Identification and Functional Characterization of a Novel Δ12 Fatty Acid Desaturase Gene from Haematococcus pluvialis
ZHANG Lin, CHEN Wenbi, YANG Shuping, ZHANG Yuanbo, XU Jilin, *,YANG Dongjie, WU Zuyao, LIU Tong, and CAO Jiayi
Identification and Functional Characterization of a Novel Δ12 Fatty Acid Desaturase Gene from
ZHANG Lin1), CHEN Wenbi1), YANG Shuping1), ZHANG Yuanbo1), XU Jilin1), *,YANG Dongjie1), WU Zuyao2), LIU Tong2), and CAO Jiayi1)
1),,315211,2),,315211,
Thefreshwater microalgaaccumulates large amounts of fatty acids in response to adverse conditions. However, the key fatty acid desaturase genes inremain unknown. In this study, we cloned and functionally characterized a Δ12 fatty acid desaturase gene, and designated it as. The open reading frame ofconsisted of 1137 base pairs and encoded 378 amino acids. The deduced polypeptide showed 70% identity to other endoplasmic reticulum Δ12 fatty acid desaturases, whereas it had only 44% identity to plastid Δ12 fatty acid desaturases. The PSORT algorithm and phylogenetic analysis further confirmed its affiliation to the endoplasmic reticulum Δ12 fatty acid desaturases. Heterologous expression was performed incells transformed with the recombinant plasmid pYES2-HpFAD2. Two additional fatty acids (C16:2 and C18:2) were detected in the yeast transformants. The results indicated Δ12 desaturation activity and substrate preference for C18:1 over C16:1. The transcriptional levels ofat different growth stages were measured by quantitative polymerase chain reaction (PCR), indicating that thetranscriptional levels were significantly higher in red cells than those in green cells. Our study brings more insight into the fatty acid biosynthetic pathway of.
Δ12 fatty acid desaturase; fatty acid;;; transcriptional level
1 Introduction
(class Chlorophyceae) is a uni- cellular freshwater microalga with various cellular forms at different growth stages.The green motile flagellated cells (macrozooids) ofrapidly transform into a non-motile palmella form (hematocyst) under stress con- ditions. During this process, astaxanthin is accumulated in large quantities as an anti-stress mechanism, giving thecells red color (Han., 2013).At the same time,accumulates large amounts of fatty acids and glyce- rolipids in response to adverse conditions (Saha., 2013).
It is well known that polyunsaturated fatty acids (PUFAs) play important roles in maintaining membrane fluidity and integrity (Kamat and Roy, 2016). They are also required in several essential physiological processes, such as neonatal growth, brain development, and signal transduction (Marventano., 2015). PUFAs have attracted attentions for years because of their health benefits (Jiang., 2016; Zhang., 2019). For example, PUFAs are helpful for preventing major chronic diseases, such asthrombosis and cerebrovascular disease (Marventano.,2015). Such benefits have led to an increase in demand for PUFAs (Chen., 2016). However, global fish stocks, the primary supplier of PUFAs in the human diet, are depleted due to over-fishing. Therefore, an alternative source for PUFA production is needed. Many microalgae can synthesize various PUFAs, such as C18:3 bysp. (Lu., 2017), C18:2 and C18:3 by, C20:5 and C22:6 bysp. (Jiang., 2016; Gao., 2019), C22:6 by(Otero., 2017), and C20:5 byandsp. (Haas., 2016). Many of these PUFAs have potential for large-scale production.
biosynthesis of PUFAs involves multiple steps of desaturation and elongation of the carbon chain, catalyzed by a set of desaturases and elongases, explicitly putative Δ15, Δ12, Δ9, Δ8, Δ6, Δ5, and Δ4 desaturases and Δ9, Δ6, and Δ5 elongases, which determine the fatty acid content. The genes encoding these desaturases and elongases have been identified and characterized in a variety of microalgae, namely Δ15 desaturase in(Kotajima, 2014), Δ12 desaturases inand(Lu., 2009; Kaye., 2015), Δ9 desaturase in(Xue., 2016), Δ6 desaturase insp. (Thiyagarajan., 2018), Δ9 elongase in(Petrie., 2010), and a long-chain fatty acid elongase insp. (Guo., 2019). Desaturases in- clude distinct catalytic positions and conserved histidine re- gions, and can be classified into four major subfamilies, including the First Desaturase subfamily, the Front-End Desaturase subfamily, the Omega Desaturase subfamily, and the Sphingolipid Desaturase subfamily (Hashimoto., 2008). Δ12 desaturase, as a member of the Omega Desaturase subfamily, catalyzes the conversion of oleic acid (OA, C18:1Δ9;-9) to linoleic acid (LA, C18:2Δ9,12;-6), which is the first and probably the most important step of PUFA biosynthesis.
