Evaluation of Related Traits of GmTST2.1 and ZmGIF1 Genes in Soybean (Glycine Max)
Dong Hai-ran, Chang Hong, Wang Jun, Bao Dong-fang, Zhao Xue, and Han Ying-peng
Key Laboratory of Soybean Biology in Chinese Ministry of Education Key Laboratory of Soybean Biology and Breeding/Genetics of Chinese Agriculture Ministry), Northeast Agricultural University, Harbin 150030, China
Abstract: Soybean (Glycine max) is one of the most important food crops and oil crops in the world. According to the role of sucrose transporter in sugar accumulation, GmTST2.1 (Glyma.04G000300) and ZmGIF1 of sugar transport related genes were separately overexpressed in the soybean cultivar Heihe 43 from the perspective of regulatory source to library relationship in the study. The function of soluble sugar accumulation in grains layed a theoretical foundation for the cultivation of new varieties of high-yield genetically modified soybeans. The results showed that the height and 100-seed weight of the over-expressed GmTST2.1 gene were increased with 7% and 17.7% and the soluble sugar content was increased with 1.575 times as much as that of the wild-type soybean. The overexpressed ZmGIF1 gene was found to be 10% higher than that of plant height, 1.8% higher than that of 100-seed weight and larger seed size and 1.3 times higher than that of soluble sugar content. Biological yields were increased in both GmTST2.1 and ZmGIF1 genes.
Key words: Glycine max, soluble sugar, GmTST2.1 gene, ZmGIF1 gene
Introduction
Soybean originates from China and belongs to the annual herbaceous plant of the genusLeguminosae. It has a history of cultivation for 5000 years in China. It was an important oil and food crop and an important source of phytophagous protein and oil in the world(Zhaoet al., 2019). Tonoplast sugar transporter (TST)is a typical membrane-binding protein with highly conserved sequence which is widely distributed in the tissues and cells of plants. Tonoplast sugar transporter is located on the vacuole membrane and is mainly transported sugar from the cytoplasm to the vacuole.TSTgene has the function of transported monosaccharide and disaccharide. InTSTsubfamily,there is a long hydrophilic ring located in the cytoplasm between the 6th and the 7th transmembrane regions ofTSTgene which may be a sugar sensor(Fabio and Weber, 2011). At present, the researches on its functions and characteristics are mainly focused onArabidopsis thalianaand sugar beet. There are three homologous genes inArabidopsis thalianaof which the expression ofAtTST1 (AT1G20840) andAtTST2 (AT4G35300) is high and the expression is the highest in the pollen with high sugar accumulation(Wormitet al., 2006). Over-expression ofAtTST1 can increase the biomass and yield (Heidiet al., 2016).Beta vulgarisTST2 is the specific sucrose transport factor and the proton transporter, coupling sucrose into the vacuole and accumulating a large amount of sugar in the vacuole to increase the yield (Nieberlet al., 2017).GIF1 (OsCIN2) is a member of the cell wall transaminase subfamily and one of the eight CIN(cell-wall invertase) members in the rice genome(Choet al., 2005).GIF1 gene is originally found in rice and is mapped to the physical region of 32 kb on chromosome 4 (Sonnewaldet al., 1997).GIF1 had been studied and reported in various plants such as the rice, corn and potato (Sunet al., 2014). The results showed thatTSTandGIF1 are the key proteins responsible for sugar transport and important effects on crop yield.
Soybean seeds contain not only protein and lipids,but also about 33% carbohydrates, of which the content of soluble sugar is as high as 16.6%. Soluble sugar is mainly divided into three categories, sucrose (Suc),fructose (Fru) and glucose (Glu). Sucrose is accountes 41.3% of the total soluble sugar and 67.5% of the total soluble sugar (Eldridgeet al., 1979; Yazdisamadiet al., 1977). Soluble sugar is not only the main photosynthetic product of higher plants, but also the main form of carbohydrate transformation, storage and reuse in plants, which plays the important role in plant metabolism (Liet al., 2012). Genes related to glucose metabolism in soybean can be controlled the yield by regulating source-sink relationship. In other crops, the expression ofTSTgene is positively correlated with sugar content and the abundance expression ofTSTgene can be regulated the yield or quality (Li, 2016;Ren, 2016). It is similarly proves that over-expression ofGIF1 gene can be increased the cell wall invertase content, grain yield and branch growth, corn cob, seed size and weight (Biet al., 2018).
In the study, Heihe 43 was used as the experimental material from the point of view of the relationship between the source and pools, according to the role of sucrose transporters in sugar accumulation. The role ofGmTST2.1 gene andZmGIF1 gene were studied in increasing yield by over-expression method.Compared with wild-type, the yield increasing ability of two kinds of sugar transporters was evaluated. The purpose of the study was to provide genetic resources and material basis for the cultivation of new high-yield transgenic soybean varieties.
Materials and Methods
Plant materials and experiment
The experiment was completed in 2018 in the drought resistant shed of Soybean Research Institute of Northeast Agricultural University. Heihe 43 and transgenic materials were planted in red round pot(pot mouth diameter 30 cm and pot height 27 cm).Select the seeds before sowing, four seeds per pot,appropriate amount of watering after sowing, proper amount of watering according to soil conditions during growth and development. Removed the impurities,prevented and controled the diseases and insect pests.After the seedling leaves were expanded, the PPT smeared leaves were screened. The plants without PPT resistance were pulled out. The plants with PPT resistance were detected by molecular biology and the phenotypic characters were observed, during growth and development.
PCR analysis of transgenic plants
Extracted DNA from transgenic leaves and wild soybean by SDS (Wanget al., 2008) small method.In order to ensure the accuracy of the test, the specific detection primers and universal bar primers designed were identified by PCR, according to the sequence of the recombinant expression vector. The program of Bar primer reaction was 94℃ denaturation for 10 min, 38 cycles of 94℃ for 30 s, 54℃ for 30 s,72℃ for 30 s, 72℃ for 10 min, 4℃ preservation. The procedure of 3300-TST2.1-nos primer reaction was 94℃ denaturation for 10 min, 38 cycles of 94℃ for 30 s, 58℃ for 30 s, 72℃ for 45s, 72℃ for 10min,4℃ preservation. The procedure of 3301-ZmGIF1-nos primer reaction was 94℃ denaturation for 10 min, 38 cycles of 94℃ for 30 s, 56℃ for 30 s, 72℃for 20 s, 72℃ for 10 min, 4℃ preservation. After PCR reaction, 5 μL product was detected by 1% agarose gel electrophoresis.
Analysis of target gene transcription level in transgenic plants
Extracted RNA by Trizol Reagent for reverse transcription (Wanget al., 2019). Fluorescence quantitative PCR amplification using the leaf tissue cDNA of transgenic plants withGmTST2.1 gene andZmGIF1 gene was the templates (qTST2.1-F: 5'-ACAGAAACAC TCACCCTCCA-3'; qTST2.1-R: 5'-ATAGTCGCAT TATCCCATCC-3', qGIF1-F: 5'-TGTGATAACAT GGTGGAGCACG-3'; qGIF1-R: 5'-GCGAAACTTA CGACTACGAGGG-3'). The system of fluorescence quantitative PCR reaction was 2XTB Green Premix ExTaqII 8 μL, ddH2O 6 μL, forward primer 0.8 μL(10 μmol · L-1), reverse primer 0.8 μL (10 μmol · L-1),ROX reference Dye II 0.4 μL (50X), cDNA 2 μL.PCR cycling conditions were as followings: 95℃denaturation for 30 s, 40 cycles of 95℃ for 5 s, 60℃for 20 s, 72℃ for 20 s, and the final extension of 60℃for 1min. The CT value was the average number of repeated tests with three technical replicates and the relative copy number was determined using 2-ΔΔCtmethod (Livak and Schmittgen, 2001).
Determination of soluble sugar content in grain of T2 generation plants
Transgenic soybean seeds had been dried for 2 h and grounded into soybean powder. Weighed 0.05 g and extracted by 80% anhydrous ethanol solution.4 000 r · min-1centrifugal filtration removed some other components, such as insoluble sugar. After extraction with anthrone reagent (95% concentrated sulfuric acid and anthrone), the total amount of soluble sugar was determined.
Drawing of standard curve and calculation of soluble sugar content
Prepared the standard solution and configured glucose into standard solution in different concentrations which were 0, 10, 20, 30, 40, 50, 60, 70, 80, 90 and 100 μg · mL-1. The standard solution of 200 μL(100 μg · mL-1) was placed in the enzyme plate and the standard curve was drawn with excel 2003 (Fig. 1).The soluble sugar was determined by the sample solution and repeated three times. Then, the solution concentration was calculated, according to the standard curve.

