Expression and prognosis analyses of SNX8 in human gastric cancer
Bo Liu, Zi-Xiang Kou*
Expression and prognosis analyses ofin human gastric cancer
Bo Liu1, Zi-Xiang Kou2*
1Tianjin University of Traditional Chinese Medicine, Tianjin 300381, China;2The Affiliated Wuqing Hospital of traditional Chinese Medicine of Tianjin University of Traditional Chinese Medicine, Tianjin 300381, China.
In this work, we evaluated the Cancer Genome Atlas databases to explore the expression and prognosis analyses ofin human gastric cancer. It showed that the expression ofmight be a useful biomarker for prognosis of gastric cancer.
: Associated with the Alzheimer's disease, the, reported as a β-amyloid toxicity enhancer, is a PX-BAR domain sub-family of sorting nexins. Nonetheless, though the specific role ofwas evident in cases of gastric cancer, very little is known about the function and expressionin gastric cancer.: Using The Cancer Genome Atlas and the Oncomine databases, the methylation and theexpression were evaluated. With the Kaplan-Meier Plotter and UALCAN database, the relationship between the several clinical parameters and, along with the survival information was revealed.:was upregulated in various subtypes of gastric cancer compared with the matched normal individuals. The expression ofwas also overexpressed regardless of cancer stage (S1, S2, S3, and S4), tumor grade (G1, G2, and G3), gender (male and female), race (Caucasian, African-American, and Asian), age (20–40, 41–60, 61–80, and 81–100 Yrs), helicobacter pylori infection, and histological subtype. And we revealed that theexpression level may be correlated with DNA methylation and copy number alterations in the gastric cancer. Besides, a positive correlation betweenandwas confirmed.: In the prognosis of gastric cancercould be used as a predictive biomarker. To investigate the molecular mechanism of the value ofin gastric cancer, detailed experiments are needed.
Biomarker, Gastric cancer, Prognosis,
Background
Worldwide, one of the most prevalent causes of death has been gastric cancer (GC) [1]. Especially in China and most parts of East Asia, the GC is still one of the most common types of cancer, despite the decrease in the number of cases of GC in recent years [2]. Over the years, patient survival has been encouraging due to the improvements in the early detection and treatment of GC/SC. Nonetheless, many deaths are still caused by GC/SC [3, 4]. A novel approach is needed to predict the outcome and the treatment response due to limitations of the molecular, clinical and pathological features in the individualized therapy of tumors. Hence, it becomes imperative to identify certain effective and available markers as surrogates to these features [5].
Associated with the Alzheimer's disease and reported to be a β-amyloid (Aβ) toxicity enhancer, theis a PX-BAR domain sub-family of sorting nexins (SNXs) [6]. From early endosomes to the trans-Golgi network, it takes a significant part in the transport of intracellular protein [7]. It is reported thatmight be a novel target gene for Neuropathic pain in patients with head and neck cancer [8]. The possible contribution to a signaling pathway in GC/SC and potentially close expression correlation betweenand TTYH3 were identified by Saha et al [3]. Nonetheless, despite the evidence of the specific role of thein GC, there is very little knowledge regarding its functions and expressions.
Hence, by performing the bioinformatics analysis of several large online databases, in the recent study, we evaluated the significance ofgene expression in GC.
Materials and methods
Oncomine database analysis
Oncomine database (http://www.oncomine.org), an integrated data-mining platform along with a cancer microarray database were used to find out the level ofin the patients with GC, which when compared with the normal individuals, were found to be as, fold change ≥ 1.5,-value ≤ 1E-4, and gene rank ≥ top 10% [9, 10].
Ualcan database
The survival analyses and the tumor subgroup gene expression were facilitated by a portal, the Ualcan database (http://ualcan.path.uab.edu) [11]. We comparedmRNA expression and DNA methylation with the clinical indicators in GC.
GEPIA database
The GEPIA ((http://gepia.cancer-pku.cn), a web server for cancer and normal gene expression profiling and interactive analyses [12]. We identifiedmRNA expression in gastric tissues compare to normal samples, and we use correlation module to identify the correlation betweenand.
UCSC Xena
We use the UCSC Xena (http://xena.ucsc.edu/) to construct the heat map betweenandand the heat map of BIRC5 expression and DNA methylation status [13].
cBioPortal
According to the cBioPortal web (http://www.cbioportal.org/) [14, 15], we identified the mutations and copy number alterations (CNAs) of thegene. The samples of the six studies on GC in the cBioPortal enabled the estimation of the frequency and the location of the mutations.
Kaplan-Meier Plotter
The Kaplan-Meier Plotter (http://kmplot.com/analysis/), a integrative data analysis tool to confirm the prognostic power [16]. The prognostic value ofgene in the post-progression survival (PPS), the progression-free survival (FP), and the overall survival (OS) could be determined using this tool.
