双翅目昆虫基因组研究进展
彭威,冯蒙洁,陈皓,韩宝瑜
综 述
双翅目昆虫基因组研究进展
彭威,冯蒙洁,陈皓,韩宝瑜
中国计量大学,浙江省生物计量及检验检疫技术重点实验室,杭州 310018
双翅目(Diptera)是完全变态昆虫中种类最多样化的昆虫,也是第一个基因组已完整测序的昆虫。目前共有110种双翅目昆虫具有公开的基因组,其中黑腹果蝇()和冈比亚按蚊()包含数百个种群基因组。比较基因组学阐明了双翅目昆虫的多种生物学问题,为基因组结构变异、遗传机制以及基因、物种、种群的进化速率和进化模式的研究提供了新思路。尽管双翅目昆虫基因组资源丰富,但仍有许多物种缺乏基因组信息。双翅目昆虫基因组研究对于揭示吸血、寄生、授粉和噬菌性等重要行为的多重起源具有重要价值。本文主要介绍了双翅目昆虫基因组的分布和不同物种基因组的特性,以及双翅目昆虫基因组中功能基因如细胞色素P450、免疫、性别决定和分化相关基因的研究进展,对双翅目昆虫比较基因组学中的重大发现进行了总结,以期在快速发展的基因组组学时代为其他物种进行基因组测序提供指导和借鉴,为开发基于基因组的害虫防治和治理提供理论基础。
双翅目昆虫;基因组特性;功能基因;比较基因组学;系统进化
昆虫是动物界种类最丰富的古老类群。目前地球上已知的昆虫有100万种左右,估计全世界昆虫总数在1000万种以上。其中,双翅目(Diptera)昆虫分布广、数量大、种类多样化,大约包含180个属,总计158,000个种,分为5个主要的下目,即大蚊下目(Tipulomorpha)、蚊下目(Culicomorpha)、蛾蚋下目(Psychodomorpha)、毛蚊下目(Bibionomorpha)和短角下目(Brachycera)[1~3]。短角下目包括约20个总科,总计80,000个物种。其中包括起源于1.8亿年前的短角亚目(Lower Brachycera)和起源于0.65亿年前的环裂亚目(Cyclorrhapha)。环裂亚目超过78个科,习性多样,包括植食性、寄生性、食真菌、哺乳动物寄生性、蛆病、吸血以及幼虫取食腐烂有机质的腐食性。另外,重要的传粉昆虫如食蚜蝇科(Syrphidae)和蜂虻科(Bombyliidae)也主要分布在环裂亚目。在传粉昆虫和开花植物互作中,适应和提高传粉的能力是双翅目昆虫形态多样性、物种多样性和生态多样性的重要驱动力[4~6]。双翅目既包括造成巨大生产损失的农业害虫如地中海实蝇()、麦瘿蚊()和丝光绿蝇(),又包括危害健康的卫生害虫如家蝇()、埃及伊蚊()和冈比亚按蚊()。其中蚊虫叮咬传播的疾病每年可导致200万人死亡。双翅目昆虫中也有为农业生态系统中的开花植物提供授粉的传粉昆虫如食蚜蝇科和蜂虻科。双翅目昆虫生活史、行为习性、取食习性和形态适应性具有多样性[1]。
模式物种黑腹果蝇(),媒介昆虫如埃及伊蚊、冈比亚按蚊和采采蝇(),农业害虫如地中海实蝇、麦瘿蚊和丝光绿蝇是双翅目昆虫中早期完成基因组完整测序的物种。同时,非模式物种基因组测序物种的数量也在增加[7,8]。目前双翅目昆虫中有多达110个物种已完成且可公开获取完整的基因组序列信息(http:// i5k.github.io/arthropod_genomes_at_ncbi)。双翅目昆虫基因组测序数量的稳步增长、以及系统发育基因组学和比较基因组学的发展为研究种间和种内水平的昆虫遗传机制和进化过程提供了新的视角。双翅目昆虫基因组测序样本覆盖率的增加为评估果蝇属和蚊子外物种进化提供了极为重要的参考。双翅目昆虫种间和种内的系统发育基因组学和比较基因组学已经在基因调控和修复[9~12]、发育[13,14]、神经生物学[15,16]、性别决定[17]、昆虫抗药性[18,19]、营养专化[20]和生态适应[21~23]等方面产生了重大的研究成果。毫无疑问,通过下一代基因测序技术和更加完善的基因组数据库,双翅目昆虫基因组研究将推动昆虫基因组学的发展,从系统生物学的角度来解决昆虫学研究中的问题,为农业害虫和病媒昆虫绿色防控提供新策略。本文综述了双翅目昆虫基因组在不同物种中的分布和研究现状,介绍了双翅目昆虫基因组的特性和双翅目昆虫基因组中功能基因如细胞色素P450、免疫、性别决定和分化相关基因研究进展,总结了双翅目昆虫比较基因组学中的重大发现,以期为了解双翅目昆虫多样性、生物学特性以及基于基因组的害虫防治和治理提供参考。
1 双翅目基因组研究现状
分子进化、系统发育和化石等证据将双翅目昆虫的起源定于2.6亿年前的二叠纪晚期,大约与其他主要的全变态昆虫同时开始出现[2,24]。由于双翅目物种间巨大的形态差异、遗传多样性和快速进化的历史进程,对充分阐明双翅目昆虫生命进化构成了挑战。但是系统进化基因组学研究有助于促进我们对双翅目生命进化的理解[1,25]。目前,双翅目亚目已完成110个物种基因组测序,主要分布在蚊科和果蝇科(表1)。(1)蚊科:按蚊科共完成27个物种基因组测序,鉴定了冈比亚按蚊吸血生理适应性和免疫相关基因表达,为了解吸血性媒介昆虫的生理适应机制及疟疾的发病机理提供了理论依据[26];发现致倦库蚊()嗅觉和味觉受体、唾液腺基因和杀虫剂解毒作用相关基因家族数目增加[27];分析了基因漂流和种群历史演变[28];利用Hi-C技术更新了埃及伊蚊基因组染色体读长[29];利用长读长测序方法对白纹伊蚊()基因组重测序,发现其N50>3 Mb[30];对16种按蚊科蚊虫的基因组比较鉴定出基因倒置和参与病媒竞争基因的快速进化[31]。(2)果蝇科:共完成33个物种基因组测序,主要是Brachycera、Cyclorrhapha、Schizophora、Ephydroidea。其中分析了黑腹果蝇基因组结构,其2/3为常染色质,1/3为异染色质,异染色质主要包括简单重复序列、中度重复元件和一些单拷贝DNA,鉴定了与DNA复制、染色体行为、转录和基因调控等相关的蛋白家族[32~37];研究了染色体倒置现象[38];对12种果蝇、、、、、、、、、、和基因组测序比较分析,发现其在基因组大小、基因数量、转座子分布等方面表现出高度保守性,与环境互作和生殖相关蛋白编码基因、非编码RNA、顺式调节区出现变异[39]。对变色伏绕眼果蝇()等10中果蝇性染色体差异的进化模式进行了研究,发现不同果蝇间性染色体组型存在极大地差异性[7,25]。
随着高通量测序技术的发展,越来越多的非模式双翅目昆虫基因组信息得以公布[40~42]。已完成的医学或农业重要性物种的基因组测序可为广大科研工作者探索潜在的害虫防控机制提供重要参考。双翅目农业重要性物种基因组测序包括多种作物或果蔬害虫,如小麦害虫麦瘿蝇和10种实蝇科(Tephritidae)害虫,如地中海实蝇、橄榄果实蝇()。另外,丽蝇科如丝光绿蝇和铜绿蝇()是绵羊蝇蛆病的重要载体,其基因组测序工作具有极其重要的价值[43]。地中海实蝇基因组鉴定超过1800个与入侵和寄主适应相关基因家族发生扩张[44];瓜实蝇()基因组筛选出多个用于害虫防治研究的候选靶标基因;鉴定了防治铜绿蝇的靶标基因[43];麦瘿蚊基因组鉴定出426个效应家族基因和2个抵御寄主植物抗性基因[45]。双翅目医学重要性物种基因组测序包括多种吸血媒介昆虫的基因组,如沙蝇3个毛蠓科(Psychodidae)物种、采采蝇6个舌蝇科(Glossinidae)物种和螯蝇1个蝇科(Muscidae)物种;鉴定了摇蚊科唾液腺相关基因表达和蛋白激酶相关基因表达[46,47];伏蝇()基因组可以应用于法医鉴定[48];舌蝇科总共完成6个物种基因组测序,鉴定了泌乳特异蛋白和卵胎生发育过程[49];家蝇科中家蝇基因组基因拷贝数增加,免疫系统识别和效应基因多样[19],厩螫蝇()基因组主要用于采采蝇基因组的比较分析;蚤蝇科蛆症异蚤蝇()基因组起初被用作低覆盖率基因组分析检测[50];由于难以获取足够高质量长须罗蛉()和巴氏白蛉()DNA,导致毛蠓科基因组测序困难。最近完成超过35个物种基因组测序工作显著提高了双翅目昆虫非模式物种测序覆盖率和基因组学及性染色体差异的进化模式研究,包括潜蝇科班潜蝇()、食虫虻科、丽蝇科红头丽蝇()和丝光绿蝇、萤蚊科和、摇蚊科、突眼蝇科和、长足蝇科、果蝇科和、实蝇科橄榄果实蝇等[7,8](表1)。
2 双翅目昆虫基因组特性
