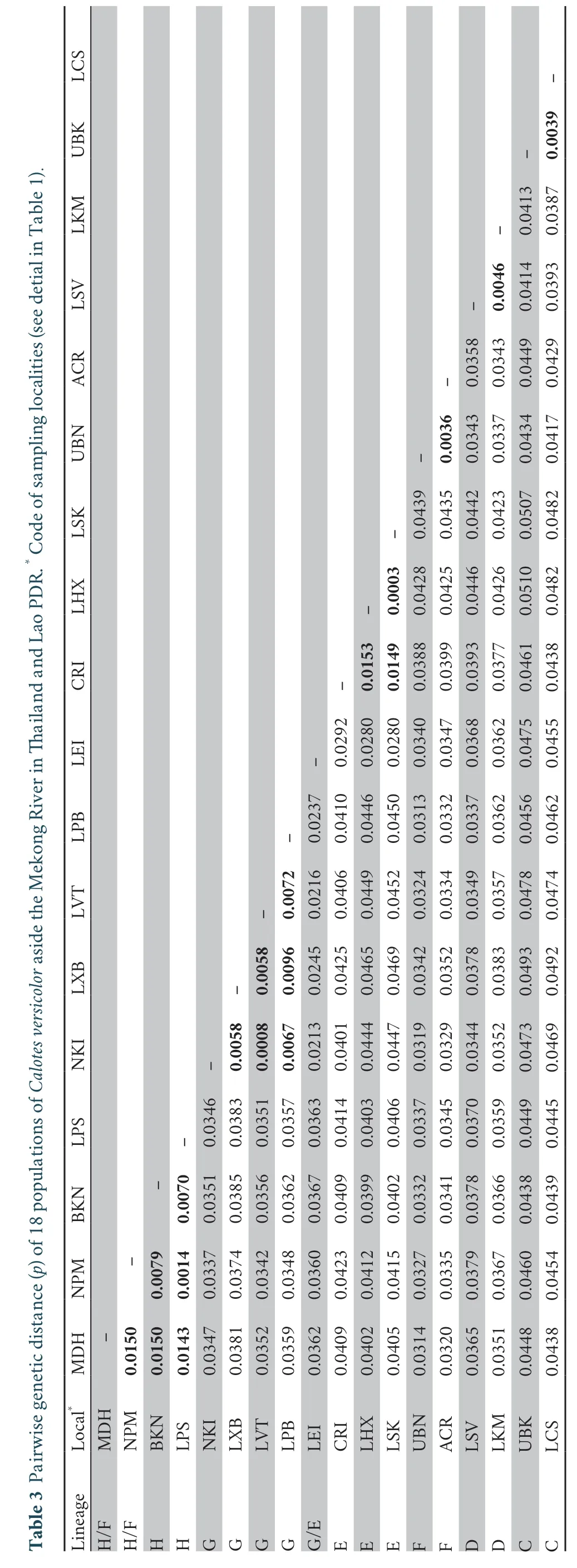
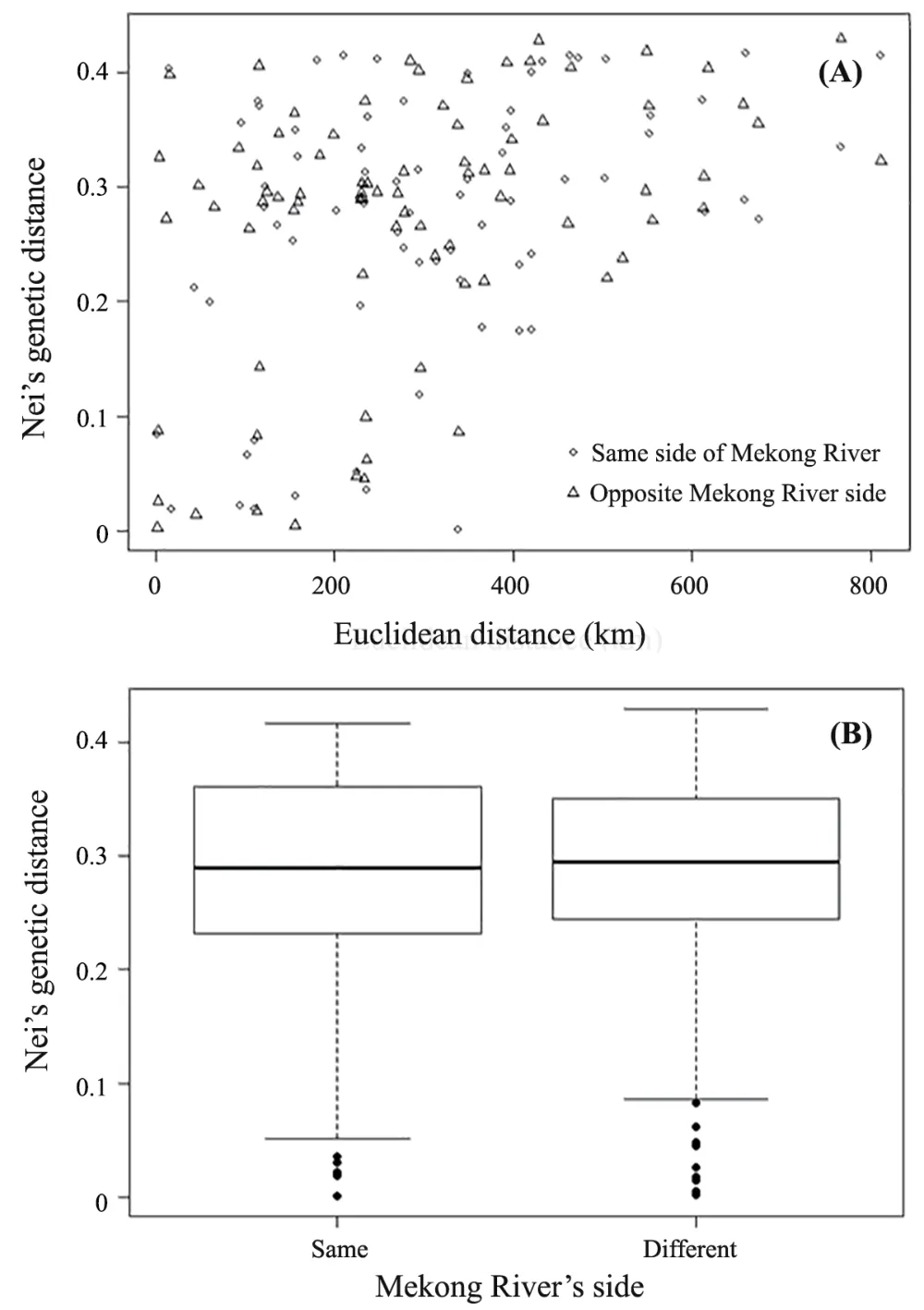
Figure 3 Graphs demonstrating the pairwise relationship between Euclidean geographic distance and Edward’s genetic distance of every sample pair of Calotes versicolor,based on CO1 sequences.The graph shows a proportional increase in genetic distance as the geographic distance increases,so called isolation by distance (IBD) (A).Another graph shows the difference between pairwise genetic distance categorizing whether the pairs were the same (Same) or different (Different) from the Mekong River’s side,recognizing Mekong River as the natural barrier of gene flow,so called isolation by barrier (IBB) (B).
The IBD test revealed that there was a spatial distance effect on the genetic differences betweenC.versicolor
populations examined in this study.This is likely to be due to limitedC.versicolor
movement/migration between spatial distance areas as previously evidenced by their related speciesCalotes mystaceous
Duméril &Bibron,1837 (Saijunthaet al.
,2017) andCalotes emma
schmidt,1925 (Saijunthaet al.
,2020).In addition,unique haplotypes were commonly found in all localities and high genetic distances (p
-distance) between the populations from different lineages were observed.A similar finding was previously reported inC.versicolor
populations from Hainan Island and adjacent mainland China,as well as Vietnam,where two mitochondrial lineages were defined and separated by putative nonphysical or ecological barriers (Huanget al.
,2013).These reports and the current study support the hypothesis that geographical distance and ecological barriers play significant roles in the genetic differences inCalotes
populations (Huanget al.
,2013;Saijunthaet al.
