Morphology and Phylogeny of Taeniacanthus yamagutii Shiino, 1957 (Hexanauplia: Taeniacanthidae), a Copepod Infecting the Gills of Rosy Goatfish Parupeneus rubescens (Mullidae) in the Arabian Gulf
ABDEL-GABER Rewaida, AL-QURAISHY Saleh, DKHIL Mohamed A., 3),ALGHAMDI Masheil, ALGHAMDI Jawahir, and KADRY Mohamed
Morphology and Phylogeny ofShiino, 1957 (Hexanauplia: Taeniacanthidae), a Copepod Infecting the Gills of Rosy Goatfish(Mullidae) in the Arabian Gulf
ABDEL-GABER Rewaida1), 2), *, AL-QURAISHY Saleh1), DKHIL Mohamed A.1), 3),ALGHAMDI Masheil1), ALGHAMDI Jawahir1), and KADRY Mohamed2)
1),,,11451,2),,,12613,3),,,11794,
The present study was to investigate the copepodid species infecting rosy goatfish,one of the most economically important fishes in the Arabian Gulf.A copepodid species identified from the examined fish specimens belongs to the Taeniacanthidae family and is labeled asShiino, 1957, mainly depending on its morphological, morphometric, and ultrastructural characteristics, in particular the presence of maxilliped claw with a conspicuous digitiform process at the base, the terminal process of the second maxilla stout, and a setiform element at the tip of each exopod spine of legs 2–4. In order to ensure the accurate identification and exact taxonomic characterization of this species, thegene sequence was analyzed.The result revealed that the presentcopepodid species belong to the Taenicanthidae family and has a close relationship with(gb| KR048852.1) in the same taxon.The present study demonstrated that the rosy goatfishis a host forspecies, which will be helpful to prevent this parasitic infection.
parasitic copepods; Taeniacanthidae;spp.; Arabian Gulf
1 Introduction
Taeniacanthidae is a unique family within the copepod order Cyclopoida, comprising members either parasitic to marine fish or associated with sea urchins (Dojiri andHu- mes, 1982; Boxshall and Halsey, 2004). Taeniacanthids ex- hibit a high degree of host specificity at both the generic and specific levels (Boxshall and Halsey, 2004). This fa- mily, along with Bomolochidae Sumpf, 1871, Tuccidae Ver- voort, 1962, and Tegobomolochidae Avdeev, 1978, are mem- bers of the bomolochiform complex (Dojiri and Cressey, 1987; Boxshall and Halsey, 2004). They are characterized by the presence of an indistinctly four-segmented antenna; bearing two pectinate processes, claw-like spines, and se- tae; a mandible with two subequal spinulated blades; a maxilla bearing spinulated elements; a concave ventral sur- face of the cephalothorax; and a lamelliform leg 1. There are 25 genera with more than 91 species in Taeniacanthi- dae (Tang and Izawa, 2005; Tang and Johnston, 2005; Ho and Lin, 2006; Walter and Boxshall, 2019). These genera includeKrøyer, 1837;;Sumpf, 1871;Brian, 1906;Wilson, 1911;Wilson, 1911;Wilson, 1924;Wilson, 1935;Yamaguti, 1939 (Accepted asSumpf, 1871);Yamaguti and Yamasu, 1959;Hu- mes and Cressey, 1961;Humes and Cressey, 1961;Pillai, 1963;Ho, 1969;Cressey, 1969;Humes, 1970;Dojiri and Cressey, 1987;Do- jiri and Cressey, 1987;Tang and Izawa, 2005;Tang and Johnston, 2005;Ho, Ohtsuka, and Nakadachi, 2006;Uyeno, Tang, and Nagasawa, 2013;Kim and Moon, 2013 (Ac- cepted asVenmathi Maran, Moon, Ad- day, and Tang, 2016);Venmathi Maran, Moon, Adday, and Tang, 2016;Venma- thi Maran, Moon, Adday, and Tang, 2016.
Research on ectoparasitic copepods has so far been very limited. As the number of copepodologists is relatively low (Ho, 2001), the morphological and anatomical fea- tures are not enough to recognize and classify copepods (Hamza., 2007; Ramdane, 2009). Several molecular approaches have recently been used to check the taxo- nomic status of copepods independently (Fls., 2006; Ferrari and von Vaupel Klein, 2019). In copepod taxon- omy, DNA barcoding was used to identify intra- and inter- specific morphological and molecular distinctions to help their phylogenetic relationship (Yazawa., 2008). How- ever, very few attempts have been made to use molecular data to restore interordinal associations within Copepoda, as a result of the insufficient taxon sampling or sequenc- ing uncertainty (Kim and Kim, 2000; Ferrari., 2010). Huys. (2007) found that nuclear ribosomal genesandinclude semi-conserved domains that intersperse with divergent regions, allowing for a wide range of taxonomic levels of phylogenetic reconstruction.
There is a need for more extensive work to gain a better understanding of the parasitic copepods infecting the Ara- bian Gulf fish in general and those off Saudi Arabia in particular. The purpose of this research is to provide com- plete data on parasitic copepods and their indices in the rosy goatfishfrom the Arabian Gulfin Saudi Arabia. In addition, partial sequences ofgene have been developed and used to evaluate the phy- logenetic status of this species within Taeniacanthidae.
2 Materials and Methods
2.1 Fish Collection
A total of 80 specimens of the rosy goatfish,(F: Mullidae), were collected from land- ing sites on the coasts of the Arabian Gulf off Dammam City in Saudi Arabia. They were immediately transported to the Laboratory of Parasitology Research, Zoology De- partment, College of Science, King Saud University, Ri- yadh, Saudi Arabia. All procedures comply with the ethi- cal standards approved by the Institutional Review Board (IRB) of King Saud University, Riyadh, Saudi Arabia. Sam- ples of fish were classified according to the standards of the database of fishbase.org. They were thoroughly check- ed for the presence of parasitic crustacean infections on their body surface, fins, head, gill filaments, oral cavities and other tissues, based on the method described previ- ously by Ravichandran. (2007).
2.2 Parasitological Studies
2.2.1 Light microscopic examination
Parasitic copepods were removed using a special nee- dle. They were washed several times in a saline solution, preserved in 70% ethyl alcohol, dehydrated in a 2-h glyc- erin ethanol series, and mounted as temporary preparations in lactophenol as described by Pritchard and Kruse (1982). Prepared samples were analyzed and photographed using the Leica DM 2500 microscope (NIS ELEMENTS soft- ware, ver. 3.8).