TCOA database
In the exploration of the TCGA resource a useful tool suppling several new and unique functions complementary to the existing tools was the TCOA database (http://tcoa.cpu.edu.cn) [17]. We use correlation module to identify the correlation betweenandby using the TCOA database.
Results
SNX8 mRNA and protein expression in GC patients
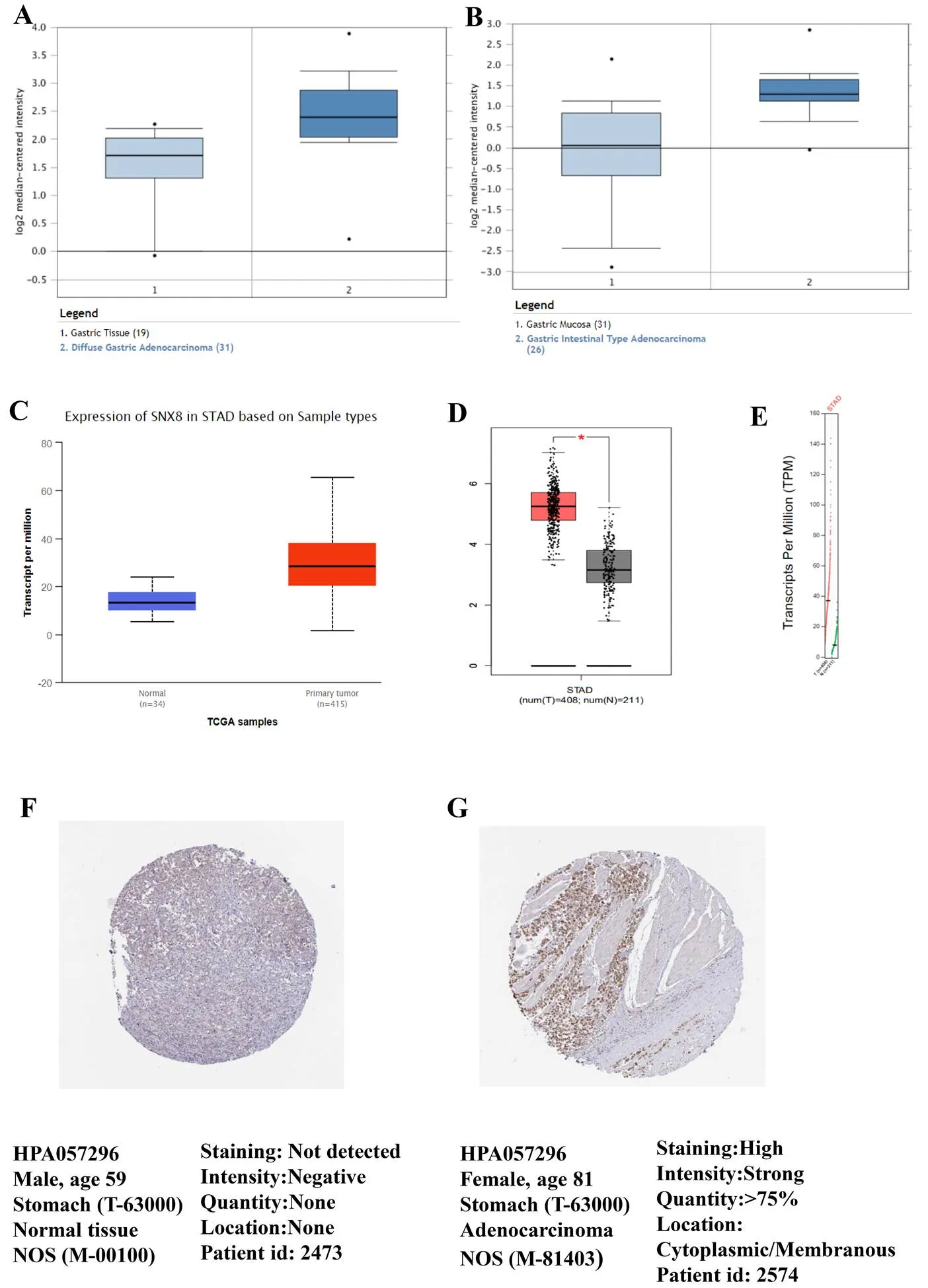
To compare the transcriptional levels ofin cancer with those in normal samples, the ONCOMINE database was utilized (Figure1). The mRNA expression levels ofwere upregulated in patients with GC in two datasets(Figure 2A–B). In Cho gastric statistics [18],was overexpressed in diffuse gastric adenocarcinoma versus normal sample with a fold change of 1.836 (Table 1). In DErrico gastric statistics [19],was found to be higher expressed in gastric intestinal type adenocarcinoma (fold change = 2.922) (Table 1). Interestingly, the higher protein expression ofwas also detected in GC tissues by UALCAN cancer database (Figure 2C) and GEPIA database (Figure 2D–E). Compare to normal tissue, we identify this trend at the protein level in stomach adenocarcinoma (tumor tissue) by using the human protein atlas project (Figure 2F–G). Of the total 37 samples of the GC patients, 33 indicated moderate or weak staining signals, while 2 showed high signals of staining. Moreover, no detectableexpression was noticed in the normal glandular cells in the healthy stomach. In a word, these results showed thatexpression could be overexpressed in gastric tissues compare to normal samples.