双翅目昆虫基因组测序始于环裂亚目的黑腹果蝇[32]和蚊下目的冈比亚按蚊[26]和埃及伊蚊[52]。黑腹果蝇、冈比亚按蚊和埃及伊蚊基因组的完成不仅催生了基因组数据库、注释参考文库以及生物信息学分析的成功建立和发展,而且极大地推动了国际合作组织对12种果蝇属和16种按蚊属双翅目昆虫的基因组测序和组装工作[31,39]。果蝇科种群基因组计划(population genomics project, DPGP)已收录超过1121种果蝇科野生种群基因组序列[38]。果蝇基因参考图谱(genetic reference panel, DPGP)包含205种黑腹果蝇品系全基因组关联分析(genome-wide association study, GWAS)数据[53~55]。因此,双翅目昆虫基因组的差异性主要来自蚊子和果蝇这两个分化水平显著不同的分支。蚊子和果蝇是双翅目现存世系中最古老的两个分支,其共同的祖先来自大约2.4亿年前[2]。果蝇属物种分支进化跨度最近为24万年前,最远为2200万年前至5500万年前之间[56];按蚊属物种分支进化跨度最近为54万年前,最远为180万年前至1亿年前之间[57]。蚊子与果蝇间、双翅目其他昆虫间以及双翅目与其他目昆虫间的比较基因组学揭示了双翅目昆虫基因组进化速率显著加快[58],使得双翅目昆虫相对于其他昆虫而言是名副其实的“长枝”进化物种。而蚊子和果蝇的比较基因组学表明这两类昆虫以显著较大的速率从彼此分化出去,进而进化为双翅目中两大类[59,60]。
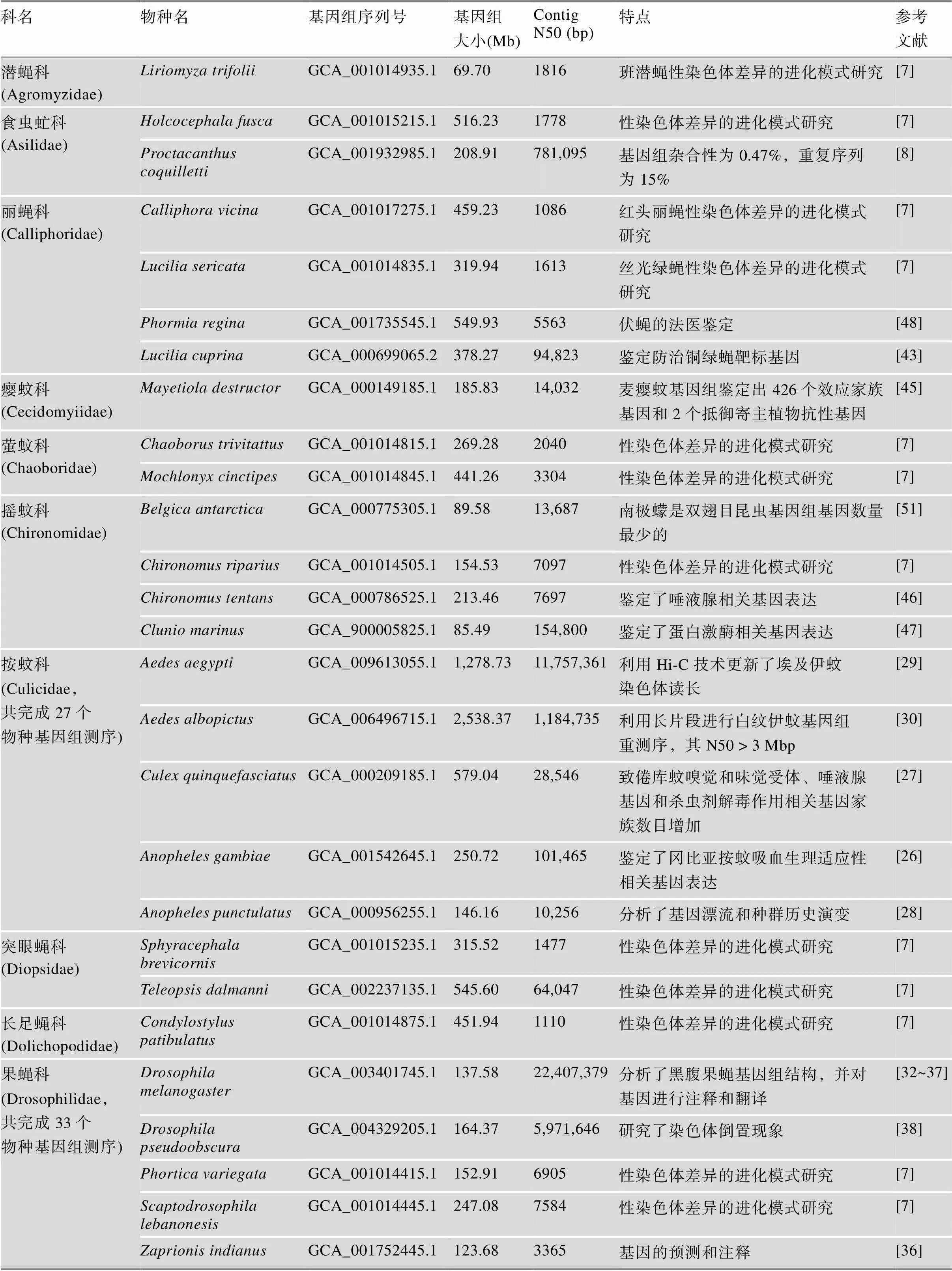
表1 双翅目昆虫基因组信息汇总
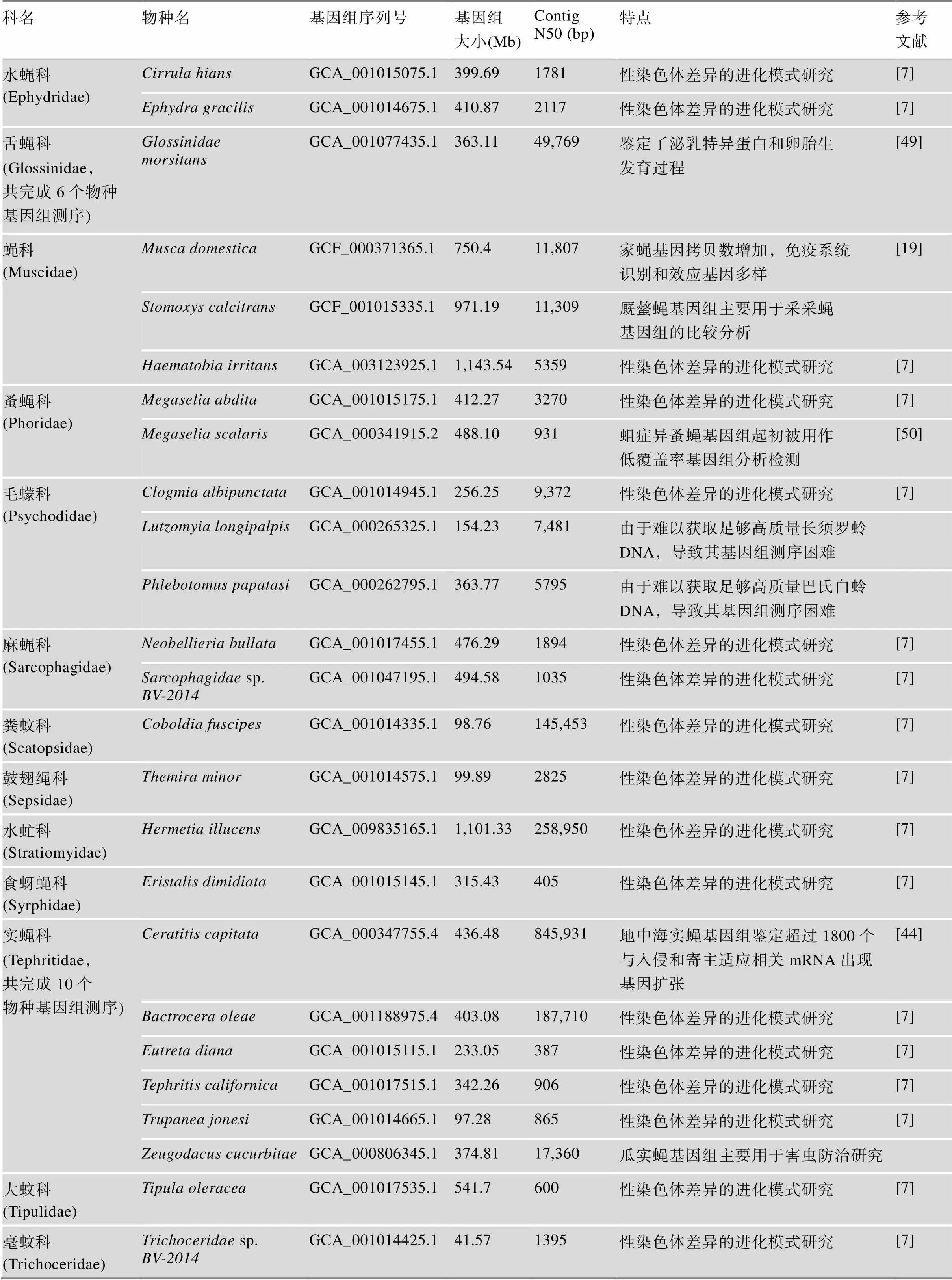
续表1
双翅目昆虫基因组大小差异巨大,从毫蚊科的41.57 Mb到白纹伊蚊的2538.37 Mb不等(表1)。即使在同一个科中,基因组的大小也差异很大,按蚊科基因组大小从146.16~ 2538.37 Mb,而果蝇科基因组大小变化相对较小,从117~386 Mb[61]。双翅目昆虫基因组大小差异巨大的原因可能是其转座子(TEs)和其他重复非编码DNA的差异导致[62,63]。TEs不仅介导物种的进化和新基因的形成,而且参与基因组的表观调控以及异染色质结构的形成。双翅目昆虫基因组中存在的大量非编码DNA是产生遗传变异的重要来源,影响基因组大小的进化方向。双翅目昆虫基因组包含的基因数量差异很大。黑腹果蝇基因组总共有13,920个基因,致倦库蚊基因组总共有18,955个基因。双翅目昆虫基因组基因数量最少的是南极蠓()中的13,517个[51],最多的是家蝇中的23,884个[64]。南极蠓是南极大陆上唯一一种真正意义上的昆虫,也是南极大陆特有的物种。测序发现其基因组规模高度简化,大约只包含9900万个碱基对,基因组中重复的基因序列很少,但与代谢功能、生长发育相关的基因却足够多。南极蠓在漫长的进化过程中,通过剔除非必须基因序列不断调整遗传信息从而适应严酷环境。这为研究生物在极端环境下的进化方向等提供了重要参考[51]。家蝇以人类和动物的排泄物为生,是包括肺结核、伤寒等多种疾病的载体。基因组测序分析发现家蝇基因组多样性高,存在大量与免疫相关基因和特殊的解毒基因,揭示了家蝇对人类疾病产生免疫力和分解废弃物的机制,这为害虫综合防治、废弃物的分解利用和人类疾病的治疗提供了一定的线索和思路[19]。地中海实蝇是一种毁灭性的果蔬害虫,现已分布于80多个国家和地区,危害包括柑桔、苹果、梨等水果和蔬菜在内的250多种寄主,其基因组大小为479 Mb,基因组注释获得14,547个基因,有1608个进化的新基因。黑腹果蝇、家蝇和地中海实蝇基因组比较分析,发现地中海实蝇多个基因、基因家族出现扩张现象,这可能是导致地中海实蝇具有较高的适应性和入侵性的原因[44]。
3 双翅目昆虫基因组中功能基因研究进展
3.1 细胞色素P450基因
P450酶系包括多功能氧化酶和细胞色素P450 ()单加氧酶。其功能高度多样,包括合成昆虫发育和繁殖所需的重要激素和化学代谢物质,从而促进昆虫对寄主植物的适应性和对环境中有毒物质如杀虫剂的解毒作用。黑腹果蝇细胞色素P450家族共鉴定出90个基因,分属25个家族,其中和家族的成员最多,占P450基因总数的一半[32]。地中海实蝇细胞色素P450家族包含103个基因和9个假基因,相较于黑腹果蝇的88个基因和3个假基因,地中海实蝇细胞色素P450家族显著扩张,主要集中于和基因家族,其扩张性却低于家蝇和基因家族[44]。地中海实蝇家族由40个基因和4个假基因组成,是黑腹果蝇家族23个基因的几乎两倍。其中、和亚家族出现显著扩张,包含14个基因、包含9个基因、包含5个基因[44]。这3个亚家族基因和双翅目昆虫杀虫剂抗性相关,其中家族通过基因簇复制快速扩张[65]。另外,在地中海实蝇基因组中发现18个连续的基因形成一个基因簇(13个属于亚家族),其中基因的过表达和氯氟氰菊酯抗性相关[66]。在黑腹果蝇基因组中发现2个和9个连续的基因形成两个基因簇。地中海实蝇基因家族出现复制表明其参与环境响应如细胞色素P450调控的抗性。家蝇和黑腹果蝇基因家族和杀虫剂抗性相关[65]。此外,细胞色素P450家族还包含蜕皮激素合成途径相关基因,在地中海实蝇基因组中发现4个P450基因()、()、()、()能够活化蜕皮激素。
3.2 免疫相关基因
免疫反应包括黑化作用、吞噬作用、包埋、凝血和脂肪体合成抗菌肽和抗菌蛋白[67]。涉及病菌识别和防御反应的四条主要信号途径是:Toll、IMD、JAK/STAT和JNK[68]。昆虫主要通过模式识别受体(PRRs)和肽聚糖识别蛋白(PGRPs)家族基因识别细菌,革兰氏阴性细菌结合蛋白(GNBPs)通过结合细菌配体从而激活免疫途径[69~71]。黑腹果蝇基因组鉴定出379个假定的免疫基因,地中海实蝇基因组鉴定出413个假定的免疫基因,家蝇基因组鉴定出771个假定的免疫基因。家蝇基因组中免疫基因数量巨大、免疫识别和受体基因的拷贝数和基因多样性显著增加的原因可能和其生活在富含病原菌的环境相关[19]。家蝇免疫识别受体Nimrods和thioester- containing proteins (Teps)拷贝数出现显著扩张。家蝇具有17个Nimrods蛋白、19个Teps蛋白。黑腹果蝇具有11个Nimrods蛋白、6个Teps蛋白。在已测序果蝇属物种中,Nimrods基因家族的拷贝数差异较大[72]。由于地中海实蝇极其多样的寄主选择性导致其免疫基因数量较多,从而应对寄主和环境条件中多种多样的病原菌[44]。革兰氏阴性细菌和真菌诱导免疫响应因子基因激活Toll信号途径,地中海实蝇由于在不同果实上产卵接触到的真菌感染导致家族基因和Toll受体家族基因出现高度扩张。地中海实蝇有17个Toll受体家族基因,而黑腹果蝇和家蝇只有9个。基因活化所必需的丝氨酸蛋白酶基因家族在地中海实蝇中也出现显著扩张,相较于黑腹果蝇的45个和家蝇的28个,地中海实蝇具有50个丝氨酸蛋白酶基因[44]。
3.3 性别决定和分化相关基因
地中海实蝇基因组已鉴定出35个直接或者间接参与性别决定和性别分化基因,其中25个基因包括()、()、()基因,6个性别特异剪切基因和4个基因具有躯体性别特异功能如剂量补偿[44,73,74]。通过比较家蝇雌成虫和雄成虫基因的表达量,已鉴定出113个雄性偏向性表达基因和81个雌性偏向性表达基因[19]。而在黑腹果蝇中10%~20%的基因具有性别偏向性表达的特性,比家蝇观察到的明显增多[75,76]。近年来双翅目昆虫基因组测序很大一部分是关于性染色体差异的进化模式研究(表1),而基于基因组测序的策略已鉴定出多种雄性性别决定因子。埃及伊蚊染色体性别决定系统缺少Y染色体,Hall等[77]基于雌雄基因组测序发现埃及伊蚊雄性决定因子基因位于1号染色体的非重组区域,处于性别决定级联反应的顶端,通过调控下游基因mRNA前体雄性特异剪切和表达,促进雄性发育。Krzywinska等[78]对冈比亚按蚊雌雄胚胎基因序列比较,在Y染色体上鉴定出一个仅在雄性早期转录表达基因,发现调控基因的雄性特异剪切和表达,从而实现雄性发育。家蝇有一个与众不同的多态性别决定系统,雄性携带一个显性的雄性决定因子,这个因子可以位于X或者Y或者任意5条常染色体上。基于家蝇基因组序列信息,Sharma等[79]阐明其性别决定系统由雄性决定因子()的存在与否来决定。Meccariello等[80]通过对地中海实蝇雄虫构建长读长基因组文库,筛选出性别决定基本信号是位于Y染色体上的雄性决定因子--()基因,通过阻止合子中基因活化,导致基因进行雄性特异剪切,引起雄性发育。此外,基因作为雄性决定因子在双翅目实蝇科其他物种如橄榄果实蝇和橘小实蝇()中也是Y染色体连接,且功能保守[80]。
4 双翅目昆虫比较基因组学研究进展