2.2.2 Scanning electron microscopic examination
Buffered glutaraldehyde (3%, pH 7.2) was used to fix recovered copepod parasites. After 4h, fixed parasites were dehydrated in the ascending ethanol series (50%, 60%, 70%, 80%, 90%, 100%) for 10min, respectively. The sam- ples were then processed at the critical point drier ‘LEICA,EM CPD300’ with Freon 13, sputter-coated with gold-pal- ladium in Auto fine coater (JEOL, JEC-3000FC), and fi- nally examined and photographed under the Etec Auto- scan at 10-kV Jeol scanning electron microscope(JSM- 6060LV) at the Molecular Biological Unit in Prince Naif Health Research Center, King Saud University, Riyadh, Saudi Arabia.
The different body parts of the recovered copepods were measured using the Olympus ocular micrometer and the average values with the range given in parentheses were employed for analysis. Sewell’s style (1949) was adopted for the armature formula of the swimming legs, in which the spines and setae are denoted by Roman and Arabic numerals, respectively.
2.3 Molecular Analysis
2.3.1 DNA extraction and polymerase chain reaction (PCR)
Genomic DNA was extracted from ethanol-contained samples using the DNeasy tissue kit© (Qiagen, Hilden, Germany) following the manufacturer’s instructions. DNA quality and purity were quantified with the NanoDrop ND-1000 spectophotometer (Thermo Fischer Scientific, Inc., Wilmington, DE, USA) and 20ng of genomic DNA was used for PCR amplification. Thegene re- gion was amplified by PCR and subsequently sequenced. The PCR for this region was produced in a total of 20µL of reaction volume containing 4μL 5× FIREPol® Master Mix (Solis BioDyne), 2μL of genomic DNA, 0.6μL of each primer, and completed to the required volume of 20µL by nuclease-free water. PCR amplification was per- formed using the following primers: 28SF, 5’-ACA ACT GTG ATG CCC TTA G-3’ and 28SR, 5’-TGG TCC GTG TTT CAA GAC G-3’ previously designed by Song. (2008). The amplification technique of the PCR thermo- cycler profile and the primer combination used to amplify this genetic marker was done according to Song. (2008). All PCR products were verified on 1% agarose gel in 1× Tris-acetate-EDTA (TAE) fixed with 1% ethidium bromide and then visualized with a UV trans-illuminator. The PCR products of the expected size were gel-excised, purified and cloned using the manufacturer’s instructions for the PureLinkTMQuick Gel Extraction Kit (Qiagen, Hilden, Germany).
2.3.2 Sequence alignment and phylogenetic analysis
Sequencing was performed on 3130×l Genetic Analy- zer (Biosystems® 3130, Thermo Fisher Scientific, USA) using the Dye Terminator Cycle Sequencing Ready Reac- tion Kit (Perkin Elmer). A BLAST search was carried out in the NCBI database to identify related sequences. The resulting sequences were aligned directly with CLUSTAL- X Multiple Sequence Alignment using other gene region sequences available from GenBankTM. The alignment was updated manually using BIOEDIT 4.8.9 software. Phy- logenetic analyses were conducted using maximum par- simony (MP) and Bayesian inference (BI). MP analysis was performed with MEGA version 7.0 (Kumar., 2016) using a maximum likelihood method focused on the Tamura-Nei model (Tamura and Nei, 1993). BI analy- sis was carried out with MrBayes ver. 2.01 (Huelsenbeck and Ronquist, 2001) using GTR+I+Γ nucleotide substi- tution model. The tree was drawn to scale, and the branch lengths in the same units were applied as the evolution distances to deduce the phylogenetic tree.
3 Results
Twenty five out of eighty (31.25%) specimens of the ex- amined rosy goatfishwere found to be naturally infected by a copepodid parasite known asShiino, 1957 in the gill region of infected fish.
3.1 Description of Female Specimens (Figs.1 (A–L), 2 (A–H))
Total body length (excluding setae on caudal rami) 2.20µm (2.07–2.43) and width 0.99 (0.87–1.03). Prosome com- posed of broad cephalothorax (1st pedigerous somite fusedwith cephalosome) and 3 narrower pedigerous somites. Ce- phalothorax bears a highly sclerotized transverse bar and a marginal hyaline membrane. Urosome includes the 5th pedigerous somite, genital double-somite and free abdo- minal somites. Cephalothorax wider than length, approxi- mately 0.410×1.03, comprising less than 20% of the over- all body length. Thoracic segments bearing legs 2, 3, and 4 relatively large and decreasing in width posteriorly. Ge- nital somite wider than long, 0.210×0.295, armed with spinules. Abdomen 4-segmented; from anterior to poste- rior 0.157×0.298, 0.138×0.275, 0.072×0.223,and 0.199×0.203; anal somite bearing patches of spinules. Caudal ramus 0.159×0.061 and bearing 6 setae.
Rostral area with a sclerotized ventromedian part. An- tennule (antenna 1) 6-segmented; armature formula: 5, 15, 8, 4, 2+1 aesthetasc, and 7+1 aesthetasc. Antenna (anten- na 2) 4-segmented first segment with 1 seta; second seg- ment with 1 short, broad, leaf-like seta; third segment with 2 processes and 1 claw; fourth segment short armed with 4 claws and 2 setae. Postantennal process relatively short tine. Labrum with posterior spinulose margin. Mandible with 2 unequal spinulate blades. Maxillule (maxilla 1) lo- bate and apically with 5 setae. Maxilla (maxilla 2) 2-seg- mented; basal segment naked; elongated distal segment with 2 small setae in the middle and distal claw-like apex. Maxilliped 3-segmented; first segment very broad with 1 naked seta; second segment very slender, with 1 pair of setae proximally; third segment with terminal claw, dis- tally with 2 rows of spinules and proximally with 2 setae and 1 short process.
Legs 1–4 biramous. Leg 1 with 2-segmented exopod and endopod. Legs 2–4 with 3-segmented exopod and endo- pod. Spines on exopods of legs 2–4 rod-shaped, with blunt tip. Armature of legs 1–4 as follows:
Leg 1: coxa 0–1; basis 1–1; exopod 1–0; 9; endopod 0– 1; 7
Leg 2: coxa 0–0; basis 1–0; exopod I–0; I–1; II,I,4; en- dopod 0–1; 0–1; III,3
Leg 3: coxa 0–0; basis 1–0; exopod 1–0; 1–1; II,I,5; en- dopod 0–1; 0–1; III,2
Leg 4: coxa 0–0; basis 1–0; exopod I–0; I–1; II,I,4; en- dopod 0–1; 0–1; I,I,1
Leg 5 uniramous, 2-segmented; basal segment with 1 dorsolateral seta and spinules on distal border; second seg- ment 0.175×0.082; inner margin straight, with 4 patches of spinules; distal margin with 3 spines and 1 seta. Leg 6 represented by 3 setae in the area of egg sac attachment.