Fig. 1 Standard curve of soluble sugar concentrationDesign formulas: y=ax+b; a=0.0025, b=0.0931, R2=0.9988
Determination of physiological indexes of agronomic characters of T2 generation plants
The growth period and T2generation which wasGmTST2.1 gene plants andZmGIF1 gene plants were observed (Renet al., 2018). The biological yield and 100-seed weight of transgenic positive plants were analyzed after mature plants (mainly plant height,node number, branch number, pod number and seed weight). The contents of protein and oil in transgenic soybean and wild-type Heihe 43 seeds were detected by near infrared spectroscopy (NIR). The test data were analyzed by SPSS data.
Results
PCR detection of over-expression GmTST2.1 in T2 generation plants
The recombinant expression vector containingGmTST2.1 gene was transformed into Heihe 43 by agrobacterium-mediated transformation of soybean stem tip to extract DNA. PCR detection was carried out with 3300-Bar-nos primers. The target band was 402 bp and carried on further PCR detected with 3300-TST2.1-nos primers. The target band was 804 bp which was ensured the accuracy of the experiment.Twenty-four soybean plants which were positive were obtained by both primers (Fig. 2).
Detection of transcription level of transgenic GmTST2.1 soybean in T2 generation plants
Both PCR primers were extracted from the positive T2plants. The qRT-PCR was performed with GmActin as the internal reference after the reverse transcription (Fig. 3). The results showed thatGmTST2.1 gene could be normally expressed and the expression level was different among different transgenic lines.