Figure 1 The expression of SNX8 at transcription level in pan-cancer by ONCOMINE (fold change ≥ 1.5,-value ≤ 1E-4, gene rank ≥ top 10%)
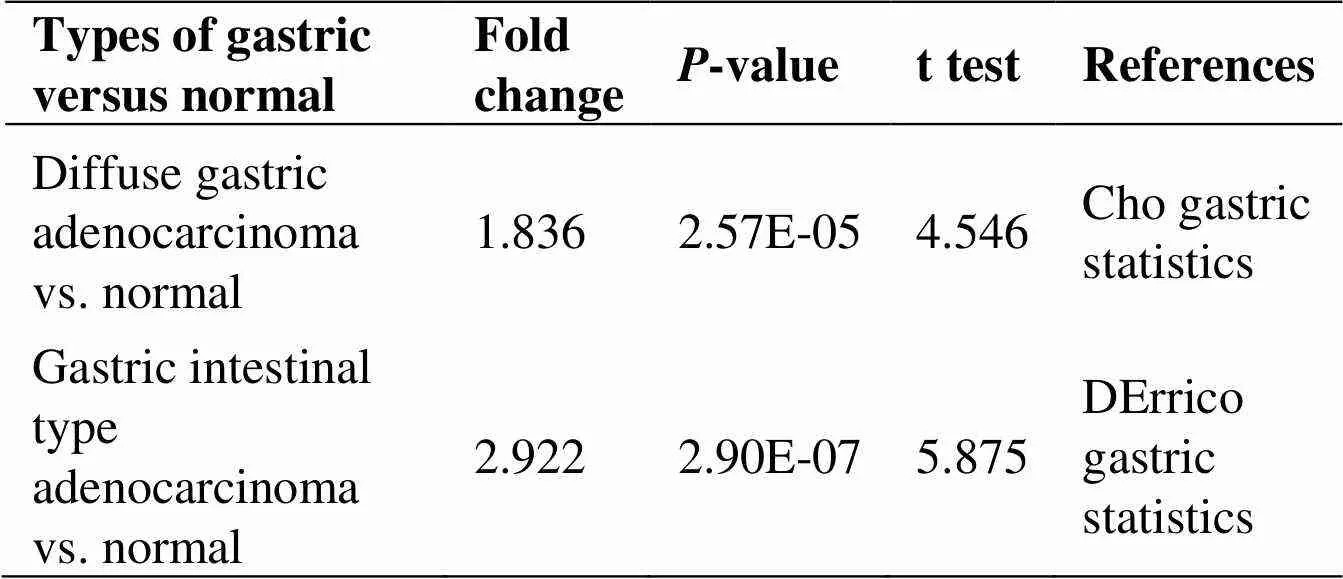
Table 1 The significant changes of SNX8 expression in transcription level between various types of gastric cancer and normal gastric tissues (ONCOMINE database)
Association between SNX8 expression and clinical features in GC patients
We comparedmRNA expression with the clinical indicators in GC patients by using the UALCAN cancer database. The expression of SNX8was upregulated regardless of cancer stage (S1, S2, S3, and S4), gender (male and female), tumor grade (G1, G2, and G3), race (Caucasian, African-American, and Asian), age (20–40, 41–60, 61–80, and 81–100 Yrs),infection, and histological subtype (Figure 3, Table 2). Compare to another age group,was higher expressed in the early age group (21–40 years old) (Figure 3D). In terms of individual cancer stages,was overexpressed in stage 2 compare to any other stages (Figure 3A).expression was also significantly upregulated in all tumor grade compared to normal tissues (Figure 3E).was also higher expressed in patient with other clinical features including patient’s gender, H.pylori infection status, histological subtypes and patient’s race (Table 2). Gene promoter methylation is a common epigenetic event which also has a great potential as a diagnostic and prognostic cancer biomarker [20]. Lower promoter methylation level ofwas observed in GC tissues compared to normal tissues regardless of cancer stage, gender, tumor grade, race and age (Figure 4A–E). Heat map and DNA methylation status indicated thatexpression might be negatively associated with DNA methylation in GC (Figure 5). The results showed that the gene expression might be negatively related with some CpG sites. Therefore, it showed that the mRNA expression ofincreased and promoter methylation reduced in GC.
CNAs and mutations of the SNX8 gene in GC
We identified the CNAs and mutations ofin GC by using cBioPortal web. The results showed that it has nine mutations and one truncation inprotein, most of which involved the PX domain (Figure 6A). Besides, the mutation frequencies were about 2% and 1% in TCGA datasets and Pfizer UHK (University of Hong Kong), separately (Figure 6B). In about 4–6% of the patients with CNAs, amplification was found to be the leading change (Figure 6C). The higherexpression was found to be correlated with gain and amplification (Figure 6D). Therefore, these results showed that theexpression level may be correlated with CNAs in GC.