3.2 Description of Male Specimens (Figs.3 (A–I),4 (A–G))
Total body length (excluding setae on caudal rami) 1.71 µm (1.65–1.79) and width 0.61 (0.54–0.72). Cephalotho- rax 0.321×0.634andcomprising about 20% of the total body length. Genital somite spinulated and slightly wider than long 0.212×0.251. Abdomen 3-segmented; segmentsfrom anterior to posterior 0.121×0.185,0.118×0.171, and 0.175×0.151; anal somite armed with spinules as in female. Caudal ramus as female but smaller in size, 0.123×0.049.
Appendages as in females, except for the features men- tioned below. Terminal segment of the second maxilla is more slender than that of the female. Maxilliped 4-seg- mented; first segment large, with 1 naked distal seta; se- cond segment with row of spinules along the inner mar- gin and a semicircular group of spinules; third segment small; fourth segment with a curved claw and 2 anterior and 1 posterior setae. Leg 5 is similar to that in females ex- cept for the second segment 0.108×0.045.
3.3 Molecular Analysis
A total of 477bp with 54.08% GC content was evalu- ated and deposited in GenBank under the accession num- ber MN413507.1 for thegene region of the existing copepoda species. Phylogenetic analyses were per- formed by aligning the partial and complete sequences ofgene with 22 taxa of three copepodid orders (Cyclopoida, Calanoida, and Harpacticoida) (Table 1, Figs.5,6). The results showed thatof this species re- vealed sequence identities 95.35%–80.13% with Cyclo- poida, 91.47%–91.05% with Harpacticoida, and 91.02%–90.62% with Calanoida (Table 1). Among Cyclopoida, the present speciesis 95.35%–94.91% identified with Taeni- canthidae family taxa, 94.04%–92.73% with Bomolochi- dae, 90.43% with Cyclopettidae, 89.46%–80.13% with Cy- clopidae, and 82.56%with Clausidiidae. Among Cyclopoida, the maximum identity (95.45%) with the lowest divergent value was recorded between the present copepodid species and(gb| KR048852.1), followed by(94.91%, gb| KR048834.1),(94.04%, gb| KR048838.1),(92.73%, gb| KR048820.1),(90.43%, gb| KR048803.1),(90.29%, gb| KR048819.1),s(89.36%, gb| MK370246.1),sp(89.32%, gb| MK370278.1),(82.56%,gb| KR048817.1),(82.16%, gb| KR 048812.1),(81.07%, gb| KR048802.1),(81.07%, gb| KR048801.1),(80.76%, gb| KF153696.1),(80.74%, gb| KF153697.1), and(80.13%, gb| KR048797.1) (Table 1). The phylogenetic trees showed that the cluster containing all of the copepodid species was clearly divided into two separate clades (Figs.5,6). The first clade contains some copepodid species belonging to the Taenicanthidae and Bo- molochidae families of the order Cyclopoida, and the other species belonging to the order of Calanoida.The second clade consisted of other copepodid species belonging to the Cyclopoidae and Cyclopettidae families of the order Cyclopoida, and other species belong to the order of Har- pacticoida. Dendrograms revealed the species studied in this research is closely related to(gb| KR048852.1) in the same taxon.

Fig.1 Photomicrographs of the adult female Taeniacanthus yamagutii infecting Parupeneus rubescens. (A, B) Whole mount preparation of: (A) Ventral view; (B) Lateral view; (C–L) High magnifications for different body parts showing: (C) Rostral area; (D) Anterior region of cephalothorax; (E) Postoral appendages; (F) Thoracic zone with legs; (G) First thoracic leg; (H) Thoracic legs from second to fifth leg; (I) Fourth and fifth thoracic legs with appearance of abdomen and egg sac; (J–L) Urosome with abdomen ended with caudal rami and setae that surrounded with egg sac. AB, abdomen; ANT 1, 1st antenna; ANT 2, 2nd antenna; AS, anal somite; AE, aesthete; CR, caudal rami; ESC, egg sac; GS, genital somite; L1, 1st leg; L2, 2nd leg; L3, 3rd leg; L4, 4th leg; L5, 5th leg; L6, 6th leg; LB, labrum; MAN, mandible; MAP, maxilliped; MAX1, maxilla 1; MAX2, maxilla 2; PR, prosome; RA, rostral area; S, setae; SP, spinule; UR, urosome.
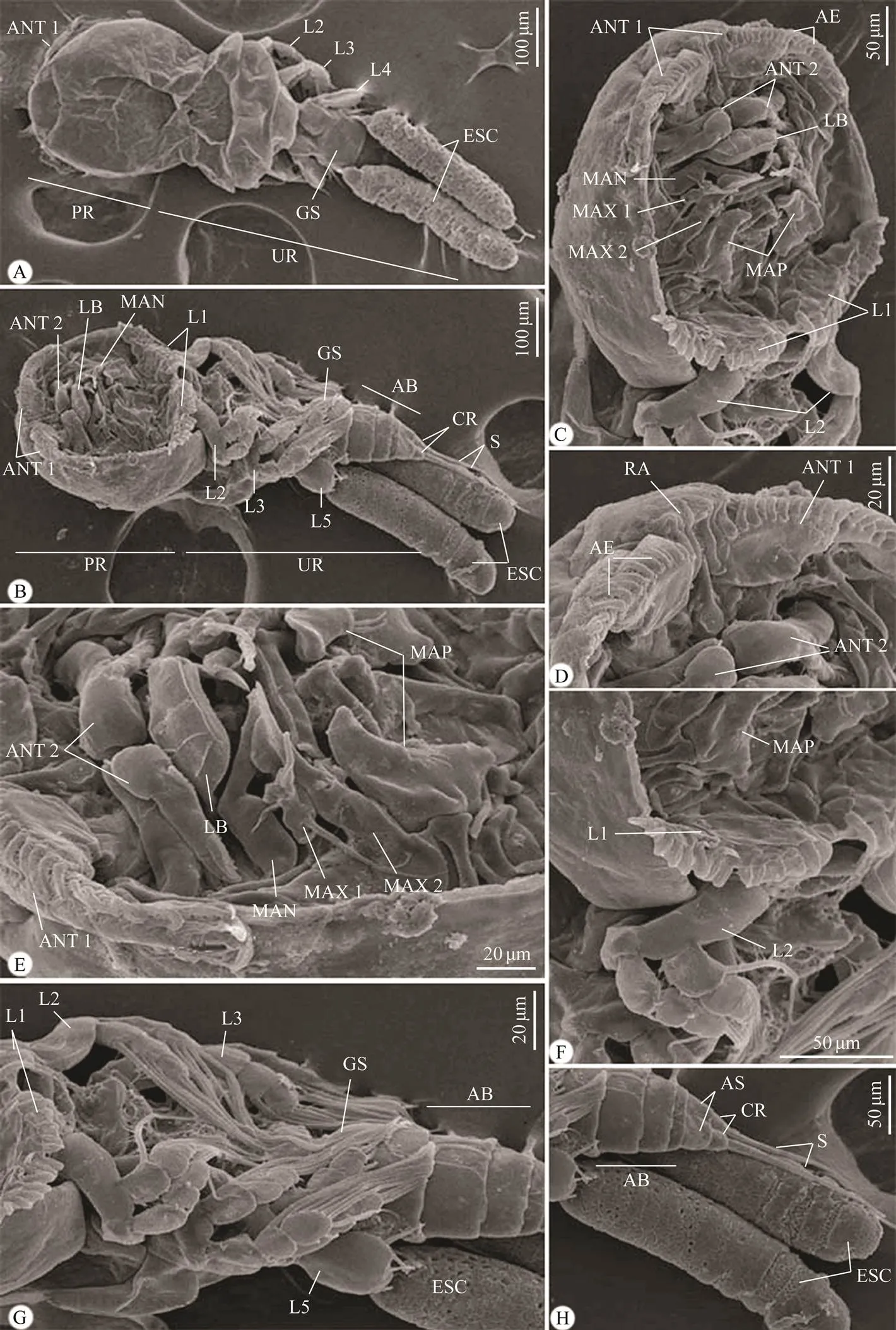