,2017,2020).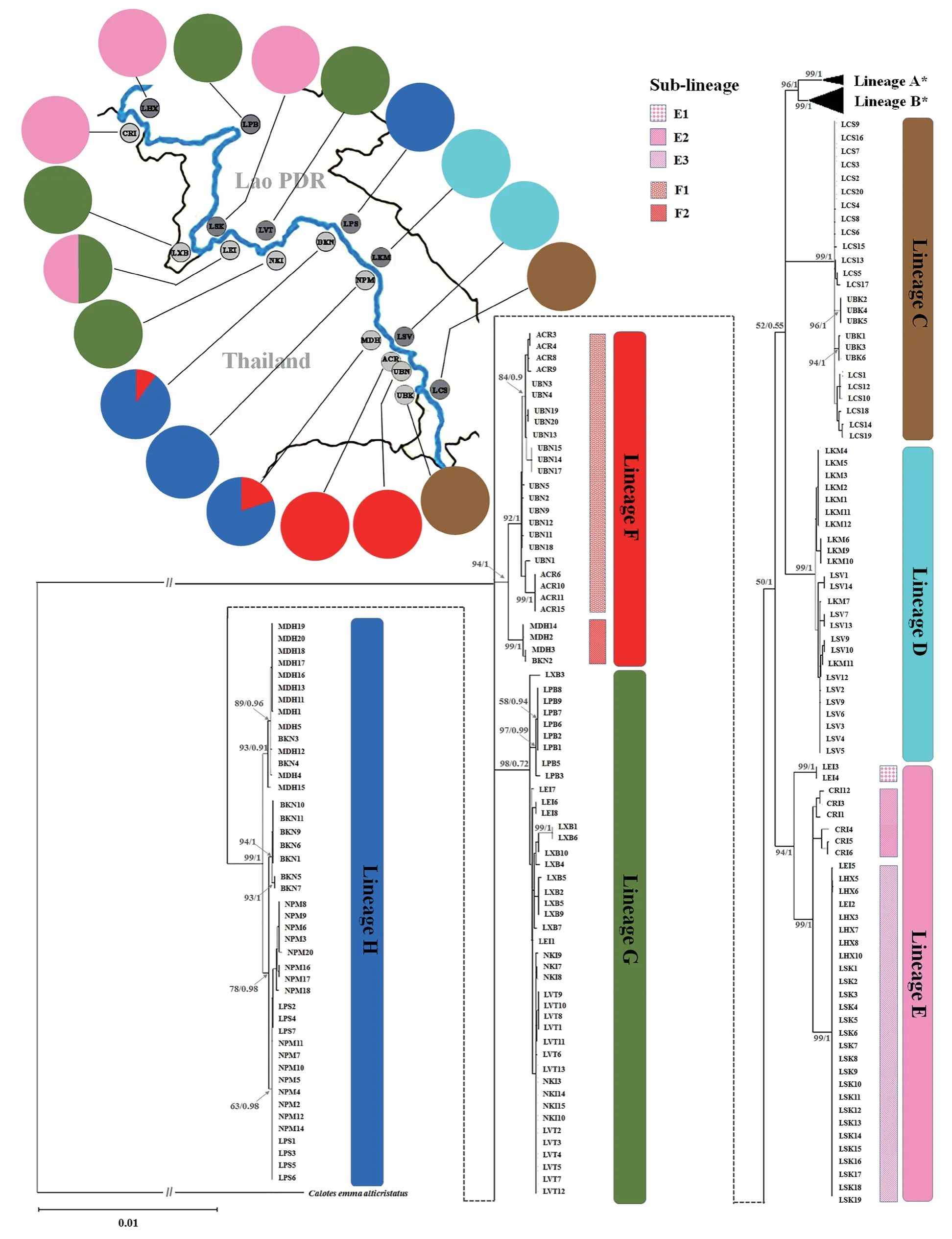
Figure 4 Phylogenetic tree constructed based on 1254 bp of CO1 sequence of Calotes versicolor 91 haplotypes along the Mekong River in Thailand and Lao PDR including the 11 sequences lineage A,namely KC875637,KC875647-KC875649,KC875741-KC875744,KC875802-KC875804) and the 24 sequences of lineage B,namely KC875610,KC875660,KC875680,KC875687,KC875690,KC875695,KC875717,KC875747,KC875749,KC875751,KC875753,KC875754,KC875759,KC875762,KC875772,KC875773,KC875775,KC875780,KC875784,KC875786,KC875809,KC875810,KC875813,KC875815 from China,KC875765 and KC875771 from Vietnam.