SNX8 expression and prognosis in GC patients
Next, we investigated the relationship betweenexpression and the clinical prognosis of patients with GC. We conducted prognosis analysis forusing Kaplan-Meier analysis in GC. In all the patients with GC, the increasedmRNA levels were found to be significantly associated with the PPS (< 0.05), the FP, and the OS (Figure 7). Hence, in case of high mRNA levels of the, the patients afflicted with GC were found to have poor PPS, FP, and OS.
Figure 2 (A) Diffuse gastric adenocarcinoma vs. normal by ONCOMINE. (B) Gastric intestinal type adenocarcinoma vs. normal by ONCOMINE. (C) Higher mRNA SNX8 was expressed in gastric cancer by UALCAN. (D)–(E) SNX8 mRNA expression in gastric cancer by GEPIA. (F)–(G) The representative protein expression of SNX8 in normal tissue and gastric tissue from the human protein atlas project.
Figure 3 Relationship betweenexpression and clinical features in gastric cancer patients. (A) Individual cancer stages; (B) race; (C) gender; (D) age; (E) tumor grade; (F) H.pylori infection status; (G) histological subtype
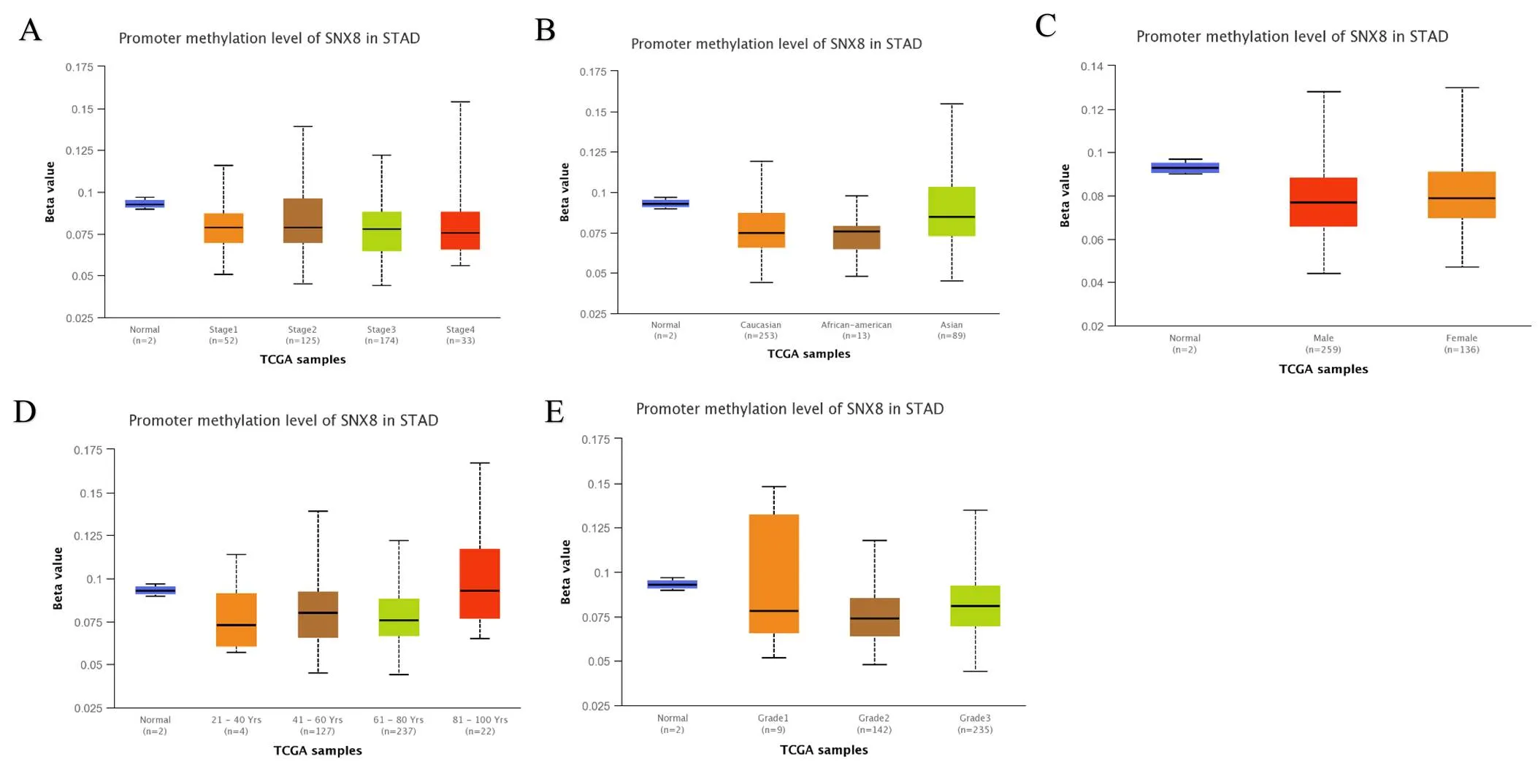
Figure 4 Promoter methylation of thegene by UALCAN. (A) Stage; (B) race; (C) gender; (D) age; (E) tumor grade.