目前,基因组结构、基因含量、共线性、染色体倒位和非编码元件进化研究是比较基因组学研究的重要领域[39,81,82]。双翅目昆虫比较基因组学研究阐明了新基因的形成[83,84]、基因和基因组互作与调控[85]和基因组塑造昆虫生物史[86,87]等分子生物学问题。利用种属水平的系统发育比较基因组学,双翅目昆虫中基因家族的进化关系逐渐得到阐述。家蝇作为世界性的卫生害虫,由于其独特的取食习性、长期暴露在杀虫剂下以及与动物病原菌之间的互作,系统发育比较基因组学已证明其与生理和行为适应性相关的细胞色素P450基因家族、化学感受受体和气味结合蛋白基因的拷贝数发生了显著变化[57]。已有研究表明,按蚊属基因组基因拷贝数的扩增和收缩比果蝇属快5倍[19]。蚊科和果蝇科中数量巨大的高质量基因组数据可用于小型调控元件如microRNA、piwi-interacting RNA、Aubergine和功能性小阅读框(smORF)的鉴定和系统进化分析[88~91]。
双翅目昆虫比较基因组学为阐明昆虫进化模式和机制、适应性和生理功能以及基因型和表型之间的联系提供了一个很好的手段。通过比较冈比亚按蚊不同染色体间的系统进化分析模式,发现其基因组中存在大量基因渗入现象,这为解释新形成物种之间常染色体至X染色体的基因转移速率差异提供了新的证据[57]。比较基因组学为计算近缘物种种群动态、种群分类排序和基因渗入在塑造昆虫遗传差异性等方面提供一个完整的研究系统[92]。蚊子间比较基因组学对于了解病原菌传播的基本生物学过程以及探索调控病媒昆虫防治的遗传机制具有越来越重要的价值[93,94]。根据果蝇科已测序基因组建立的系统发育进化树已被用来研究种间基因、基因组、调控网络、发育途径和生态适应等分子生物学问题的进化框架[95,96]。目前,总共有30种果蝇科昆虫完成基因组组装,其中23种来自水果果蝇亚属(),另外7种来自果蝇亚属()。果蝇科昆虫间的比较基因组学有助于阐明DNA结合蛋白的基因调控机制,并鉴定出塑造双翅目发育、行为和生理过程的保守直系同源调控基因结构[97,98]。
双翅目昆虫功能基因组学和比较基因组学是研究昆虫与植物互作的重要手段。植物寄生性麦廮蝇的基因组研究表明,有多种基因产物充当效应蛋白抑制植物防御,并调节宿主细胞诱导植物产生五倍子[45]。地中海实蝇功能基因组学鉴定出多种气味结合蛋白、水通道蛋白和免疫反应基因,参与调控宿主植物适应性协同进化[44]。果蝇科昆虫在发育进程中的植食性已出现多次进化,对斑翅果蝇()和黄果蝇()的比较基因组学研究发现,取食受损植物组织和取食正常植物组织前后会导致基因表达出现显著性变化,主要包括与营养、规避植物防御和寄主定位相关基因的表达[99~101]。鉴于双翅目昆虫测序成本相对较低,大量果蝇科和蚊子种群基因组测序工作得以完成。果蝇科种群基因组计划和果蝇基因参考图谱是研究定量遗传学的重要参考文库,可获得和测定特定品系的定量表型,并可鉴定其与先前基因组序列的关联性[53~55]。利用DPGP已实现果蝇科昆虫48种定量表型的遗传学分析[42]。此外,双翅目昆虫具有高丰度和高耐受的染色体内倒位现象,拟暗果蝇() 54个种群基因组学研究对3号染色体倒位多态性进行了鉴定[38]。对分布在非洲的765种冈比亚按蚊和个体进行测序发现,相较于黑腹果蝇0.5%的个体多态性和人类0.5%个体多态性,蚊子个体多态性为3%[102]。冈比亚按蚊种群基因组测序不仅推动了某些假定基因驱动(gene drive)的应用,还鉴定出远距离基因漂流现象和物种间基因渗入与抗性等位基因的传播有关。
5 结语与展望
目前虽然有大量双翅目昆虫完成基因组测序工作,但是测序样本范围极度失衡,已有基因组主要集中在果蝇科和蚊科,双翅目其它科物种基因组测序还比较缺乏,许多常见科中的昆虫尚未进行测序[103]。首先,通过实验室饲养、区域生物调查合作和全球基因组计划等可以实现双翅目昆虫基因组测序样本的多样化。双翅目中取食习性和行为习性多样的物种或者模式物种可继续充当未来基因组测序工作的主要对象。眼蕈蚊(Sciaridae)就是其中一个很好的候选对象:多数眼蕈蚊是腐生或以真菌为食,但少数也能侵入活体植物组织。因而眼蕈蚊是研究发育调控基因扩增、性别决定、细胞凋亡、免疫以及染色体结构多态性遗传机制的模式物种[104]。对双翅目昆虫生理、生态或行为特征具有差异性的近缘物种进行基因组测序,能有效阐明昆虫生物适应的遗传和分子机制。其次,食蚜蝇科(Syrphidae)、蚤蝇科(Phoridae)、秆蝇科(Chloropidae)和家蝇科(Muscidae)昆虫有植食性、寄生性和食真菌性等多种食性,而丽蝇科(Calliphoridae)、麻蝇科(Sarcophagidae)、家蝇科(Muscidae)、虱蝇科(Hippoboscidae)和狂蝇科(Oestridae)昆虫具有哺乳动物或鸟类寄生性和无脊椎动物寄生性等。这些昆虫的比较基因组学研究将有助于阐明双翅目昆虫适应性的关键遗传调控因子。而对双翅目中医学重要性物种进行基因组测序将有助于揭示吸血和栖息地习性转变等一系列行为的遗传学基础。蚋科(Simuliidae)昆虫刺吸人畜的血液,是人畜蟠尾丝虫病的传播媒介,然而蚋科尚无完整的基因组序列信息。最后,寄生昆虫、传粉昆虫和捕食昆虫基因组信息也极其缺乏。栖息地或寄主选择具有差异性的物种间的比较基因组学研究将揭示双翅目昆虫寄主专化性、寄主寻找和规避寄主免疫系统的协同进化模式。将来对双翅目更多科昆虫进行基因组测序是了解双翅目昆虫基因和基因组,以及基因组功能如性染色体进化和多样性的重要手段[33]。因此,全基因组测序、功能基因组学、进化生物学、比较基因组学、生物信息学分析等技术是推动双翅目昆虫基因组学在害虫防治、资源昆虫利用、药物靶点开发及进化生物学等方面应用的重要手段[105~109]。
[1] Wiegmann BM, Yeates DK. Phylogeny of Diptera. 2017, Chapter 11. In Manual of Afrotropical Diptera. Volume 1. Introductory Chapters and Keys to Diptera Families Suricata. Edited by Kirk-Spriggs AH, Sinclair BJ. SANBI Graphics & Editing.
[2] Wiegmann BM, Trautwein M, Winkler IS, Barr NB, Kim J, Lambkin CL, Bertone MA, Cassel BK, Bayless KM, Heimberg AM, Wheeler BM, Peterson KJ, Pape T, Sinclair BJ, Skevington JH, Blagoderov V, Caravas J, Kutty SN, Schmidt-Ott U, Kampmeier GE, Thompson FC, Grimaldi DA, Beckenbach AT, Courtney GW, Friedrich M, Meier R, Yeatesd DK. Episodic radiations in the fly tree of life., 2011, 108(14): 5690–5695.
[3] Bertone MA, Courtney GW, Wiegmann BM. Phylogenetics and temporal diversification of the earliest true flies (Insecta: Diptera) based on multiple nuclear genes., 2008, 33(4): 668–687.
[4] Labandeira CC. Fossil history and evolutionary ecology of Diptera and their associations with plants. The evolutionary biology of flies., 2005, 217–273.
[5] Ssymank A, Kearns CA, Pape T, Thompson FC. Pollinating flies (Diptera): A major contribution to plant diversity and agricultural production., 2008, 9(1–2): 86–89.
[6] Wiens JJ, Lapoint RT, Whiteman NK. Herbivory increases diversification across insect clades., 2015, 6(1): 8370–8370.