Fig.2 Scanning electron micrographs of the adult female Taeniacanthus yamagutii infecting Parupeneus rubescens. (A, B) Whole mount preparation of: (A) Dorsal view; (B) Ventral view; (C–H) High magnifications for different body parts of: (C–E) Anterior region of cephalothorax with postoral appendages; (F, G) Thoracic legs; (H) Abdomen and egg sac. AB, abdomen; ANT 1, 1st antenna; ANT 2, 2nd antenna; AS, anal somite; AE, aesthete; CR, caudal rami; ESC, egg sac; GS, genital somite; L1, 1st leg; L2, 2nd leg; L3, 3rd leg; L4, 4th leg; L5, 5th leg; L6, 6th leg; LB, labrum; MAN, mandible; MAP, maxilliped; MAX1, maxilla 1; MAX2, maxilla 2; PR, prosome; RA, rostral area; S, setae; Sp, spinule; UR, urosome.
4 Discussion
Taeniacanthidae Wilson, 1911 is a family that flourish as fish parasites.Sumpf, 1871 is the largest genus of the Taeniacanthidae family, comprising 62 spe- cies of poecilostome copepods parasitic on both cartila- ginous and bony fish (Walter and Boxshall, 2019). The present copepodid species is consistent with otherspecies with special respect to the previously de- scribed, which are closely link-ed to each other with the distinctive generic features and identical body proportions. The current study ofis the first report of this parasitic spe- cies as a rosy goatfish ectoparasite.
was originally described as ‘’ (Bassett-Smith, 1898). Shiino (1957) pro- posed that the specimens of Yamaguti were not conspe- cific to the species described by Bassett-Smith in 1898, and thus established a new species,. This spe- cies was later transferred to the genusby Yamaguti and Yamasu (1959).is the most closely related species to bothYamaguti and Ya- masu, 1959, andYamaguti and Yamasu, 1959.The most distinctive characters ofare the ter-minal process of the second maxilla stout, maxilliped claw with a conspicuous digitiform process at the base, a seti- form element at the tip of each exopod spine of the legs 2–4.

Fig.3 Photomicrographs of the adult male Taeniacanthus yamagutii infecting Parupeneus rubescens. (A, B) Whole mount preparation of: (A) Ventral view; (B) Lateral view; (C) Rostral area with antenna; (D) Postoral appendages; (E–H) Thoracic legs; (F) Abdomen with caudal rami. AB, abdomen; ANT 1, 1st antenna; ANT 2, 2nd antenna; AS, anal somite; AE, aesthete; CR, caudal rami; GS, genital somite; L1, 1st leg; L2, 2nd leg; L3, 3rd leg; L4, 4th leg; L5, 5th leg; LB, labrum; MAN, mandible; MAP, maxilliped; MAX1, maxilla 1; MAX2, maxilla 2; PR, prosome; RA, rostral area; S, setae; SP, spi- nule; UR, urosome.

Fig.4 Photomicrographs of the adult adult male Taeniacanthus yamagutii infecting Parupeneus rubescens. (A, B) Whole mount preparation of: (A) Ventral view; (B) Dorsal view; (C–G) High magnifications for different body parts showing: (C, D) Anterior region of cephalothorax with postoral appendages; (E–G) Thoracic legs. AB, abdomen; ANT 1, 1st antenna; ANT 2, 2nd antenna; AS, anal somite; CR, caudal rami; GS, genital somite; L1, 1st leg; L2, 2nd leg; L3, 3rd leg; L4, 4th leg; L5, 5th leg; LB, labrum; MAN, mandible; MAP, maxilliped; PR, prosome; RA, rostral area; S, setae; SP, spinule; UR, urosome.

Table 1 Copepod species used in phylogenetic analyses of Taeniacanthus yamagutii specimens obtained in this study
()
()

Parasite speciesOrder/familyAccession no.Percent identity (%)GC content Halicyclops sp.Cyclopoida/CyclopidaeMK370278.189.32%63.2% Hemicyclops ctenidisCyclopoida/ClausidiidaeKR048817.182.56%56.8% Acanthocyclops vernalisCyclopoida/CyclopidaeKR048812.182.16%60.5% Tropocyclops ishidaiCyclopoida/CyclopidaeKR048802.181.07%58.7% Thermocyclops taihokuensisCyclopoida/CyclopidaeKR048801.181.07%58.7% Macrocyclops albidusCyclopoida/CyclopidaeKF153696.180.76%59.6% Diacyclops bicuspidatusCyclopoida/CyclopidaeKF153697.180.74%59.7% Mesocyclops pehpeiensisCyclopoida/CyclopidaeKR048797.180.13%60.6%

Fig.5 Maximum parsimony analysis based on the 28S rRNA gene sequence demonstrating the position of the present co- pepodid species.