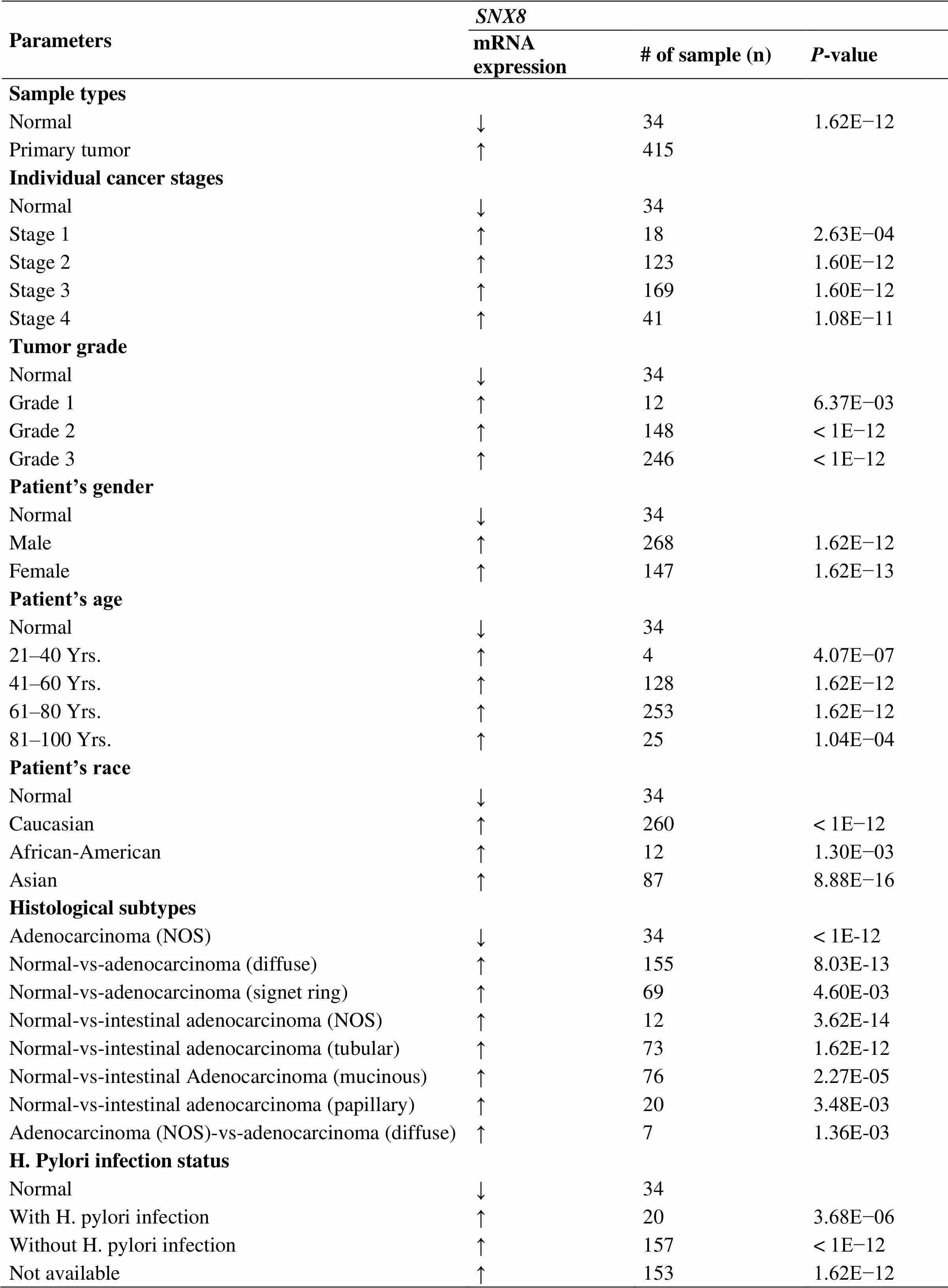
Table 2 Association between SNX8 and clinical parameters in gastric cancer
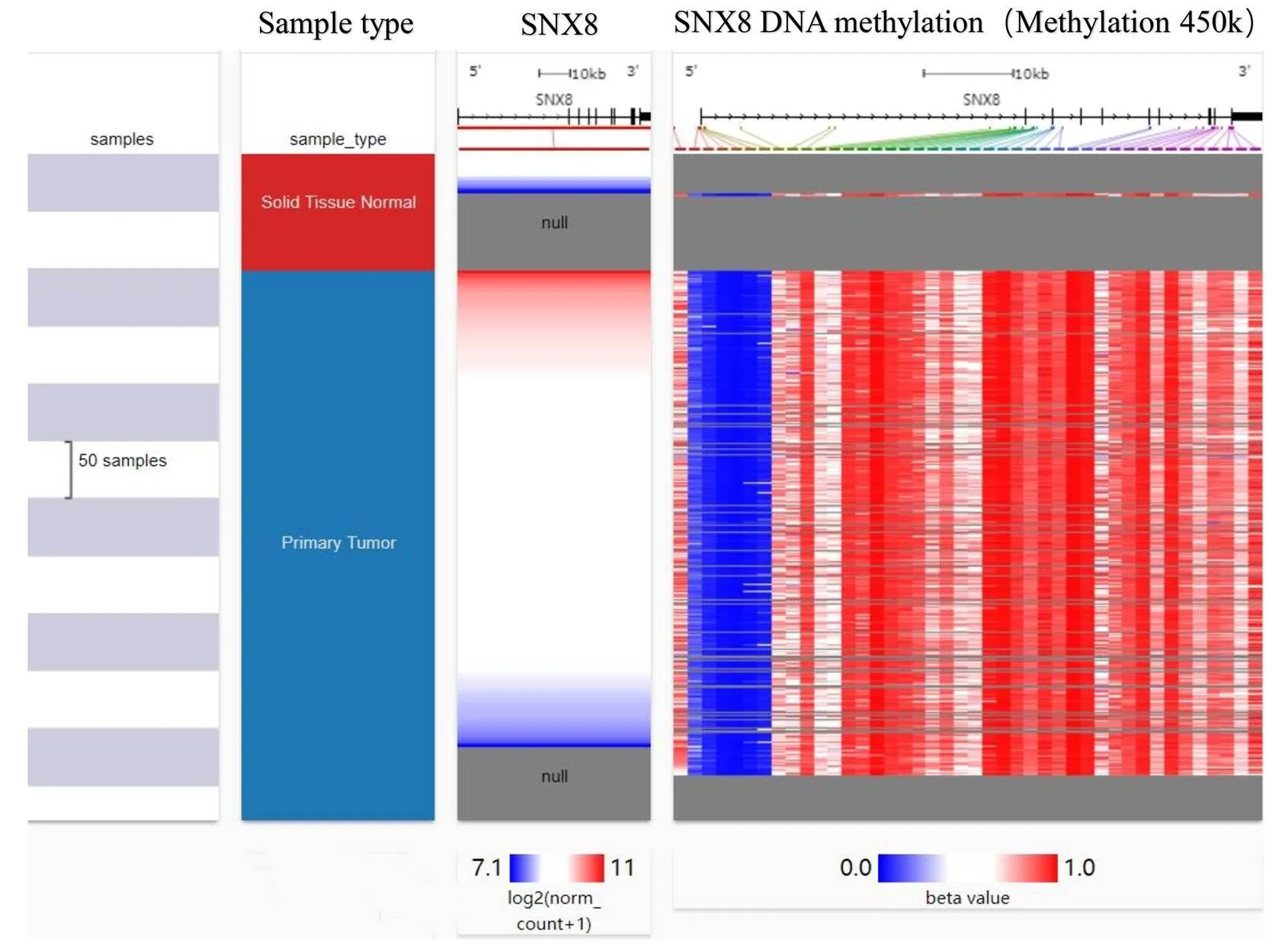
Figure 5 Heat map ofexpression and DNA methylation status by UCSC
Figure 6 Mutation and CNAs inin gastric cancer by cBioPortal. (A) Nine mutations and one truncation in theprotein. (B)–(C)mutation frequencies in gastric cancer. (D) The correlation betweenexpression and CNAs in gastric cancer. CNAs, copy number alterations.
Co-expression of SNX8 gene in GC patients
We finally conducted the co-expression ofgene by using the Oncomine database and palanisamy gastricdataset. It showed that several genes were positively co-expressed within GC, and the most highly correlated gene was(R = 0.979) (Figure 8A).was positively correlated withby using GEPIA database (-value = 0, R = 0.51) and TCOA database (< 0.001, R = 0.28) (Figure 8B–C). The heat map revealed thatwas also closely related withby using the UCSC Xena (Figure 8D). Our observation indicated that expression of SNX8 and CHST12might be positively correlated andcould be involved in thesignaling pathways in GC.
Discussion
As the fifth most common form of cancer in the world, GC is ranked as the third leading cause of cancer related deaths [21, 22]. In the patients with advanced stage GC, the clinical outcome has been quite unsatisfactory, in spite of the numerous advancements in the novel targeted therapy, radiotherapy and chemotherapy in recent decades [23]. Hence, it is crucial to explore the prediction of the clinical progress and prognosis with valuable biomarkers in the patients with GC [24, 25].
is a PX-BAR domain sub-family of SNXs, which is reported as a Aβ toxicity enhancer and associated with Alzheimer's disease [26, 27].It suggested that extreme changes in cholesterol reduceexpression and that overexpression ofexacerbates aberrant handling of neuronal cholesterol [6]. Additionally,plays vital roles in diverse cellular functions [28], which is involved in endocytosis and endosomal sorting [29, 30]. It was also reported thatis a positive regulator of the RNA virus-triggered induction of downstream effector genes and innate immune response [31]. However, the significance ofexpression in the prognosis of GC remains unclear.