[7] Vicoso B, Bachtrog D. Numerous transitions of sex chromosomes in Diptera., 2015, 13(4): e1002078.
[8] Dikow RB, Frandsen PB, Turcatel M, Dikow T. Genomic and transcriptomic resources for assassin flies including the complete genome sequence of(Insecta: Diptera: Asilidae) and 16 representative transcriptomes., 2017, 5: e2951.
[9] Richards S, Liu Y, Bettencourt BR, Hradecky P, Letovsky S, Nielsen R, Thornton K, Hubisz MJ, Chen R, Meisel RP, Couronne O, Hua SJ, Smith MA, Zhang PL, Liu J, Bussemaker HJ, van Batenburg MF, Howells SL, Scherer SE, Sodergren E, Matthews BB, Crosby MA, Schroeder AJ, Ortiz-Barrientos D, Rives CM, Metzker ML, Muzny DM, Scott G, Steffen D, Wheeler DA, Worley KC, Havlak P, Durbin KJ, Egan A, Gill R, Hume J, Morgan MB, Miner G, Hamilton C, Huang YM, Waldron L, Verduzco D, Clerc-Blankenburg KP, Dubchak I, Noor MAF, Anderson W, White KP, Clark AG, Schaeffer SW, Gelbart W, Weinstock GM, Gibbs RA. Comparative genome sequencing of: Chromosomal, gene, and cis-element evolution., 2005, 15(1): 1–18.
[10] Bellen HJ, Levis RW, He YC, Carlson JW, Evans-Holm M, Bae E, Kim J, Metaxakis A, Savakis C, Schulze KL, Hoskins RA, Spradling AC. Thegene disruption project: progress using transposons with distinctive site specificities., 2011, 188(3): 731–743.
[11] Gratz SJ, Ukken FP, Rubinstein CD, Thiede G, Donohue LK, Cummings AM, O’connor-Giles KM. Highly specific and efficient CRISPR/Cas9-catalyzed homology- directed repair in., 2014, 196(4): 961–971.
[12] Xue ZY, Wu MH, Wen KJ, Ren MD, Long L, Zhang XD, Gao GJ. CRISPR/Cas9 mediates efficient conditional mutagenesis in., 2014, 4(11): 2167–2173.
[13] Jory A, Estella C, Giorgianni MW, Slattery M, Laverty TR, Rubin GM, Mann RS. A survey of 6,300 genomic fragments for cis-regulatory activity in the imaginal discs of., 2012, 2(4): 1014–1024.
[14] Geib SM, Calla B, Hall B, Hou SB, Manoukis NC. Characterizing the developmental transcriptome of the oriental fruit fly,(Diptera: Tephritidae) through comparative genomic analysis withutilizing modENCODE datasets., 2014, 15(1): 942–942.
[15] Grönke S, Clarke DF, Broughton S, Andrews TD, Partridge L. Molecular evolution and functional characterization of Drosophila insulin-like peptides., 2010, 6(2): e1000857.
[16] Neely GG, Hess A, Costigan M, Keene AC, Goulas S, Langeslag M, Griffin RS, Belfer I, Dai F, Smith SB, Diatchenko L, Gupta V, Xia CP, Amann S, Kreitz S, Heindl-Erdmann C, Wolz S, Ly CV, Arora S, Sarangi R, Dan D, Novatchkova M, Rosenzweig M, Gibson DG, Truong D, Schramek D, Zoranovic T, Cronin SJF, Angjeli B, Brune K, Dietzl G, Maixner W, Meixner A, Thomas W, Pospisilik JA, Alenius M, Kress M, Subramaniam S, Garrity PA, Bellen HJ, Woolf CJ, Penninger JM. A genome-widescreen for heat nociception identifiesas an evolutionarily conserved pain gene., 2010, 143(4): 628–638.
[17] Mahajan S, Bachtrog D. Convergent evolution of Y chromosome gene content in flies., 2017, 8(1): 785–785.
[18] Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance., 2014, 45: 89–110.
[19] Scott JG, Warren WC, Beukeboom LW, Bopp D, Clark AG, Giers SD, Hediger M, Jones AK, Kasai S, Leichter CA, Li M, Meisel RP, Minx P, Murphy TD, Nelson DR, Reid WR, Rinkevich FD, Robertson HM, Sackton TB, Sattelle DB, Thibaud-Nissen F, Tomlinson C, van de Zande L, Walden KK, Wilson RK, Liu NN. Genome of the house fly,L., a global vector of diseases with adaptations to a septic environment., 2014, 15(10): 466–466.
[20] Groen SC, Whiteman NK. Usingto study the evolution of herbivory and diet specialization., 2016, 14: 66–72.
[21] Bergland AO, Tobler R, González J, Schmidt PS, Petrov D. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in., 2016, 25(5): 1157–1174.