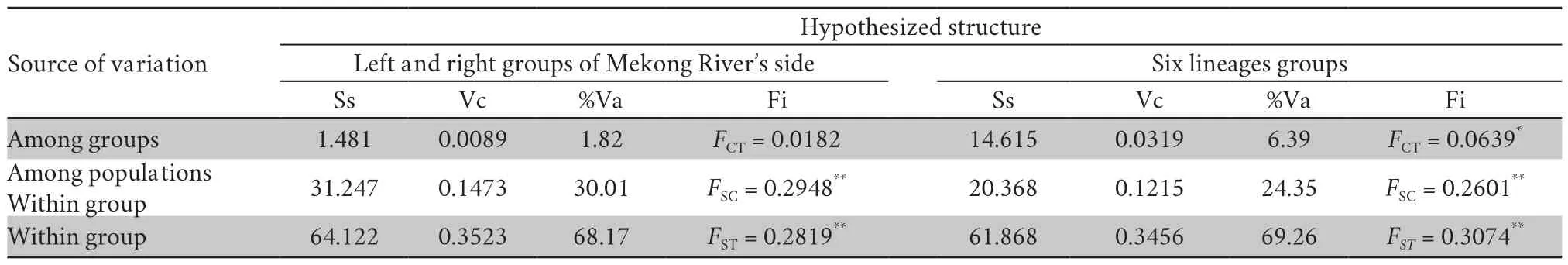
Table 4 Analysis of molecular variance (AMOVA) of Calotes versicolor classified into two groups defined by the left and right side of the Mekong River,as well as the six groups defined by six lineages (lineage C -lineage H) classified in this study.
The Mekong River can act as a significant natural barrier blocking gene flow between populations of several organisms on different banks of the river,including several freshwater and terrestrial animals,such asBrachytrupes portentosus
(Lichtenstein 1796) (Tantrawatpanet al.
,2011) andBithynia siamensis goniomphalos
(Morelet,1866) (Tantrawatpanet al.
,2020).However,several reptile species in this region such asGekko gecko
(Linneasus,1758) (Saijunthaet al.
,2019) show no significant genetic difference between the populations from different sides of the Mekong River.Our results also indicate that the variation found inC.versicolor
populations from different sides(left and right) of the river is not related to genetic distances indicated by the IBB test.These findings suggest that the Mekong River does not play a significant role as a natural barrier to block the gene flow of endemic reptiles.In addition,morphological variation ofC.versicolor
from different localities along the Mekong River in Thailand and Lao PDR has been recorded,and the high variation observed by morphometric analyses has been used to define seven major groups (Thongnetret al.
,2015).Thus,the results from present and previous studies suggest that cryptic lineages ofC.versicolor
exist in the Mekong River Basin in Thailand and Lao PDR that are genetically distinct from the Chinese and Vietnamese isolates.Interestingly haplotype 24 (two specimens)from LEI was genetically very distinct from the other F lineage observed in a haplotype network analysis.This finding revealed that another cryptic lineage ofC.versicolor
may exist in this geographical area.This needs to be further evaluated by expanding sampling sites from this locality.Our findings,together with previous studies,suggest thatC.versicolor
in Asia is a species complex,which may contain more genetic structuring than that determined in this and previous studies (Huanget al.
,2013).More specimens from different distribution areas should be collected for a more comprehensive study of the genetic diversity,genetic structure and morphometric analyses ofC.versicolor
throughout Southeast Asia,as well as their other distribution localities worldwide.Acknowledgements
This research was supported by Mahasarakham University in FY2015 to W.SAIJUNTHA.We would like to thank Dr.Adrian R.Plant for English improving. Asian Herpetological Research2021年1期
Asian Herpetological Research2021年1期
- Asian Herpetological Research的其它文章
- An Integrative Taxonomy of Amphibians of Nepal:An Updated Status and Distribution
- A New Species of the Gekko japonicus Group (Squamata:Gekkonidae) from Southwest China
- Species Diversity,Distribution,and Microhabitats of Anurans on Mt.Kalo-Kalo of the Mt.Kalatungan Range Natural Park,Bukidnon,Philippines
- Species Account of Anurans from the Western Slope of Mt.Kitanglad,Mindanao Island,Philippines
- Behavioral and Neurogenomic Responses to Acoustic and Visual Sexual Cues are Correlated in Female Torrent Frogs
- The Influence of Environmental Factors on the Behavior and Mortality Risk of Polypedates megacephalus Tadpoles