Fig.6 Phylogeneticposition ofthe recovered copepod speciesbased on Bayesian analysis of the 28S rRNA gene sequence.
In addition, it has some similarities to otherspecies, such asRichiardi, 1870,Lewis, 1967,Tang., 2011, andTang., 2011. The similarities include the same antennule armature; the presence of spinules row on the ventromedian surface and along the posterior margin of the labrum; the lobate maxillule carrying semi-pinnate seta, anterior knob-like process and 2 long and 2 short naked setae; the third exopodal segment of leg 3 bearing 5 setae; and a terminal rod-shaped spines on each exopo- dal spine of legs 2–4. It is quite similar toTang., 2011, andTang, 2011 as they all have a spinulate terminal process on the basis of maxilla, and their endopod of maxilliped are elongated, claw-like and strongly curved. Additionally, they all bear a large thorn- like process on the proximal anterior surface, a minute basal seta and a small spiniform process on the posterior surface. It is also similar toTang. 2011 as they both have the same antennule armature, endopodal segment of maxilliped with a terminal curved claw.
There are some similarities toYama- guti, 1939,Yamaguti and Yamasu, 1959, andSebastian, 1964. They all have a lateral border of cephalothorax provided with a thin flexible membrane originating from the ventral side.The maxillule has three setae, the outer seta is stout and the middle one is rela- tively small. It is also quite similar toby the presence of a mandible tipped with 2 unequal blades bearing marginal serrations along the posterior margin; the same maxillule structure; the third segment with max- illiped with a curved claw carrying a tiny seta. It is simi- lar withDevi & Shyamasundari, 1980, andPearse, 1952, as they all have slightly pro- truded anteriorly rostral area with one sclerite in the ven- tro-median area of the flask-shaped, and 2 unequal blades with marginal serrations on the mandible. Additionally, it is similar to, andClaus, 1864, andby having a curved claw on the distal seg- ment of maxilliped.
It has some similarities toYamaguti, 1939,,Avdeev, 1977,Dojiri and Cressey, 1987, having a rostrum with cir- cular ventral sclerotization; the same structure of the an- tennule and maxillule; the terminal segment of the maxil- liped with 2 small setae proximally and strongly curved distal claw. It is also similar toRangnekar and Murti, 1960, as they both have the terminal exopod segment of leg 4 with formula II,I,4.
There are, however, some differences with otherspecies, such asClaus, 1864 hav- ing an antennule armature of 5,11,7,4,2+1 aesthetasc, 7+1 aesthetasc; sharp curved tine of post-antennal process; and the presence of sickle-shaped claw of maxilliped;Yamaguti, 1954 having 5-segmented antennule with armature of 12,6,3,1+2,7 aesthetasc; 2-segmented antenna with proximal segment bears 3 simple and 2 pectinate claws; third exopodal segment of 1st leg bears 6 plumose setae;Yamaguti, 1954 getting the armature of a 5-segmented antennule 13+3+2, 6+3, 1+3, 3+1+2, 7 setae; and exopod of 1st leg with 8 plumose setae.
It also distinguished from,,, with a maxillary hook has a stout base and a curved claw; the third segment of the antenna has two spine-setae and four strong claws. With regard to,,, there are some struc- tural changes as the presence of two sclerotized structures on each side of T-shaped plate of rostral area; and caudal rami bears 7 setae. Lewis (1967) noted some differences withhaving a cephalothorax with a transverse dor- sal striae; and a distomedial corner of a maxilliped basis with a bilobed protrusion;having a flagellum at the distal end of each exopodal spine of legs 2–4; andYamaguti and Yamasu, 1959 having multiple rows of spinules on large pectinate process of antenna; and a subterminal flagellum on each spine of free exopo- dal segment of leg 5.
In addition, it differs from, andhaving a highly protuberant rostral area that lacks a scle- rotized structure on the ventral surface; the second endo- podal segment of antenna bears 2 unequal pectinate pro- cesses and a claw-like spine. It is different from, having 14 setae on the second antennulary segment. It’s different fromIzawa, 1986, having the setal formula of the antennule as 15,5,3,4,2+1 aesthete, 7+1 aesthete; and maxilliped with a long claw on 5th segment that bears 2 proximal setae and ridge-like teeth.
In the case of a comparison to, there are some differences, such as the rostral area broadly pro- truded anteriorly but without sclerite on its ventral surface; and the distal segment of maxilla tipped with 2 pinnate spines in addition to a tiny subterminal seta. In addition, it differs fromhaving post-antennal process witha curved, sharply pointed process; and labrum fringed with spinules. It differed from, andhaving an armature formula of antennule as 5,15,4,3,4,2+1 aesthete, and 7+1 aesthete;having inclusion of 5 setae on the third exopodal segment of legs 2 and 4, and the presence of 8 setae on the distal segment of the endopod of leg 1.
In addition,Wilson, 1924 has a diffe- rence of an apical spine that is at least twice as long as proximal outer spine on terminal exopodal segment of leg 2. It is different fromDojiri and Cressey, 1987, andTang., 2016 having caudal ramus bears seven setae; and terminal segment of maxilla with spinulated terminal process;Tang., 2016 hav- ing a maxillipedal claw with bristles on a large proximal seta;Kim and Moon, 2013 having maxilli- ped with short terminal claw;Shiino, 1957 having leg 5 exopod about 3 times as long as wide; andYamaguti, 1939 having a ventral surface of anal somite with 2 pairs of spinule rows. Dojiri and Cressey (1987) described a discrepancy between our para- sitic species and,,,,and,of rounded process tipped with a nipple-like knob on the first maxilla.
Accurate and reliable identification of parasite species is necessary for biodiversity assessment (Cepeda., 2012). DNA sequence analysis of target genes offers in- valuable information for such analyses. This research in- vestigated the variation of a portion of thegene as a marker for the identification and classification of eco- logically significant cyclopoid copepod genusin the Arabian Gulf fish. In thefragment, D1–D2 region was chosen to analyze, which was suggest- ed by Sonnenberg. (2007) as a taxonomic marker due to its heterogeneity. Previous studies have used this mar- ker for the analysis of copepods and other taxa (Huys., 2006, 2012; Brown., 2010; Blanco-Bercial., 2011; Hayward., 2011; Yeom., 2018).