[22] Ramasamy S, Ometto L, Crava CM, Revadi S, Kaur R, Horner DS, Pisani D, Dekker T, Anfora G, Rota-Stabelli O. The evolution of olfactory gene families inand the genomic basis of chemical-ecological adaptation in., 2016, 8(8): 2297–2311.
[23] Cicconardi F, Di Marino D, Olimpieri PP, Arthofer W, Schlick-Steiner BC, Steiner FM. Chemosensory adaptations of the mountain fly Drosophila nigrosparsa (Insecta: Diptera) through genomics’ and structural biology’s lenses., 2017, 7(1): 43770.
[24] Misof B, Liu SL, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, Niehuis O, Petersen M, Izquierdo-Carrasco F, Wappler T, Rust J, Aberer AJ, Aspöck U, Aspöck H, Bartel D, Blanke A, Berger S, Böhm A, Buckley TR, Calcott B, Chen JQ, Friedrich F, Fukui M, Fujita M, Greve C, Grobe P, Gu SC, Huang Y, Jermiin LS, Kawahara AY, Krogmann L, Kubiak M, Lanfear R, Letsch H, Li YY, Li ZY, Li JG, Lu HR, Machida R, Mashimo Y, Kapli P, McKenna DD, Meng GL, Nakagaki Y, Navarrete- Heredia JL, Ott M, Ou YX, Pass G, Podsiadlowski L, Pohl H, von Reumont BM, Schütte K, Sekiya K, Shimizu S, Slipinski A, Stamatakis A, Song WH, Su X, Szucsich NU, Tan MH, Tan XM, Tang M, Tang JB, Timelthaler G, Tomizuka S, Trautwein M, Tong XL, Uchifune T, Walzl MG, Wiegmann BM, Wilbrandt J, Wipfler B, Wong TKF, Wu Q, Wu GX, Xie YL, Yang SZ, Yang Q, Yeates DK, Yoshizawa K, Zhang Q, Zhang R, Zhang WW, Zhang YH, Zhao J, Zhou CR, Zhou LL, Ziesmann T, Zou SJ, Li YR, Xu X, Zhang Y, Yang HM, Wang J, Wang J, Kjer KK, Zhou X. Phylogenomics resolves the timing and pattern of insect evolution., 2014, 346(6210): 763–767.
[25] Kutty SN, Wong WH, Meusemann K, Meier R, Cranston P. A phylogenomic analysis of(Diptera) resolves the relationships among the eight constituent families., 2018, 43(3): 434– 446.
[26] Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JMC, Wides R, Salzberg SL, Loftus B, Yandell M, Majoros WH, Rusch DB, Lai ZW, Kraft CL, Abril JF, Anthouard V, Arensburger P, Atkinson PW, Baden H, de Berardinis V, Baldwin D, Benes V, Biedler J, Blass C, Bolanos R, Boscus D, Barnstead M, Cai S, Center A, Chatuverdi K, Christophides FK, Chrystal MA, Clamp M, Cravchik A, Curwen V, Dana A, Delcher A, Dew I, Evans CA, Flanigan M, Grundschober-Freimoser A, Friedli L, Gu ZP, Guan P, Guigo R, Hillenmeyer ME, Hladun SL, Hogan JR, Hong YS, Hoover J, Jaillon O, Ke ZX, Kodira C, Kokoza E, Koutsos A, Letunic I, Levitsky A, Liang Y, Lin JJ, Lobo NF, Lopez JR, Malek JA, McIntosh TC, Meister S, Miller J, Mobarry C, Mongin E, Murphy SD, O'Brochta DA, Pfannkoch C, Qi R, Regier MA, Remington K, Shao HG, Sharakhova MV, Sitter CD, Shetty J, Smith TJ, Strong R, Sun JT, Thomasova D, Ton LQ, Topalis P, Tu ZJ, Unger MF, Walenz B, Wang A, Wang J, Wang M, Wang XL, Woodford KJ, Wortman JR, Wu M, Yao A, Zdobnov EM, Zhang HY, Zhao Q, Zhao SY, Zhu SC, Zhimulev I, Coluzzi M, della Torre A, Roth CW, Louis C, Kalush F, Mural RJ, Myers EW, Adams MD, Smith HO, Broder S, Gardner MJ, Fraser CM, Birney E, Bork P, Brey PT, Venter JC, Weissenbach J, Kafatos FC, Collins FH, Hoffman SL. The genome sequence of the malaria mosquito., 2002, 298(5591): 129–149.
[27] Arensburger P, Megy K, Waterhouse RM, Abrudan JL, Amedeo P, Antelo BG, Bartholomay LC, Bidwell SL, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Fraser-Liggett C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC, Kodira CD, Lobo NF, Mao CH, Mayhew G, Michel K, Mori A, Liu NN, Naveira H, Nene V, Nguyen N, Pearson MD, Pritham EJ, Puiu D, Qi YM, Ranson H, Ribeiro JMC, Roberston HM, Severson DW, Shumway M, Stanke M, Strausberg RL, Sun C, Sutton G, Tu ZJ, Tubio JMC, Unger MF, Vanlandingham DL, Vilella AJ, White O, White JR, Wondji CS, Wortman J, Zdobnov EM, Birren B, Christensen BM, Collins FH, Cornel A, Dimopoulos G, Hannick LI, Higgs S, Lanzaro GC, Lawson D, Lee NH, Muskavitch MAT, Raikhel AS, Atkinson PW. Sequencing ofestablishes a platform for mosquito comparative genomics., 2010, 330(6000): 86–88.
[28] Logue K, Small ST, Chan ER, Reimer LJ, Siba P, Zimmerman PA, Serre D. Whole-genome sequencing reveals absence of recent gene flow and separate demographic histories formosquitoes in Papua New Guinea., 2015, 24(6): 1263–1274.
[29] Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, Aiden EL. De novo assembly of thegenome using Hi-C yields chromosome-length scaffolds., 2017, 356(6333): 92–95.
[30] Artemov GN, Peery AN, Jiang XF, Tu ZJ, Stegniy VN, Sharakhova MV, Sharakhov IV. The physical genome mapping of Anopheles albimanus corrected scaffold misassemblies and identified interarm rearrangements in genus Anopheles., 2017, 7(1): 155–164.
[31] Neafsey DE, Waterhouse RM, Abai MR, Aganezov S, Alekseyev MA, Allen JE, Amon J, Arca B, Arensburger P, Artemov GN, Assour LA, Basseri H, Berlin A, Birren BW, Blandin SA, Brockman AI, Burkot TR, Burt A, Chan CS, Chauve C, Chiu JC, Christensen M, Costantini C, Davidson VL, Deligianni E, Dottorini T, Dritsou V, Gabriel SB, Guelbeogo WM, Hall AB, Han MV, Hlaing T, Hughes DS, Jenkins AM, Jiang XF, Jungreis I, Kakani EG, Kamali M, Kemppainen P, Kennedy RC, Kirmitzoglou IK, Koekemoer LL, Laban N, Langridge N, Lawniczak MK, Lirakis M, Lobo NF, Lowy E, MacCallum RM, Mao C, Maslen G, Mbogo C, McCarthy J, Michel K, Mitchell SN, Moore W, Murphy KA, Naumenko AN, Nolan T, Novoa EM, O'Loughlin S, Oringanje C, Oshaghi MA, Pakpour N, Papathanos PA, Peery AN, Povelones M, Prakash A, Price DP, Rajaraman A, Reimer LJ, Rinker DC, Rokas A, Russell TL, Sagnon N, Sharakhova MV, Shea T, Simão FA, Simard F, Slotman MA, Somboon P, Stegniy V, Struchiner CJ, Thomas GW, Tojo M, Topalis P, Tubio JM, Unger MF, Vontas J, Walton C, Wilding CS, Willis JH, Wu YC, Yan G, Zdobnov EM, Zhou XF, Catteruccia F, Christophides GK, Collins FH, Cornman RS, Crisanti A, Donnelly MJ, Emrich SJ, Fontaine MC, Gelbart W, Hahn MW, Hansen IA, Howell PI, Kafatos FC, Kellis M, Lawson D, Louis C, Luckhart S, Muskavitch MA, Ribeiro JM, Riehle MA, Sharakhov IV, Tu ZJ, Zwiebel LJ, Besansky NJ. Highly evolvable malaria vectors: The genomes of 16 Anopheles mosquitoes., 2015, 347(6217): 1258522–1258522.
[32] Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides P, Scherer S, Li PW, Hoskins RA, Galle R, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris ML, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, Woodage T, Worley KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of., 2000, 287(5461): 2185–2195.
[33] Zhou Q, Bachtrog D. Sex-specific adaptation drives early sex chromosome evolution in., 2012, 337(6092): 341–345.
[34] Chen ZX, Sturgill D, Qu JX, Jiang HY, Park S, Boley N, Suzuki AM, Fletcher AR, Plachetzki DC, Fitzgerald PC, Artieri CG, Atallah J, Barmina O, Brown JB, Blankenburg KP, Clough E, Dasgupta A, Gubbala S, Han Y, Jayaseelan JC, Kalra D, Kim YA, Kovar CL, Lee AL, Li MM, Malley JD, Malone JH, Mathew T, Mattiuzzo NR, Munidasa M, Muzny DM, Ongeri F, Perales L, Przytycka TM, Pu LL, Robinson G, Thornton RL, Saada N, Scherer SE, Smith HE, Vinson C, Warner CB, Worley KC, Wu YQ, Zou XY, Cherbas P, Kellis M, Eisen MB, Piano F, Kionte K, Fitch DH, Sternberg PW, Cutter AD, Duff MO, Hoskins RA, Graveley BR, Gibbs RA, Bickel PJ, Kopp A, Carninci P, Celniker SE, Oliver B, Richards S. Comparative validation of themodENCODE transcriptome annotation., 2014, 24(7): 1209–1223.
[35] Sanchez-Flores A, Peñaloza F, Carpinteyro-Ponce J, Nazario-Yepiz NO, Abreu-Goodger C, Machado CA, Markow TA. Genome evolution in three species of cactophilic., 2016, 6(10): 3097–3105.