Fig. 2 PCR detection for transgenic soybeans T2 generation plantsM, DL2 000 Maker; A, PCR detection of Bar primer; B, PCR detection of specific primer; 1-12, PCR product; 13, Negative control; 14, H2O;15, Positive control.

Fig. 3 qRT-PCR detection of GmTST2.1 from T2 transgenic positive soybean1-5, T2 transgenic positive plants; 6, Wild-type.
PCR detection of over-expression ZmGIF1 in T2 generation plants
DNA of leaves of T2positive plants was extracted and detected by PCR with 3301-Bar-nos primers.The target band was 468 bp. To ensure the detection accuracy, the 559 bp target band was further detected by PCR with 3301-ZmGIF1-nos primers (Fig. 4). The results showed thatZmGIF1 gene had been integrated into soybean genome and could be inherited to the offspring.
Detection of transcription level of transgenic ZmGIF1 soybean in T2 generation plants
After reverse transcription of RNA in leaves of T2plants with positive PCR primers, GmActin was used as the internal reference gene for qRT-PCR detection(Fig. 5). The results showed thatZmGIF1 gene could be normally expressed, but the expression levels were different among different transgenic lines. The expression levels of transgenic plants were significantly higher than those of wild-type.
Significant difference in pod setting status between transgenic soybean and wild soybean
The pod number of per plant, the seed number of per plants, the seed weight of per plant and 100-seed weight of transgenicGmTST2.1 gene, transgenicZmGIF1 gene and wild-type Heihe 43 were detected.The pod number of per plant, the seed number of per plant, the seed weight of per plant and 100-seed weight of per plant of T2transgenic plants were significantly higher than those of wild-type in Table 1.