Previously, Huys and Boxshall (1991) summarized ten taxonomic orders of copepods, nine of which had marine representatives. Huys. (2002) and Ho. (2003) reported that Copepoda are classified into three infraclasses:Progymnoplea Lang, 1948 (or Platycopioida Fosshagen and Iliffe, 1985); Gymnoplea Giesbrecht, 1892 (or Calanoi- da Sars, 1903); and Podoplea Giesbrecht, 1892. The pre- sent phylogenetic trees include two clades representing the last two infraclasses. Ho (1990), Ho. (2003) and Blanco-Bercial. (2011) stated that Calanoida is the most basal taxon of Neocopepoda, and the sister-group of Podoplea. This conclusion is in agreement with the pre- sent results. Dahms (2004), Jenner (2009), and Schizas. (2015) reported that Podoplea had been divided into two main clades. The first clade is named ‘MHPSM-clade’, in- cluding Siphonostomatoida Thorell, 1859; Poecilostoma- toida Thorell, 1859; Monstrilloida Sars, 1901; Harpacticoi- da Sars, 1903; Mormonilloida Boxshall, 1979. The second is named as ‘MCG-clade’, comprising Cyclopoida Burmeis- ter, 1835; Misophrioida Gurney, 1933; and Gelyelloida Huys, 1988. This phylogenetic classification has been ob- served herein. In fact, Harpacticoida is a basal-branch group of Podoplea and strongly linked to Cyclopoida, which has been accepted by Eyun (2017). The current study reveal- ed the paraphyletic status of Cyclopoida, which is consis- tent with the results of Martínez-Arbizu (2000).
Previously, Dole-Olivier. (2000) and Kim and Moon (2013) reported that Cyclopoida is made up of more than 12 families, 80 genera, and 450 marine species. In the pre- sent study, this order was expressed by 5 families, inclu- ding Taeniacanthidae, Bomolochidae, Cyclopettidae, Cyclo- pidae, and Clausidiidae. In addition, our molecular analy- sis provided nodal support for a monophyletic bomolo- chiform complex consisting of the first two very closely related families. It was based on the evidence gathered by Ho. (2006) and Huys. (2012). Furthermore, this phylogeny has shown that the bomolochiform family and the Clausidiidae family form a monophyletic group, which is in agreement with the data obtained by Huys. (2012). Herein, Taeniacanthidae includes three genera,, and. By compa- ringgene sequences, the species in this study was detected to be a distinct species with a very closely relationship with(KR048852.1).
It could be concluded that the present study provided valuable information of a copepodid species named as, Shiino, 1957, which can infectrosy goatfishand in the future, other genes will be studied to provide more information about this species, which will be helpful to prevent the infection of the fishes in the local area.
Acknowledgement
This study was supported by Researchers Supporting Project (No. RSP-2020/25), King Saud University, Riyadh, Saudi Arabia.
Avdeev, G. V., 1977. Two new and one known species of para- sitic copepods of theBrian, 1906 genus (Cyclo- poida, Taeniacanthidae) from the Indian Ocean., 101: 132-138.
Avdeev, G. V., 1978. Sistematicheskoe polozhenie roda Tegobo- molochus Izawa, 1976 (Copepoda, Cyclopoida)., 102: 119-122.
Bassett-Smith, P. W., 1898. Some new parasitic copepods found on fish at Bombay.,7, 1: 1-17.
Blanco-Bercial, L., Bradford-Grieve, J., and Bucklin, A., 2011. Molecular phylogeny of the Calanoida (Crustacea: Copepoda)., 59: 103-113.
Boxshall, G. A., 1979. The planktonic copepods of the north- eastern Atlantic Ocean: Harpacticoida, Siphonostomatoida and Mormonilloida., 35 (3): 201-264.
Boxshall, G. A., and Halsey, S. H., 2004... The Ray Society, London, 966pp.
Brian, A., 1906. Copepodi parassiti dei Pesci d’Italia. Stab. Tipo- Litografico R. Istituto Sordomuti,Genova, 1-191.
Brown, L., Bresnan, E., Graham, J., Lacaze, J. P., Turrell, E., and Collins, C., 2010. Distribution, diversity and toxin composi- tion of the genus(Dinophyceae) in Scottish wa- ters., 45: 375-393.
Burmeister, H., 1835. Beschreibung einiger neuen oder weniger bekannten Schmarotzerkrebse, nebst allgemeinen Betrachtun- gen über die Gruppe, welcher sie angehören., 17 (1): 269-336.
Cepeda, G. D., Blanco-Bercial, L., Bucklin, A., Berón, C. M., and Viñas, M. D., 2012. Molecular systematic of three species of(Copepoda, Cyclopoida) from the Atlantic Ocean: Comparative analysis using 28S rDNA., 7 (4): e35861.
Claus, C., 1864. Beiträge.,, 14: 365-382.
Cressey, R. F., 1969. Five new parasitic copepods from California inshore fish.,82: 409-428.
Dahms, H. U., 2004.thefrom Harpac- ticoidabranch of the Co- pepoda (Arthropoda, Crustacea)., 1 (1): 29-51.
Devi, D. V. U., and Shyamasundari, K., 1980. Studies on the Co- pepod pararsites of fishes of the Waltair Coast: Family Tae- niacanthidae., 39 (2): 197-208.
Dojiri, M., and Cressey, R. F., 1987. Revision of the Taeniacan- thidae (Copepoda: Poecilostomatoida) parasitic on fishes and sea urchins., 447: 1-250.
Dojiri, M., and Humes, A. G., 1982. Copepods (Poecilostomata: Taeniacanthidae) from sea urchins (Echinoidea) in the south- west Pacific., 74: 381- 436.
Dole-Olivier, M. J., Galassi, D. M. P., Marmonier, P., and Creu- zé Des Châtelliers, M., 2000. The biology and ecology of lo- tic microcrustaceans., 44: 63-91.
Eyun, S. I., 2017. Phylogenomic analysis of Copepoda (Arthro- poda, Crustacea) reveals unexpected similarities with earlier proposed morphological phylogenies., 17: 23.
Ferrari, F. D., and von Vaupel Klein, J. C., 2019 Rhabdomoplea, a new superorder for the thaumatopsylloid copepods: The con- sequence of an alternative hypothesis of copepod phylogeny., 92: 177-188.
Ferrari, F. D., Ivanenko, V. N., and Dahms, H. U., 2010. Body ar- chitecture and relationships among basal copepods., 30: 465-477.
Fls, R. H., Llewellyn-Hughes, J., Olson, P. D., and Nagasawa, K.,2006. Small subunit rDNA and Bayesian inference reveal() as highly trans- formed Mytilicolidae, and support assignment of Chondracan- thidae and Xarifiidae to Lichomolgoidea (Cyclopoida)., 87: 403-425.
Fosshagen, A., and Iliffe, T. M., 1985. Two new generea of Ca- lanoida and a new order of Copepoda, Platycopioida, from ma- rine caves on Bermuda., 70: 345-358.