Δ12 Desaturases contain three conserved histidine boxes, including HXXXH, HXXHH, and HXXHH, which are considered the di-iron center of the active site and are critical for desaturase activity (Avelange-Macherel., 1995).Two types of Δ12 desaturases with distinct substrate specificities and electron donors have been identified in the endoplasmic reticulum (ER) and plastids of plants (Shanklin and Cahoon, 1998; Chodok., 2013). The microsomal Δ12 desaturases in the ER recognize phosphatidylcholines as acyl substrates, with NADH and cytochrome b5 as electron donors. In contrast, the plastid Δ12 desaturases in the chloroplast mainly act upon glycolipids as substrates, with NADPH and ferredoxin as electron donors (Los., 1998).
Fatty acid biosynthesis in microalgae has been gaining increasing attention and numerous desaturase genes have been characterized (Domergue., 2003; Lu.,2009; Iskandarov., 2010). However, to the best of our knowledge, the key fatty acid desaturase genes inremain unknown. In this study, a cDNA encoding Δ12 desaturase was isolated fromand named, and its function was tested in the yeast. Moreover,transcriptional levels and the fatty acid composition ofat different growth stages were also investigated. This re- port will deepen our understanding of the biosynthesis and physiological function of fatty acids in.
2 Materials and Methods
2.1 Strain, Medium, and Growth Conditions
was provided by the Algal Collection Lab of Ningbo University (China). A combination of antibiotics, including ampicillin, gentamycin sulfate, kanamycin, and chloramphenicol (100mgL–1each) was applied to obtain axenic cultures.was cultivated at 23℃±1℃ under 30μmolphotonsm–2s–1fluorescent light with a 12h:12h light:dark cycle. The algae were incubated in NBU3# medium composed of KNO3(100mgL–1), MnSO4(0.25mgL–1), K2HPO4(10mgL–1), FeSO4·7H2O (2.5mgL–1), Na2EDTA (20mgL–1), vitamin B1(5×10−6mgL–1), and vitamin B12(5×10−7mgL–1).
2.2 Isolation of a Putative Δ12 Fatty Acid Desaturase Gene
at the mid-logarithmic growth phase was harvested by centrifugation at 6500×for 5min. The cell pellet was frozen in liquid nitrogen and ground into a fine powder. Total RNA was extracted using the E.Z.N.A. Plant Total RNA Kit (Omega, Madison, WI, USA). Genomic DNA was extracted using the CTAB method (Po- rebski, 1997). The OD260/OD280, OD260/OD230, and concentrations of RNA and genomic DNA were mea- sured with the Nanodrop ND1000 (Nanodrop Technologies, Wilmington, DE, USA).
AncDNA library was constructed in our previous study (data not shown). A fragment from this library was predicted to be a putative Δ12 fatty acid de- saturase gene and named. The gene-specific pri- mers FAD-F1 and FAD-R1 were designed according to the known fragment. The first-strand cDNA was synthesized as the template for the polymerase chain reaction (PCR) to obtain the full-length open reading frame (ORF) of. Then, another PCR was carried out to obtain theDNA sequence with the genomic DNA tem- plate and the FAD-F1 and FAD-R1 primers. Both PCRs were performed with®Plus DNA Polymerase (TransGen, Beijing, China). The resulting PCR fragments were subcloned into the pMD19-T vector (TaKaRa, Dalian, China) and sequenced.