Figure 7expression and prognosis in gastric cancer patients. (A) OS; (B) FP; (C) PPS. PPS, post-progression survival; FP, progression-free survival; OS, overall survival.
Figure 8 Co-expression profile of thegene in gastric cancer. (A) Co-expression profile ofidentified using the Oncomine database. (B) Correlation betweenandexpression in breast cancer analyzed using the GEPIA. (C) Co-expression analysis betweenandmRNA expression in gastric cancer determined using TCOA. (D) Heat map ofandexpression by UCSC Xena web-based tool.
First, we indicated that the mRNA expression levels ofwere upregulated in GC patients compare to normal samples by using Oncomine database, UALCAN cancer database and GEPIA database. Besides, in Oncomine database, it showed thatwas significantly upregulated in diffuse gastric adenocarcinoma and gastric intestinal type adenocarcinoma with respect to normal tissues. We also used the human protein atlas project to reveal thatprotein was overexpressed in GC patients compared to normal glandular cells in healthy stomach.
Second, we comparedmRNA expression with the clinical indicators in GC patients by using the UALCAN cancer database. The expression ofwas upregulated regardless of cancer stage (S1, S2, S3, and S4), gender (male and female), tumor grade (G1, G2, and G3), race (Caucasian, African-American, and Asian), age (20–40, 41–60, 61–80, and 81–100 Yrs), helicobacter pylori infection, and histological subtype.
Then, we analyzed the mechanisms ofdysregulation in GC. A lower level of the promoter methylation ofwas found in the GC tissues, with its expression being negatively related to the DNA methylation. Besides, in the CNAs, amplification and gain were correlated withhigher expression. Thus, theexpression level may be correlated with DNA methylation and CNAs in the GC.
Next, we analyzed the prognostic significance ofin GC. The results showed that high mRNA levels of thewas predicted to have poor OS, FP and PPS in GC patients by using the Kaplan-Meier Plotter. So the expression ofmight be a useful biomarker for prognosis of GC.
Finally, we conducted the co-expression ofgene by using the Oncomine, TOCA, GEPIA and UCSC Xena web-based tools.is the most highly correlated gene and it was positively correlated withexpression.is a protein coding gene, and it regulates endosome to chondroitin sulfate biosynthesis [32].has been reported in various malignant tumors and exhibits an important role in carcinogenesis and progression [33],mRNA expression is associated with breast pericanalicular fibroadenoma [34]. Besides, higher mRNA expression ofwas also measured in ovarian cancer samples in comparison to non-malignant ones [35]. After co-expression and correlation analysis in the present study, we conducted thatmight be adopted as a promising predictive biomarker and potential therapeutic target with co-expressedgene.
Conclusion
The higherwas identified to be associated with worse survival rate in GC patients in our study. Through CNAs and promoter methylation, the elevated expression ofcould be regulated. Nevertheless, to investigate the molecular mechanism of these results, in-depth experiments would be needed.
1. Wu JY, Lee CY, Graham DY, et al. The eradication of helicobacter pylori to prevent gastric cancer: a critical appraisal, Expert Rev Gastroenterol Hepatol 2019, 13: 17–24.
2. RaFiei E, Mohammadian-Hafshejani A, Towhidi F, et al. Lack of any relationship of stomach cancer incidence and mortality with development in Asia. Asian Pac J Cancer Prev 2016, 17: 3777–3783.
3. Saha SK, Biswas PK, Gil M, et al. High expression of is related to poor clinical outcomes in human gastric cancer. J Clin Med 2019, 8: 23–28.
4. Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014, 23: 700–713.
5. Duffy MJ, Walsh S, McDermott EW, et al. Biomarkers in breast cancer: where are we and where are we going? Adv Clin Chem 2015, 71: 338–345.
6. Muirhead G, Dev KK. The expression of neuronal sorting nexin 8 () exacerbates abnormal cholesterol levels. J Mol Neurosci 2014, 53: 125–134.
7. Vanzo RJ, Martin MM, Sdano MR, et al.: a candidate gene forcardiac malformations including tetralogy of fallot. Am J Med Genet 2014, 164: 554–556.
8. Reyes-Gibby CC, Wang J, Yeung SCJ, et al. Genome-wide association study identifies genes associated with neuropathy in patients with head and neck cancer. Sci Rep 8789, 8: 8789.
9. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004, 6: 1–6.
10. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia 2007, 9: 166–180.
11. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19: 649–658.