Giesbrecht, W., 1892. Systematik und Faunistik der pelagischen Copepoden des Golfes von Neapel., 19: 1-831.
Gurney, R., 1933.. Ray Society, London, 384pp.
Hamza, F., Boxshall, G., and Kechemir-Issad, N., 2007. A new species ofNune-Ruivo, 1954 (Copepoda: Hat- schekiidae) parasitic on(Cadenat) of Al- geria., 67: 119-124.
Hayward, C. J., Svane, I., Lachimpadi, S. K., Itoh, N., Bott, N., and Nowak, B. F., 2011. Sea lice infections of wild fishes near ranched southern bluefin tuna () in South Australia., 320: 178-182.
Ho, J. S., 1969. Copepods of the family Taeniacanthidae (Cyclo- poida) parasitic on fishes in the Gulf of Mexico., 19: 111-130.
Ho, J. S., 1990. Phylogenetic analysis of copepod orders.Jour- nal of Crustacean , 10: 528-536.
Ho, J. S., and Kim, I. H., 2001. New species ofPo- che, 1902 (Copepoda: Hatschekiidae) parasitic on marine fish- es of Kuwait., 49: 73-79.
Ho, J. S., and Lin, C. L., 2006. Two species ofgen. nov. (Copepoda: Taeniacanthidae) parasitic on the laced mo- ray (Bloch and Schneider) in Tai- wan., 45: 578-585.
Ho, J. S., Dojiri, M., Gordon, H., and Deets, G. B., 2003. A new species of Copepoda (Thaumatopsyllidae) symbiotic with a brittle star from California, USA, and designation of a new order Thaumatopsylloida.Journal of Crustacean , 23: 582-594.
Ho, J. S., Ohtsuka, S., and Nakadachi, N., 2006. A new family of poecilostomatoid copepods (Umazuracolidae) based on speci- mens parasitic on the black scraper () in Japan., 23: 483-496.
Huelsenbeck, J. P., and Ronquist, F., 2001. MrBayes: Bayesian inference of phylogenetic trees., 17: 754-755.
Humes, A. G., 1970.gen. et sp. n., a cyclopoid copepod parasitic on an echinoid at Eniwetok Atoll., 56 (3): 575-583.
Humes, A. G., and Cressey, R. F., 1961. Copepodes Taeniacan- thides parasites d’un oursin a Madagascar., 3: 1-24.
Huys, R., 1988.,. Boxshall, G. A., and Sch- minke, H. K., eds., Springer Netherlands, 485-495.
Huys, R., and Boxshall, G. A., 1991.. The Ray Society, London, 468pp.
Huys, R., Fatih, F., Ohtsuka, S., and Llewellyn-Hughes, J., 2012. Evolution of the bomolochiform superfamily complex (Co- pepoda: Cyclopoida): New insights from ssrDNA and morpho- logy, and origin of umazuracolids from polychaete-infesting ancestors rejected., 42: 71-92.
Huys, R., Llewellyn-Hughes, J., Conroy-Dalton, S., Olson, P. D., Spinks, J. N., and Johnston, D. A., 2007. Extraordinary host switching in siphonostomatoid copepods and the demise of the Monstrilloida: Integrating molecular data, ontogeny and antennulary morphology., 43: 368-378.
Huys, R., Llewellyn-Hughes, J., Olson, P. D., and Nagasawa, K., 2006. Small subunit rDNA and Bayesian inference reveal(Copepoda incertae sedis) as highly trans- formed Mytilicolidae, and support assignment of Chondra- canthidae and Xarifiidae to Lichomolgoidea (Cyclopoida)., 87: 403-425.
Huys, R., Lopez-Gonzalez, P. J., Roldan, E., and Luque, A. A., 2002. Brooding in cocculiniform limpets (Gastropoda) and fa- milial distinctiveness of the Nucellicolidae (Copepoda): Mis- conceptions reviewed from a chitonophilid perspective., 75 (2): 187-217.
Izawa, K., 1986. On the development of parasitic copepoda. III Taeniacanthus lagocephali Pearse (Cyclopoida: Taeniacanthi- dae).Publications of the , 31 (1): 37-54.
Jenner, R. A., 2009. Higher-level crustacean phylogeny: Consen- sus and conflicting hypotheses.,39 (2): 143-153.
Kim, I. H., and Moon, S. Y., 2013. Ten new species of parasitic cycloopoid copepods (Crustacea) belonging to the families Bo- molochidae, Philichthyidae, and Taeniacanthidae from marine fishes in Korea., 48 (4): 361-398.
Kim, J., and Kim, W., 2000. Molecular phylogeny of poecilo- stome copepods based on the 18S rDNA sequences., 4: 257-261.
Krøyer, H., 1837. Om Snyltekrebsene, isaer med Hensyn til den danske Fauna., 1 (2): 172-208.
Kumar, S., Stecher, G., and Tamura, K., 2016. MEGA7: Mole- cular evolutionary genetics analysis version 7.0 for bigger datasets.Molecular and Evolution,33: 1870-1874.
Lang, K., 1948. Copepoda ‘Notodelphyoida’ from the Swedish west-coast with an outline on the systematics of the Copepods.14): 1-36.
Lewis, A. G., 1967. Copepod crustaceans parasitic on teleost fishes of the Hawaiian Islands., 121: 1-204.
Martínez-Arbizu, P., 2000. The paraphyly of Cyclopinidae Sars, 1913, and the phylogenetic position of poecilostome families within Cyclopoida Burmeister, 1835 (Copepoda: Crustacea). PhD thesis. University of Oldenburg.
Pearse, A. S., 1952. Parasitic crustacea from the Texas Coast.,2: 5-42.
Pillai, N. K., 1963. Copepods of the family Taeniacanthidae pa- rasitic on South Indian fishes., 6: 110-128.
Pritchard, M. H., and Kruse, G. O. W., 1982.. Uni- versity of Nebraska Press,Lincoln, 141pp.
Ramdane, Z., 2009. Identification et écologie des ectoparasites Crustacés des poissons Téléostéens de la côte Est algérienne. Thèse de doctorat de l’Université Badji Moktar Annaba, 235pp.
Rangnekar, P. G., and Murti, N. N., 1960. Two new copepods from the fishes of Bombay.,29 (3): 206-210.
Ravichandran, S., Ajith Kumar, T. T., Ronald Ross, P., and Mu- thulingam, M., 2007. Histopathology of the infestation of pa- rasitic isopodof the host fish., 2 (1): 68-71.