[36] Khanna R, Mohanty S. Whole genome sequence resource of Indian., 2017, 17(3): 557–564.
[37] Mohanty S, Khanna R. Genome-wide comparative analysis of four Indianspecies., 2017, 292(6): 1197–1208.
[38] Fuller ZL, Haynes GD, Richards S, Schaeffer SW. Genomics of natural populations: Evolutionary forces that establish and maintain gene arrangements in., 2017, 26(23): 6539–6562.
[39] Clark AG, Eisen MB, Smith D, Bergman CM, Oliver B, Markow TA, Kaufman TC, Kellis M, Gelbart WM, Iyer VN, Pollard DA, Sackton TB, Larracuente AM, Singh ND, Abad JP, Abt DN, Adryan B, Aguade M, Akashi H, Anderson WW, Aquadro CF, Ardell DH, Arguello R, Artieri CG, Barbash DA, Barker D, Barsanti P, Batterham P, Batzoglou S, Begun D, Bhutkar A, Blanco E, Bosak SA, Bradley RK, Brand AD, Brent MR, Brooks AN, Brown RH, Butlin RK, Caggese C, Calvi BR, Bernardo de Carvalho A, Caspi A, Castrezana S, Celniker SE, Chang JL, Chapple C, Chatterji S, Chinwalla A, Civetta A, Clifton SW, Comeron JM, Costello JC, Coyne JA, Daub J, David RG, Delcher AL, Delehaunty K, Do CB, Ebling H, Edwards K, Eickbush T, Evans JD, Filipski A, Findeiss S, Freyhult E, Fulton L, Fulton R, Garcia AC, Gardiner A, Garfield DA, Garvin BE, Gibson G, Gilbert D, Gnerre S, Godfrey J, Good R, Gotea V, Gravely B, Greenberg AJ, Griffiths-Jones S, Gross S, Guigo R, Gustafson EA, Haerty W, Hahn MW, Halligan DL, Halpern AL, Halter GM, Han MV, Heger A, Hillier L, Hinrichs AS, Holmes I, Hoskins RA, Hubisz MJ, Hultmark D, Huntley MA, Jaffe DB, Jagadeeshan S, Jeck WR, Johnson J, Jones CD, Jordan WC, Karpen GH, Kataoka E, Keightley PD, Kheradpour P, Kirkness EF, Koerich LB, Kristiansen K, Kudrna D, Kulathinal RJ, Kumar S, Kwok R, Lander E, Langley CH, Lapoint R, Lazzaro BP, Lee SJ, Levesque L, Li RQ, Lin CF, Lin MF, Lindblad-Toh K, Llopart A, Long MY, Low L, Lozovsky E, Lu J, Luo MZ, Machado CA, Makalowski W, Marzo M, Matsuda M, Matzkin L, McAllister B, McBride CS, McKernan B, McKernan K, Mendez-Lago M, Minx P, Mollenhauer MU, Montooth K, Mount SM, Mu X, Myers E, Negre B, Newfeld S, Nielsen R, Noor MA, O'Grady P, Pachter L, Papaceit M, Parisi MJ, Parisi M, Parts L, Pedersen JS, Pesole G, Phillippy AM, Ponting CP, Pop M, Porcelli D, Powell JR, Prohaska S, Pruitt K, Puig M, Quesneville H, Ram KR, Rand D, Rasmussen MD, Reed LK, Reenan R, Reily A, Remington KA, Rieger TT, Ritchie MG, Robin C, Rogers YH, Rohde C, Rozas J, Rubenfield MJ, Ruiz A, Russo S, Salzberg SL, Sanchez-Gracia A, Saranga DJ, Sato H, Schaeffer SW, Schatz MC, Schlenke T, Schwartz R, Segarra C, Singh RS, Sirot L, Sirota M, Sisneros NB, Smith CD, Smith TF, Spieth J, Stage DE, Stark A, Stephan W, Strausberg RL, Strempel S, Sturgill D, Sutton G, Sutton GG, Tao W, Teichmann S, Tobari YN, Tomimura Y, Tsolas JM, Valente VL, Venter E, Venter JC, Vicario S, Vieira FG, Vilella AJ, Villasante A, Walenz B, Wang J, Wasserman M, Watts T, Wilson D, Wilson RK, Wing RA, Wolfner MF, Wong A, Wong GK, Wu CI, Wu G, Yamamoto D, Yang HP, Yang SP, Yorke JA, Yoshida K, Zdobnov E, Zhang PL, Zhang Y, Zimin AV, Baldwin J, Abdouelleil A, Abdulkadir J, Abebe A, Abera B, Abreu J, Acer SC, Aftuck L, Alexander A, An P, Anderson E, Anderson S, Arachi H, Azer M, Bachantsang P, Barry A, Bayul T, Berlin A, Bessette D, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Bourzgui I, Brown A, Cahill P, Channer S, Cheshatsang Y, Chuda L, Citroen M, Collymore A, Cooke P, Costello M, D'Aco K, Daza R, De Haan G, DeGray S, DeMaso C, Dhargay N, Dooley K, Dooley E, Doricent M, Dorje P, Dorjee K, Dupes A, Elong R, Falk J, Farina A, Faro S, Ferguson D, Fisher S, Foley CD, Franke A, Friedrich D, Gadbois L, Gearin G, Gearin CR, Giannoukos G, Goode T, Graham J, Grandbois E, Grewal S, Gyaltsen K, Hafez N, Hagos B, Hall J, Henson C, Hollinger A, Honan T, Huard MD, Hughes L, Hurhula B, Husby ME, Kamat A, Kanga B, Kashin S, Khazanovich D, Kisner P, Lance K, Lara M, Lee W, Lennon N, Letendre F, LeVine R, Lipovsky A, Liu XH, Liu JL, Liu ST, Lokyitsang T, Lokyitsang Y, Lubonja R, Lui A, MacDonald P, Magnisalis V, Maru K, Matthews C, McCusker W, McDonough S, Mehta T, Meldrim J, Meneus L, Mihai O, Mihalev A, Mihova T, Mittelman R, Mlenga V, Montmayeur A, Mulrain L, Navidi A, Naylor J, Negash T, Nguyen T, Nguyen N, Nicol R, Norbu C, Norbu N, Novod N, O'Neill B, Osman S, Markiewicz E, Oyono OL, Patti C, Phunkhang P, Pierre F, Priest M, Raghuraman S, Rege F, Reyes R, Rise C, Rogov P, Ross K, Ryan E, Settipalli S, Shea T, Sherpa N, Shi L, Shih D, Sparrow T, Spaulding J, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Strader C, Tesfaye S, Thomson T, Thoulutsang Y, Thoulutsang D, Topham K, Topping I, Tsamla T, Vassiliev H, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Young G, Yu Q, Zembek L, Zhong DN, Zimmer A, Zwirko Z, Jaffe DB, Alvarez P, Brockman W, Butler J, Chin C, Gnerre S, Grabherr M, Kleber M, Mauceli E, MacCallum I. Evolution of genes and genomes on thephylogeny., 2007, 450(7167): 203–218.
[40] Lack JB, Lange JD, Tang AD, Corbett-Detig RB, Pool JE. A thousand fly genomes: an expandedgenome nexus., 2016, 33(12): 3308–3313.
[41] Stanley CE, Kulathinal RJ. flyDIVaS: A comparative genomics resource fordivergence and selection., 2016, 6(8): 2355– 2363.
[42] Mackay TFC, Huang W. Charting the genotype- phenotype map: lessons from thegenetic reference panel., 2018, 7(1), e289.
[43] Anstead CA, Korhonen PK, Young ND, Hall RS, Jex AR, Murali SC, Hughes DST, Lee SF, Perry T, Stroehlein AJ, Ansell BRE, Breugelmans B, Hofmann AS, Qu JX, Dugan S, Lee SL, Chao H, Dinh H, Han Y, Doddapaneni HV, Worley KC, Muzny DM, Ioannidis P, Waterhouse RM, Zdobnov EM, James PJ, Bagnall NH, Kotze AC, Gibbs RA, Richards S, Batterham P, Gassera RB.genome unlocks parasitic fly biology to underpin future interventions., 2015, 6(1): 7344–7344.
[44] Papanicolaou A, Schetelig MF, Arensburger P, Atkinson PW, Benoit JB, Bourtzis K, Castanera P, Cavanaugh JP, Chao H, Childers CP, Curril I, Dinh H, Doddapaneni H, Dolan A, Dugan S, Friedrich M, Gasperi G, Geib S, Georgakilas G, Gibbs RA, Giers SD, Gomulski LM, González-Guzmán M, Guillem-Amat A, Han Y, Hatzigeorgiou AG, Hernández-Crespo P, Hughes DST, Jones JW, Karagkouni D, Koskinioti P, Lee SL, Malacrida AR, Manni M, Mathiopoulos K, Meccariello A, Murali SC, Murphy TD, Muzny DM, Oberhofer G, Ortego F, Paraskevopoulou MD, Poelchau M, Qu JX, Reczko M, Robertson HM, Rosendale AJ, Rosselot AE, Saccone G, Salvemini M, Savini G, Schreiner P, Scolari F, Siciliano P, Sim SB, Tsiamis G, Ureña E, Vlachos IS, Werren JH, Wimmer EA, Worley KC, Zacharopoulou A, Richards S, Handler AM. The whole genome sequence of the Mediterranean fruit fly,(Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species., 2016, 17(1): 192.