12. Tang Z, Li C, Kang B, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017, 45: 254–261.
13. Goldman M, Craft B, Swatloski T, et al. The UCSC cancer genomics browser: update 2015. Nucleic Acids Res Jan 2015, 43: 812–817.
14. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13: 2498–2504.
15. Saito R, Smoot ME, Ono K, et al. A travel guide to cytoscape plugins. Nat Methods 2012, 9: 1069–1076.
16. Györffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010, 123: 725–731.
17. Sun Q, Li M, Wang X. The cancer omics atlas: an integrative resource for cancer omics annotations. BMC Med Genomics 2018, 11: 63.
18. Cho JY, Lim JY, Cheong JH, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res 2011, 17: 1850–1857.
19. D'Errico M, de Rinaldis E, Blasi MF, et al. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer 2009, 45: 461–469.
20. Bouras E, Karakioulaki M, Bougioukas KI, et al. Gene promoter methylation and cancer: an umbrella review. Gene 2019, 710: 333–340.
21. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015, 136: E359–E386.
22. Guggenheim DE, Shah MA. Gastric cancer epidemiology and risk factors. J Surg Oncol 2013, 107: 230–236.
23. Tan AC, Chan DL, Faisal W, et al. New drug developments in metastatic gastric cancer. Therap Adv Gastroenterol 2018, 11: 1756284818808072.
24. Li J, Xu Q, Wang W, Sun S. MIR100HG: a credible prognostic biomarker and an oncogenic lncRNA in gastric cancer. Biosci Rep 2019, 39: BSR20190171.
25. Matsuoka T, Yashiro M. Biomarkers of gastric cancer: current topics and future perspective. World J Gastroenterol 2018, 24: 2818–2832.
26. Xie Y, Niu M, Ji C, et al.enhances non-amyloidogenic APP trafficking and attenuates Aβ accumulation and memory deficits in an AD mouse. Front Cell Neurosci 2019, 13: 410.
27. Rosenthal SL, Wang X, Demirci FY, et al. Beta-amyloid toxicity modifier genes and the risk of Alzheimer's disease. Am J Neurodegener Dis 2012, 1: 191–198.
28. Wei J, Lian H, Guo W, et al.modulates innate immune response to DNA virus by mediating trafficking and activation of MITA. PLoS Pathog 2018, 14: e1007336.
29. Wei J, Guo W, Lian H, et al.mediates IFNγ-triggered noncanonical signaling pathway and host defense against. Proc Natl Acad Sci U S A 2017, 114: 13000–13005.
30. Johannes L, Wunder C. The SNXy flavours of endosomal sorting. Nat Cell Biol 2011, 13: 884–886.
31. Guo W, Wei J, Zhong X, et al.modulates the innate immune response to RNA viruses by regulating the aggregation of VISA. Cell Mol Immunol 2020, 17: 1126–1135.
32. Nuytemans K, Ortel TL, Gomez L, et al. Variants in chondroitin sulfate metabolism genes in thrombotic storm. Thromb Res 2018, 161: 43–51.
33. Klüppel M, Vallis KA, Wrana JL. A high-throughput induction gene trap approach defines C4ST as a target of BMP signaling. Mech Dev 2002, 118: 77–89.
34. Fernández-Vega I, García O, Crespo A, et al. Specific genes involved in synthesis and editing of heparan sulfate proteoglycans show altered expression patterns in breast cancer. BMC Cancer 2013, 13: 24.
35. Oliveira-Ferrer L, Heßling A, Trillsch F, et al. Prognostic impact of chondroitin-4- sulfotransferasein ovarian cancer. Tumour Biol 2015, 36: 9023–9030.
This study was supported by the Wuqing District Science and Technology Project of Tianjin (WQKJ201962).
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agreed to be accountable for all aspects of the work.
GC, gastric cancer; TCGA, The Cancer Genome Atlas; Aβ, β-amyloid; SNXs, sorting nexins; PPS, post-progression survival; FP, progression-free survival; OS, overall survival; CNAs, copy number alterations.
:
The authors declare that they have no conflict of interest.
: 08 December 2020
:
Bo Liu, Zi-Xiang Kou. Expression and prognosis analyses ofin human gastric cancer. Precision Medicine Research 2020, 2 (4): 153–164.
: Xiao-Hong Sheng.
: 18 September 2020,
24 November 2020,
*Corresponding to: Zi-Xiang Kou, The Affiliated Wuqing Hospital of traditional Chinese Medicine of Tianjin University of traditional Chinese Medicine, No 10 Jichang Road, wuqing District, Tianjin 301700, China. E-mail:kouzixiang181818@126.com.
 Precision Medicine Research2020年4期
Precision Medicine Research2020年4期
- Precision Medicine Research的其它文章
- Exploring the key genes and pathways of ulcerative colitis in a mouse model using gene expression profiling
- Research progress of individualized precision treatment for pancreatic cancer
- Site-specific glycosylation characterization of SARS-CoV-2 spike protein ensures better vaccine development