Richiardi, S., 1870. Intorno ad una nuova specie del genere, 11: 47-59.
Sars, G. O., 1901. Copepoda Calanoida., 4: 1-28.
Sars, G. O., 1903.. Bergen Museum, Norway, 13-30.
Schizas, N. V., Dahms, H. U., Kangtia, P., Corgosinho, P., and Ga- lindo, A. M., 2015. A new species ofClaus, 1863 (Copepoda: Harpacticoida: Longipediidae) from Caribbean me- sophotic reefs with remarks on the phylogenetic affinities of Polyarthra., 11 (8): 789-803.
Sebastian, M. J., 1964.sp. nov., a cope- pod parasite of the fishSteindach- ner.,6 (1): 94-97.
Sewell, R. B. S., 1949. The littoral and semi-parasitic Cyclo- poida, Monstrilloida and Notodelphyoida., 9 (2): 17-199.
Shiino, S. M., 1957. Copepods parasitic on Japanese fishes, 16: Bomolochidae and Taeniacanthidae., 2 (3): 411-428.
Song, Y., Wang, G. T., Yao, W. J., Gao, G., and Nie, P., 2008. Phy- logeny of freshwater parasitic copepods in the Ergasilidae (Copepoda: Poecilostomatoida) based on 18S and 28S rDNA sequences., 102 (2): 299-306.
Sonnenberg, R., Nolte, A., and Tautz, D., 2007. An evaluation of LSU rDNA D1-D2 sequences for their use in species identification., 4: 6.
Sumpf, K., 1871. Über eine neue Bomolochiden Gattung nebst Bemerkungen über die Mundwerkzeuge der sogenannten Poe- cilostomen. Inaugural-Dissertation, Universität Göttingen, 32pp.
Tamura, K., and Nei, M., 1993. Estimation of the number of nu- cleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees.Molecular and Ev,10: 512-526.
Tang, D., 2011. A new species of(Copepoda: Tae- niacanthidae) parasitic on two pufferfish species,and.(Teleostei: Tetraodontidae), from Australia., 58 (3): 233-239.
Tang, D., and Izawa, K., 2005.(Ya- maguti, 1939) gen. n., comb. n. (Copepoda: Taeniacanthidae), and ectoparasite of flatfishes from Japanese waters., 1071: 47-60.
Tang, D., and Johnston, M. D., 2005., a new ge- nus for(Pillai, 1963) comb. nov. (Poe- cilostomatoida: Taeniacanthidae), a parasitic copepod of ba- toid fishes (Chondrichthyes: Elasmobrachii) from the Indo- West Pacific., 44: 337-346.
Tang, D., Uyeno, D., and Nagasawa, K., 2011. Parasitic cope- pods of the family Taeniacanthidae (Crustacea) from trigger- fishes (Teleostei, Balistidae) and filefishes (Teleostei, Mona- canthidae) collected in the Indo-West Pacific region., 3103: 33-56.
Tang, D., Uyeno, D., and Nagasawa, K., 2016. A review of thespecies group (Crustacea: Copepoda: Taeniacanthidae), with descriptions of two new species., 4174 (1): 212-236.
Thorell, T., 1859. Till Kannedomen om vissa parasitiskt lefvan- de Entomostraceer. Ofvers. K. VetenskAkad., 16 (8): 355-362.
Uyeno, D., Tang, D., and Nagasawa, K., 2013., a new genus and species of taeniacanthid copepod (Cyclo- poida) parasitic on a filefish (Actinopterygii: Monacanthidae) collected from Cebu Island, the Phillippines., 61 (2): 515-523.
Venmathi Maran, B. A., Moon, S. Y., Adday, T. K., and Tang, D., 2016. Cepolacanthus kimi, a new genus and species of copepod (Cyclopoida: Taeniacanthidae) parasitic on bandfish(Valenciennes, 1835) (Actinopterygii: Ce- polidae) caught off the Iraqi coast.4174: 249-258.
Vervoort, W., 1962. A review of the genera and species of the Bomolochidae (Crustacea, Copepoda), including the descrip- tion of some old and new species., 56: 1-111.
Walter, T. C., and Boxshall, G., 2019..
Wilson, C. B., 1911. North American parasitic copepods belong- ing to the family Frgasilidae., 39: 263-400.
Wilson, C. B., 1924. New North American parasitic copepods, new hosts, and notes on copepod nomenclature.,64: 1-22.
Wilson, C. B., 1935. Parasitic copepods from the Dry Tortugas., 29: 327-347.
Yamaguti, S., 1939. Parasitic copepods from fishes of Japan. Part5. Caligoida, III., 2: 443-487.
Yamaguti, S., 1954. Parasitic copepods from fishes of Celebes and Borneo., 3 (3): 137-160.
Yamaguti, S., and Yamasu, T., 1959. Parasitic copepods from fishes of Japan with descriptions of 26 new species and remarks on two known species., 5: 89-165.
Yazawa, R., Yasuike, M., Leong, J., von Schalburg, K. R., Cooper, G. A., Beetz-Sargent, M., Robb, A., Davidson, W. S., Jones, S. R. M., and Koop, B. F., 2008. EST and mitochondrial DNA sequences support a distinct pacific form of salmon louse,., 10: 741-749.
Yeom, J., Nikitin, M. A., Ivanenko, V. N., and Lee, W., 2018. A new minute ectosymbiotic harpacticoid copepod living on the sea cucumberin the East/Japan Sea., 6: e4979.
. E-mail: rewaida@sci.cu.edu.eg
February 1, 2020;
June 18, 2020;
July 7, 2020
(Edited by Qiu Yantao)
 Journal of Ocean University of China2020年6期
Journal of Ocean University of China2020年6期
- Journal of Ocean University of China的其它文章
- Numerical Simulation and Analysis of Electromagnetic Fields Induced by a Moving Ship Based on a Three-Layer Geoelectric Model
- Applying Both Chemical Liquefaction and Enzymatic Catalysis Can Increase Production of Agaro-Oligosaccharides from Agarose
- In situ Assemblies of Bacteria and Nutrient Dynamics in Response to an Ecosystem Engineer, Marine Clam Scapharca subcrenata, in the Sediment of an Aquaculture Bioremediation System
- Harvesting Microalgae Biomass Using Sulfonated Polyethersulfone (SPES)/PES Porous Membranes in Forward Osmosis Processes
- Simulation of the Internal Wave of a Subsurface Vehicle in a Two-Layer Stratified Fluid
- Effects of Different Weaning Protocols on Survival,Growth and Nutritional Composition of Pharaoh Cuttlefish (Sepia pharaonis) Juvenile