[45] Zhao CY, Escalante LN, Chen H, Benatti TR, Qu JX, Chellapilla S, Waterhouse RM, Wheeler D, Andersson MN, Bao RY, Batterton M, Behura SK, Blankenburg KP, Caragea D, Carolan JC, Coyle M, El-Bouhssini M, Francisco L, Friedrich M, Gill N, Grace T, Grimmelikhuijzen CJP, Han Y, Hauser F, Herndon N, Holder M, Ioannidis P, Jackson L, Javaid M, Jhangiani SN, Johnson AJ, Kalra D, Korchina V, Kovar CL, Lara F, Lee SL, Liu XM, Löfstedt C, Mata R, Mathew T, Muzny DM, Nagar S, Nazareth LV, Okwuonu G, Ongeri F, Perales L, Peterson BF, Pu LL, Robertson HM, Schemerhorn BJ, Scherer SE, Shreve JT, Simmons D, Subramanyam S, Thornton RL, Xue K, Weissenberger GM, Williams CE, Worley KC, Zhu DH, Zhu YM, Harris MO, Shukle RH, Werren JH, Zdobnov EM, Chen MS, Brown SJ, Stuart JJ, Richards S. A massive expansion of effector genes underlies gall-formation in the wheat pest Mayetiola destructor., 2015, 25(5): 613–620.
[46] Kutsenko A, Svensson T, Nystedt B, Lundeberg J, Björk P, Sonnhammer E, Giacomello S, Visa N, Wieslander L. The Chironomus tentans genome sequence and the organization of the Balbiani ring genes., 2014, 15(1): 819–819.
[47] Kaiser TS, Poehn B, Szkiba D, Preussner M, Sedlazeck FJ, Zrim A, Neumann T, Nguyen L, Betancourt AJ, Hummel T, Vogel H, Dorner S, Heyd F, von Haeseler A, Tessmar-Raible K. The genomic basis of circadian and circalunar timing adaptations in a midge., 2016, 540(7631): 69–73.
[48] Andere AA, Platt RN, Ray DA, Picard CJ. Genome sequence ofMeigen (Diptera: Calliphoridae): implications for medical, veterinary and forensic research., 2016, 17(1): 842.
[49] Watanabe J, Hattori M, Berriman M, Lehane MJ, Hall N, Solano P, Aksoy S, Hide W, Toure YT, Attardo GM. Genome sequence of the tsetse fly (): vector of African trypanosomiasis., 2014, 344(6182): 380–386.
[50] Rasmussen DA, Noor MA. What can you do with 01×genome coverage? A case study based on a genome survey of the scuttle fly(Phoridae)., 2009, 10: 382.
[51] Kelley JL, Peyton JT, Fiston-Lavier AS, Teets NM, Yee MC, Johnston JS, Bustamante CD, Lee RE, Denlinger DL. Compact genome of the Antarctic midge is likely an adaptation to an extreme environment., 2014, 5(1): 4611–4611.
[52] Nene V, Wortman JR, Lawson D, Haas BJ, Kodira CD, Tu ZJ, Loftus BJ, Xi ZY, Megy K, Grabherr M, Ren QH, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu JS, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao CH, Mauceli E, Menck CFM, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O'leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JMC, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng QD, Zhao Q, Zhao TM, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of, a major arbovirus vector., 2007, 316(5832): 1718–1723.
[53] Huang W, Massouras A, Inoue Y, Peiffer JA, Ramia M, Tarone AM, Turlapati L, Zichner T, Zhu DH, Lyman RF, Magwire MM, Blankenburg K, Carbone MA, Chang K, Ellis LL, Fernandez S, Han Y, Highnam G, Hjelmen CE, Jack JR, Javaid M, Jayaseelan J, Kalra D, Lee S, Lewis L, Munidasa M, Ongeri F, Patel S, Perales L, Perez A, Pu LL, Rollmann SM, Ruth R, Saada N, Warner C, Williams A, Wu YQ, Yamamoto A, Zhang YQ, Zhu YM, Anholt RR, Korbel JO, Mittelman D, Muzny DM, Gibbs RA, Barbadilla A, Johnston JS, Stone EA, Richards S, Deplancke B, Mackay TFC. Natural variation in genome architecture among 205genetic reference panel lines., 2014, 24(7): 1193–1208.
[54] Huang W, Richards S, Carbone MA, Zhu DH, Anholt RRH, Ayroles JF, Duncan LH, Jordan KW, Lawrence F, Magwire MM, Warner CB, Blankenburg K, Han Y, Javaid M, Jayaseelan J, Jhangiani SN, Muzny D, Ongeri F, Perales L, Wu YQ, Zhang YQ, Zou XY, Stone EA, Gibbs RA, Mackay TFC. Epistasis dominates the genetic architecture ofquantitative traits., 2012, 109(39): 15553–15559.
[55] Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu DH, Casillas S, Han Y, Magwire MM, Cridland JM, Richardson MF, Anholt RR, Barrón M, Bess C, Blankenburg KP, Carbone MA, Castellano D, Chaboub L, Duncan L, Harris Z, Javaid M, Jayaseelan JC, Jhangiani SN, Jordan KW, Lara F, Lawrence F, Lee SL, Librado P, Linheiro RS, Lyman RF, Mackey AJ, Munidasa M, Muzny DM, Nazareth L, Newsham I, Perales L, Pu LL, Qu C, Ràmia M, Reid JG, Rollmann SM, Rozas J, Saada N, Turlapati L, Worley KC, Wu YQ, Yamamoto A, Zhu YM, Bergman CM, Thornton KR, Mittelman D, Gibbs RA. Thegenetic reference panel., 2012, 482(7384): 173– 178.
[56] Obbard DJ, Maclennan J, Kim KW, Rambaut A, O’grady PM, Jiggins FM. Estimating divergence dates and substitution rates in thephylogeny., 2012, 29(11): 3459–3473.
[57] Fontaine MC, Pease JB, Steele A, Waterhouse RM, Neafsey DE, Sharakhov IV, Jiang X, Hall AB, Catteruccia F, Kakani EG. Extensive introgression in a malaria vector species complex revealed by phylogenomics., 2015, 347(6217): 1258524–1258524.
[58] Savard J, Tautz D, Lercher MJ. Genome-wide acceleration of protein evolution in flies (Diptera)., 2006, 6(1): 7–7.
[59] Zdobnov EM, Bork P. Quantification of insect genome divergence., 2007, 23(1): 16–20.
[60] Zdobnov EM, Von Mering C, Letunic I, Torrents D, Suyama M, Copley RR, Christophides GK, Thomasova D, Holt RA, Subramanian GM, Mueller HM, Dimopoulos G, Law JH, Wells MA, Birney E, Charlab R, Halpern AL, Kokoza E, Kraft CL, Lai ZW, Lewis S, Louis C, Barillas-Mury C, Nusskern D, Rubin GM, Salzberg SL, Sutton GG, Topalis P, Wides R, Wincker P, Yandell M, Collins FH, Ribeiro J, Gelbart WM, Kafatos FC, Bork P. Comparative genome and proteome analysis ofand., 2002, 298(5591): 149–159.
[61] Craddock EM, Gall JG, Jonas M. Hawaiiangenomes: size variation and evolutionary expansions., 2016, 144(1): 107–124.
[62] Elliott TA, Gregory TR. What's in a genome? The C-value enigma and the evolution of eukaryotic genome content., 2015, 370(1678): 20140331.
[63] Bargues N, Lerat E. Evolutionary history of LTR-retrotransposons among 20species., 2017, 8(1): 7–7.
[64] Waterhouse RM. A maturing understanding of the composition of the insect gene repertoire., 2015, 7: 15–23.
[65] Feyereisen R. Insect CYP genes and P450 enzymes. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry, Oxford: Elsevier; 2012. p. 237–316.
[66] Arouri R, Le Goff G, Hemden H, Navarro-Llopis V, M’Saad M, Castanera P, Feyereisen R, Hernández- Crespo P, Ortego F. Resistance to lambda-cyhalothrin in Spanish field populations ofand metabolic resistance mediated by P450 in a resistant strain., 2015, 71(9): 1281–1291.
[67] Lemaitre B, Hoffmann J. The host defense of., 2007, 25(1): 697–743.
[68] Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in., 2002, 3(5): 711–722.
[69] Kang DW, Liu G, Lundström A, Gelius E, Steiner H. A peptidoglycan recognition protein in innate immunity conserved from insects to humans., 95(17): 10078–10082.
[70] Kim YS, Ryu JH, Han SJ, Choi KH, Nam KB, Jang IH, Lemaitre B, Breyi PT, Lee WJ. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and β-1,3-glucan that mediates the signaling for the induction of innate immune genes incells., 275(42): 32721–32727.
[71] Lee WJ, Lee JD, Kravchenko VV, Ulevitch RJ, Brey PT. Purification and molecular cloning of an inducible gram-negative bacteria-binding protein from the silkworm,., 1996, 93(15): 7888–7893.
[72] Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. Dynamic evolution of the innate immune system in., 2007, 39(12): 1461–1468.
[73] Gomulski LM, Dimopoulos G, Xi ZY, Soares MB, Bonaldo MF, Malacrida AR, Gasperi G. Gene discovery in an invasive tephritid model pest species, the Mediterranean fruit fly,., 2008, 9(1): 243.
[74] Gabrieli P, Falaguerra A, Siciliano P, Gomulski LM, Scolari F, Zacharopoulou A, Franz G, Malacrida AR, Gasperi G. Sex and the single embryo: early deveopment in the Mediterranean fruit fly,., 10(1): 12.
[75] Zhang Y, Sturgill D, Parisi M, Kumar S, Oliver B. Constraint and turnover in sex-biased gene expression in the genus., 2007, 450(7167): 233– 237.
[76] Gnad F, Parsch J. Sebida: a database for the functional and evolutionary analysis of genes with sex-biased expression., 2006, 22(20): 2577–2579.
[77] Hall AB, Basu S, Jiang XF, Qi YM, Timoshevskiy VA, Biedler JK, Sharakhova MV, Elahi R, Anderson MA, Chen XG, Sharakhov IV, Adelman ZN, Tu ZJ. A male-determining factor in the mosquito., 2015, 348(6240): 1268–1270.
[78] Krzywinska E, Dennison NJ, Lycett G, Krzywinski J. A maleness gene in the malaria mosquito., 2016, 353(6294): 67–69.
[79] Sharma A, Heinze S, Wu Y, Kohlbrenner T, Morilla I, Brunner C, Wimmer E, van de Zande L, Robinson M, Beukeboom L, Bopp D. Male sex in houseflies is determined by, a paralog of the generic splice factor gene., 2017, 356(6338): 642– 645.