Fig. 4 PCR detection for transgenic soybeans T2 generation plantsM, DL2 000 Maker; A, PCR detection of Bar primer; B, PCR detection of specific primer; 1-13, PCR product; 14, Negative control; 15, H2O;16, Positive control.
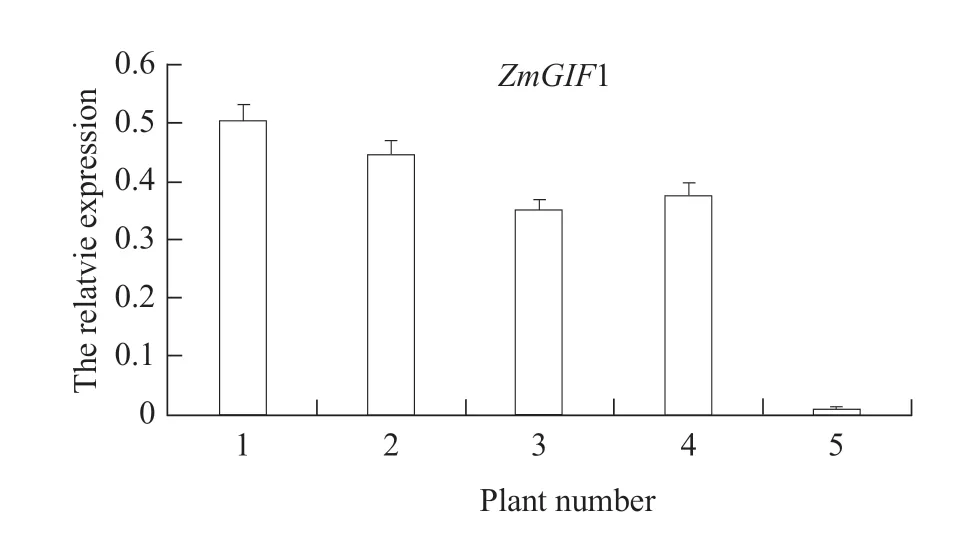
Fig. 5 qRT-PCR detection of ZmGIF1 from T2 transgenic positive soybean1-4, T2 transgenic positive plants; 5, Wild-type.

Table 1 Difference analysis of weight, number of seeds and number of pods of transgenic and wild-type by t-test
Significant difference of seed shape between transgenic soybean and wild soybean
The seed length of T2transgenic plants withGmTST2.1 gene was significantly shorter than that of the wildtype Heihe 43. The seed length, the seed width and the seed thickness of T2generation transgenic plants withZmGIF1 gene were significantly higher than those of wild-type in Table 2.

Table 2 Difference analyses of grain shape of transgenic plants and wild-type by t-test
Significant difference of seed protein and oil content between transgenic soybean and wildtype soybean
The grain quality of transgenicGmTST2.1 gene,transgenicZmGIF1 gene and wild-type Heihe 43 were obtained by near-infrared spectroscopy. The results showed that there was no significant difference in oil content and protein content between T2generation of transgenic soybean and wild-type withGmTST2.1 gene andZmGIF1 gene (Table 3).
Significant difference of soluble sugar content between transgenic soybean and wild soybean
Soluble sugar content in seeds of T2generation transgenicGmTST2.1 gene, T2generation transgenicZmGIF1 gene and wild-type Heihe 43 were measured by enzyme linked immunosorbent assay (ELISA). The results showed that the average soluble sugar content in the seeds of Heihe 43 was 0.08 g, which was higher than that of transgenicGmTST2.1 soybean seed in the control (Fig. 6), but the soluble sugar reached a very significant difference in the transgenicZmGIF1 soybean seed (Fig. 7).

Table 3 Difference analysis of seed quality of transgenic soybean and wild-type by t-test

Fig. 6 Changes of soluble sugar content in transgenic soybean1-8, Transgenic positive plants; 9, Wild-type.
Significant difference in plant height between transgenic soybean and wild soybean
The plant height of transgenicGmTST2.1 plants was 7% higher than that of wild-type. The plant height of transgenicZmGIF1 plants was 10% higher than that of wild-type. The corresponding number of main stem nodes was increased that the increasing of plant height was closely related to the elongation of internodes.

Fig. 7 Changes of soluble sugar content in transgenic soybean1-9, Transgenic positive plants; 10, Wild-type.

Table 4 Difference analyses of plant height in transgenic soybean and wild-type by t-test