2.3 Pre-Sequence Analysis
The isoelectric point and molecular weight of the pu- tative Δ12 fatty acid desaturase were determined (http:// isoelectric.ovh.org/). The transmembrane (TM) regions were predicted using the transmembrane hidden Markov model (TMHMM) (http://www.cbs.dtu.dk/services/TMH MM/). The signal peptide analysis was performed using a signal peptide prediction server (http://www.cbs.dtu.dk/ser- vices/SignalP-2.0/). The distribution of the hydrophobic amino acids was determined using the Kyte-Doolittle hy- dropathy scale. Subcellular localization was predicted with the PSORT family of programs (http://www.psort.org/). Mul- tiple sequence alignment was performed with Clustal- x1.81. The phylogram was deduced using Mega 4 software (http://www.megasoftware.net/).
2.4 Functional Characterization of Saccharomyces cerevisiae
The FAD-F2 and FAD-R2 primers were designed to createIII andI restriction sites adjacent to the start and stop codons, respectively. The Kozak consensus sequence (GCCACC) was inserted in front of the start codon to enhance translational efficiency. The PCR was carried out with the first-strand cDNA template and the FAD-F2 and FAD-R2 primers, and the PCR products were subcloned into the pMD19-T vector, and named pMD19-HpFAD2. In the next step, pMD19-HpFAD2 and the pYES2 expression vector (Invitrogen, Carlsbad, CA, USA) were synchronously double-digested withIII andI. The targeted fragments were ligated with T4 ligase to yield the recombinant plasmid pYES2-HpFAD2.
INVSc1 (MATa/MATα) was used as the heterologous host. The pYES2 empty vector and the recombinant plasmid pYES2-HpFAD2 were individuallytransferred into competent INVSc1 cells using anEasyComp Transformation Kit (Invitrogen). After selec- tion on SC-U plates deficient in uracil, three random yeast transformants harboring pYES2 or pYES2-HpFAD2 were respectivelypicked and cultivated in SC-U liquid medium containing 2% (w/v) glucose without uracil. When the cultures reached an OD600of 0.4, expression was induced by supplementation with 2% (w/v) galactose and 1% (v/v) NP-40. The cultures were harvested after another 48h cul- tivation at 28℃. The pellet was stored at −80℃ for fatty acid analysis. All assays were carried out in triplicate.
2.5 Collection of Algal Biomass at Different Growth Stages
wasinoculated with an initial OD750of 0.01 (corresponding to 1.0×104cellsmL–1) at 23℃±1℃ under 30μmolphotonsm–2s–1fluorescent light and a 12h:12h light:dark cycle. OD750values and microscopic observations were determined daily.When the culture reached the stationary stage, the light intensity was increased to 100μmolphotonsm–2s–1, and the light regime was shifted to a 14h:10h light:dark cycle to induce the transformation of green cells to red cells. Cells were harvested at six different time points (days 5, 11, 17, 22, 25, and 30) in triplicate. Days 5, 11, and 17 corresponded to the lag, logari- thmic, and stationary growth stages of the green cell stage, respectively. Days 22, 25, and 30 represented the early, middle, and late stages of the red cell stage, respectively. The cells were collected, rinsed with PBS buffer, and stored at −80℃ for transcriptional and fatty acid profiling analyses. Samples (50mL each) were collected and ly- ophilized to measure the dry biomass concentration.
2.6 Quantitative Analysis of the Transcriptional Level
Primer pairs for qPCR (Table 1) were designed for.was chosen as the internal reference gene. The qPCR was performed with 10-fold serial dilutions (105–1010copiesμL–1) and each pair of primers were detected to generate a standard curve and to estimate PCR efficiency. The PCR products were quantified continuously with Mastercycler ep realplex(Eppendorf, Mannheim, Germany) using SYBR Green fluorescence. PCR cycling was comprised of an initial step at 94℃ for 30s followed by 40 cycles at 95℃ for 5s, 58℃ for 15s, and 72℃ for 10s. Data were analyzed by the 2−ΔΔCtmethod. The experiment was performed in three biological replicates and three technical replicates of each sample.