[80] Meccariello A, Salvemini M, Primo P, Hall B, Koskinioti P, Dalikova M, Gravina A, Gucciardino MA, Forlenza F, Gregoriou M, Ippolito D, Monti SM, Petrella V, Perrotta MM, Schmeing S, Ruggiero A, Scolari F, Giordano E, Tsoumani KT, Marec F, Windbichler N, Arunkumar KP, Bourtzis K, Mathiopoulos KD, Ragoussis J, Vitagliano L, Tu ZJ, Papathanos PA, Robinson MD, Saccone G.() orchestrates male sex determination in major agricultural fruit fly pests., 2019, 365(6460): 1457–1460.
[81] Von Grotthuss M, Ashburner M, Ranz JM. Fragile regions and not functional constraints predominate in shaping gene organization in the genus., 2010, 20(8): 1084–1096.
[82] Jiang Xf, Peery A, Hall AB, Sharma A, Chen XG, Waterhouse RM, Komissarov A, Riehle MM, Shouche YS, Sharakhova MV, Lawson D, Pakpour N, Arensburger P, Davidson VLM, Eiglmeier K, Emrich S, George P, Kennedy RC, Mane SP, Maslen G, Oringanje C, Qi YM, Settlage R, Tojo M, Tubio JMC, Unger MF, Wang B, Vernick KD, Ribeiro JMC, James AA, Michel K, Riehle MA, Luckhart S, Sharakhov IV, Tu ZJ. Genome analysis of a major urban malaria vector mosquito,., 2014, 15(9): 459.
[83] Zhou Q, Zhang GJ, Zhang Y, Xu SY, Zhao RP, Zhan ZB, Li X, Ding Y, Yang S, Wang W. On the origin of new genes in., 2008, 18(9): 1446– 1455.
[84] Zhao L, Saelao P, Jones CD, Begun DJ. Origin and spread of de Novo genes inpopulations., 2014, 343(6172): 769–772.
[85] Mackay TFC. Epistasis and quantitative traits: using model organisms to study gene-gene interactions., 2014, 15(1): 22–33.
[86] Bhutkar A, Schaeffer SW, Russo S, Xu M, Smith TF, Gelbart WM. Chromosomal rearrangement inferred from comparisons of 12genomes., 2008, 179(3): 1657–1680.
[87] Corbett-Detig RB, Hartl DL. Population genomics of inversion polymorphisms in., 2012, 8(12): e1003056.
[88] Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, Ruby JG, Brennecke J; Harvard FlyBase curators; Berkeley Drosophila Genome Project, Hodges E, Hinrichs AS, Caspi A, Paten B, Park SW, Han MV, Maeder ML, Polansky BJ, Robson BE, Aerts S, van Helden J, Hassan B, Gilbert DG, Eastman DA, Rice M, Weir M, Hahn MW, Park Y, Dewey CN, Pachter L, Kent WJ, Haussler D, Lai EC, Bartel DP, Hannon GJ, Kaufman TC, Eisen MB, Clark AG, Smith D, Celniker SE, Gelbart WM, Kellis M. Discovery of functional elements in 12genomes using evolutionary signatures., 2007, 450(7167): 219–232.
[89] Ladoukakis ED, Pereira V, Magny EG, Eyre-Walker A, Couso JP. Hundreds of putatively functional small open reading frames in., 2011, 12(11): 1–17.
[90] Lewis SH, Salmela H, Obbard DJ. Duplication and diversification of Dipteran argonaute genes, and the evolutionary divergence of Piwi and Aubergine., 2016, 8(3): 507–518.
[91] Mohammed J, Flynt AS, Panzarino A, Mondal MH, Decruz M, Siepel A, Lai EC. Deep experimental profiling of microRNA diversity, deployment, and evolution across thegenus., 2018, 28(1): 52–65.
[92] Mallet J, Besansky N, Hahn MW. How reticulated are species?, 2016, 38(2): 140–149.
[93] Severson DW, Behura SK. Mosquito genomics: Progress and challenges., 2012, 57(1): 143–166.
[94] Oppenheim SJ, Rosenfeld JA, Desalle R. Genome content analysis yields new insights into the relationship between the human malaria parasiteand its anopheline vectors., 2017, 18(1): 205–205.
[95] Singh ND, Larracuente AM, Sackton TB, Clark AG. Comparative genomics on thephylogenetic tree., 2009, 40:459–480.
[96] Markow TA. The secret lives offlies., 2015, 4: e06793.
[97] He QY, Bardet AF, Patton B, Purvis J, Johnston J, Paulson A, Gogol M, Stark A, Zeitlinger J. High conservation of transcription factor binding and evidence for combinatorial regulation across sixspecies., 2011, 43(5): 414–420.
[98] Carl S, Russell S. Comparative genomics of transcription factor binding in. In: Edited by Raman C, Goldsmith MR, Agunbiade TA. Short Views on Insect Genomics and Proteomics.Springer International Publishing, 2015.
[99] Gloss AD, Vassão DG, Hailey AL, Dittrich ACN, Schramm K, Reichelt M, Rast TJ, Weichsel A, Cravens MG, Gershenzon J, Montfort WM, Whiteman NK. Evolution in an ancient detoxification pathway is coupled with a transition to herbivory in the Drosophilidae., 2014, 31(9): 2441–2456.
[100] Goldman-Huertas B, Mitchell RF, Lapoint RT, Faucher CP, Hildebrand JG, Whiteman NK. Evolution of herbivory inlinked to loss of behaviors, antennal responses, odorant receptors, and ancestral diet., 2015, 112(10): 3026–3031.
[101] Hickner PV, Rivaldi CL, Johnson CM, Siddappaji M, Raster GJ, Syed Z. The making of a pest: Insights from the evolution of chemosensory receptor families in a pestiferous and invasive fly,., 2016, 17(1): 648–648.
[102] Fontaine MC. Genetic diversity of the African malaria vector., 2017, 552(7683): 96–100.
[103] Wiegmann BM, Richards S. Genomes of Diptera., 2018, 25: 116–124.
[104] Simon CR, Siviero F, Monesi N. Beyond DNA puffs: What can we learn from studying sciarids?, 2016, 54(7): 361–378.
[105] Hou L, Zhan S, Zhou X, Li F, Wand XH. Advances in research on insect genomics in China., 2017, 54(5): 693–704.侯丽, 詹帅, 周欣, 李飞, 王宪辉. 中国昆虫基因组学的研究进展. 应用昆虫学报, 2017, 54(5): 693–704.
[106] Zhang CX. Current research status and prospects of genomes of insects important to agriculture in China., 2015, 48(17): 3454–3462.张传溪. 中国农业昆虫基因组学研究概况与展望. 中国农业科学, 2015, 48(17): 3454–3462.
[107] Ye GY, Fang Q. Entomological research in the genomic age., 2011, 48(6): 1531–1538.叶恭银, 方琦. 基因组时代的昆虫学研究. 应用昆虫学报, 2011, 48(6): 1531–1538.
[108] Zhou H, Shen J. Research on functional genomics of agriculture insects: Review and prospect., 2017, 39(2): 239–248.周行, 沈杰. 农业昆虫的功能基因组学研究: 回顾与展望. 环境昆虫学报, 2017, 39(2): 239–248.
[109] Yin CL, Li MZ, He K, Ding SM, Guo DH, Xi Y, Li F. The progress of insecg genomic research and the gene database., 2017, 39(1): 1–18.尹传林, 李美珍, 贺康, 丁思敏, 郭殿豪, 席羽, 李飞. 昆虫基因组及数据库研究进展. 环境昆虫学报, 2017, 39(1): 1–18.
Progress on genome sequencing of Dipteran insects
Wei Peng, Mengjie Feng, Hao Chen, Baoyu Han
Diptera is among the most diverse holometabolan insect orders and was the earliest order to have a genome fully sequenced. The genomes of 110 fly species have been sequenced and published and many hundreds of population- level genomes have been obtained in the model organismsand. Comparative genomics elucidate many aspects of the Dipteran biology, thereby providing insights for on the variability in genome structure, genetic mechanisms, and rates and patterns of evolution in genes, species, and populations. Despite the availability of genomic resources in Diptera, there is still a significant lack of information on many other insects. The sequencing of the genomes in Dipteran insects would be of great value to exhibit multiple origins of key fly behaviors such as blood feeding, parasitism, pollination, and mycophagy. In this review, we briefly summarize the distribution and characteristics of Dipteran genomes, introduce the progress of functional genes such as Cytochrome P450, immunity, sex determination and differentiation related genes in Dipteran genome, and highlight the significant findings generated by comparative genomics approach among Dipteran species. This paper provides the guidelines and references for choosing additional taxa for genome sequencing studies in the rapidly developing genome omics era, and offers a fundamental basis for genome-based pest control and management.
Diptera; genome characteristics; functional genes; comparative genomics; phyletic evolution
2020-05-06;
2020-09-06
联合国粮农组织和国际原子能署项目(编号:D44003),国家重点研发计划项目(编号:2018YFC1604402),浙江省重点研发计划项目(编号:2020C02026)和浙江省基础公益研究计划项目(编号:LGN18C160006,LGN20C140005)资助[Supported by the International Atomic Energy Agency’s Coordinated Research Project (No. D44003), the National Key Research and Development Program of China (No. 2018YFC1604402), the Key Research and Development Program of Zhejiang Province, China (No. 2020C02026), and the Fundamental and Public Welfare of Zhejiang Province of China (Nos. LGN18C160006, LGN20C140005)]
彭威,博士,讲师,研究方向:昆虫生物化学与分子生物学。E-mail: pengwei@cjlu.edu.cn
彭威。
韩宝瑜,博士,教授,研究方向:昆虫化学生态学。E-mail: hanby15@163.com
10.16288/j.yczz.20-130
https://kns.cnki.net/kcms/detail/11.1913.R.20201021.1052.002.html
URI: 2020/10/22 11:48:10
(责任编委: 张蔚)