Fig. 8 Height changes of transgenic positive soybean1-3, Transgenic positive plants; CK, Wild-type.
Discussion
Effect of transgenic GmTST2.1 and ZmGIF1 on plant height of soybean
The demand of C skeleton and energy depends on a large number of carbohydrates. The sugar signal can regulate the extension of stem and the sugar needed in sink tissue is supplied by cell wall invetase by decomposing sucrose in the process of plant growth.It was found that over-expressionGIF1 gene in the stem growth of plants was enhanced and the yield and quality of crops were improved (Liet al., 2013). TheAtTSTmutant ofArabidopsis thalianawas studied(Wingenteret al., 2010). It was found that the cell tissue of deficient plants was changed, the leaves become smaller and the early development of plants was accelerated. The cell wall invertase and vacuole transporter were very important to the growth of crop stems. The transgenicGmTST2.1 andZmGIF1 genes had significant effects on soybean height in the study.Data analyses showed that the number of nodes in T2generations withGmTST2.1 gene did not change, but plant height were increased by 7% compared with wild-type. The results showed that the increasing of plant height was due to the extension of node space.The plant height of T2generations transgenicZmGIF1 soybean was 10% higher than that of wild plants and the number of nodes per plant was increased. The molecular mechanism of increasing space between nodes of transgenic plants needed to be further studied.
Effects of GmTST2.1 gene transfer and ZmGIF1 gene transfer on soybean quality and yield
The content of soluble sugar, protein and fat determines the seed quality of soybean. The pyruvate and the glycolysis product of glucose are the source of fat and protein in the soybean kernels. There is a competitive relationship between them in terms of the substrate and medium that balancing their competition was pyruvate carboxylase (PEP Case). At the same time, the flowing direction of pyruvate is also regulated by pyruvate carboxylase and the ratio of fat and protein of the grain is finally controlled. There is a negative correlation between soybean protein and fat content. This correlation is overturned and transferred thediacylglycerol2Agene into soybean (Lardizabalet al., 2008). It was found that the oil content in soybean seed was increased by 1.5%. However, the protein content had not been changed a lot. Compared with wild-type, the protein and fat contents of transgenic T2generations did not change, but the soluble sugar content was significantly increased in the study.
When the content of assimilation available to crops is constant, the biological yield is proportional to the economic yield. When the photosynthetic rate of crops is constant, the content of assimilation available to crops is proportional to the economic yield.Therefore, it is not possible to try to increase the yield of soybeans by increasing the photosynthetic rate of crops or increasing the available assimilation. Sucrose synthase (Fanget al., 2017) was the key enzyme to regulate the sucrose transport. Its activity might control the unloading of sucrose and determine the strength of the reservoir. The transport of carbon was regulated by sugar transporter which converted photosynthetic products into the necessary substances of seed itself. It was the great significance to improve the formation of yield and quality.
The soluble sugar content of T2generation transgenic soybean was increased, but the content of protein and fat did not obviously change in the study. Carbon was the temporary way of soluble sugar storage which was regulated by grain metabolism and soluble sugar input. Grain metabolism had a certain degree of relationship with the change of grain content.The phenotypes of plant height, economic yield and biological yield were increased in T2generation of transgenicGmTST2.1 andZmGIF1 soybean. Increasing production of soybeans is the key problem in modern agriculture.
Conclusions
Through molecular biological detection: 24 strains ofGmTST2.1 gene and 28 strains ofZmGIF1 gene were detected in T2generation. BothGmTST2.1 gene andZmGIF1 gene could significantly increase the yields of soybean. The seed weight and soluble sugar content of transgenic plants withGmTST2.1 gene increased,plant height increased by 7%, and biological yield increased.ZmGIF1 gene could regulate seed shape.The transgenic plants withZmGIF1 gene had larger seeds, higher seed weight and soluble sugar content,and increased plant height by 10%. Presently, the cultivation of new soybean varieties with high yield was the most important breeding goal of soybean breeders.
 Journal of Northeast Agricultural University(English Edition)2020年4期
Journal of Northeast Agricultural University(English Edition)2020年4期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Soybean Leaf Morphology Classification Based on FPN-SSD and Knowledge Distillation
- CRISPR/Cas9-based Editing of Acetyl-CoA Carboxylase (ACC1) Gene in Barley
- A Bicycle Tourism Based Study on Planning and Designing of Urbanrural Recreational Greenway
—— A Case Study of Harbin City - Effects of Two Chelating Agents on Availability of Calcium and Phosphorus in Black Soil of Vegetable Fields
- Effects of Crop Rotation and Microbial Fertilizer on Nutrient Absorption and Beneficial Bacterium Abundance in Rhizosphere of Continuous Cropped Eggplant
- Effect of Chromium Propionate Substituting 25% Rumen-protected Choline on Production Performance and Blood Indicators of Perinatal Dairy Cows