Table 1 Primers sequences
2.7 Fatty Acid Analysis
Transmethylation was performed by incubating the lyophilized biomass in KOH/methanol and then in HCl/me- thanol at 80℃ (Lepage, 1984). The fatty acid methyl esters were analyzed by gas chromatography-mass spectro- metry (GC-MS) (7890B/7000C, Agilent Technologies, Palo Alto, CA, USA) coupled with a tandem quadrupole massspectrometer and a MultiPurpose polar capillary column (CD-2560 column, 100m×250μm×0.2μm). The oven tem-perature program started at 140℃ for 5min, then rose to 240℃ at a rate of 4℃min–1and was held for 20min. The injector temperature was set to 250℃ in splitless mode. Ni- trogen was used as the carrier gas at a flow rate of 0.81mLmin–1, and pressure was set at constant flow mode. The electron impact mode was used at an ionization energy of 70eV. The fatty acid peaks were assigned with the MS data and the NIST14 commercial mass spectral database.
3 Results
3.1 Identification of the ORF and DNA Sequence
A sequence in thecDNA library was predicted to be a Δ12 fatty acid desaturase geneand named. By aligning with known Δ12 fatty acid desatu- rase genes of other organisms, theORF was de- termined to be 1137bp in length with an encoded protein of 378 amino acids. The calculated isoelectric point and molecular weight of the protein were 6.74 and 43.29kDa, respectively.The distribution of hydrophobic amino acids according to the Kyte-Doolittle hydropathy scale demon- strated that HpFAD2 is a membrane protein (Kyte and Doo- little, 1982). Five transmembrane helices (amino acids 44– 66, 73–95, 105–127, 169–191, and 220–242) were obtain- ed by TMHMM corresponding to the predicted membrane- spanning domains. No signal peptide for secreted protein was obtained from the signal peptide prediction server. These results together with the PSORT analysis suggested that HpFAD2 is localized in theER.
In addition, nine introns were identified in theDNA sequence, with lengths varying from 75 to 681bp (Fig.1). The sequences of all nine introns abided by the rule of a 5’ donor site of GT and a 3’ acceptor site of AG. The nucleotide sequences ofDNA and the ORF were deposited in GenBank with accession numbers MH- 817077 and MH817076, respectively.

Fig.1 The distributions and lengths of the HpFAD2 introns.
The neighbor-joining method was adopted to conduct the phylogenetic analysis, using 17 Δ12 desaturase genesfrom different organisms. The results demonstrated that allsequences were grouped into two major groups andbelonged to the ER desaturases group(Fig.2A). In addition, pairwise alignments revealed that the predictedprotein shared only limited identities (42%– 46%) with the plastid Δ12 desaturase proteins,., CAA 37584 (42%), ABD58898 (46%), CAA55121 (44%), AAA-50158 (44%), AAW63039 (44%), AAA50157 (44%), ABI- 73993 (44%), AAA92800 (44%), and NP_194824 (44%). Nevertheless, pairwise alignments of the ER Δ12 desatu- rases produced higher identities (61%–78%),., XP_ 001691669 (78%), ACF98528 (73%), BAB78716 (73%), AAO37754 (70%), ABA41034 (69%), AAA32782 (61%), and AAF78778 (67%). These results strongly suggest thatencoded an ER Δ12 desaturase, which was consistent with the PSORT subcellular localization ana- lysis.

Fig.2 (A) Neighbor-joining tree of HpFAD2 and other Δ12-desaturases. (B) HpFAD2 amino acid sequences compared with the plastidial and ER microsomal homologs from other species. The alignment was generated by the ClustalX program and Mega 4. The sequences shown are: CAA37584 Synechocystis sp., ABD58898 Mesostigma viride, CAA55121 Spinacia oleracea, AAA50158 Glycine max, AAW63039 Olea europaea, AAA50157 Brassica napus, ABI73993 Descurainia Sophia, AAA92800 Arabidopsis thaliana, NP_194824 A. thaliana, XP_001691669 Chlamydomonas reinhardtii, ACF98528 C. vulgaris, BAB78716 C. vulgaris, AAO37754 Punica granatum, ABA41034 Jatropha curcas, AAA32782 A. thaliana, and AAF78778 B. napus. HpFAD2 from H. pluvialis is marked with a black triangle. The three histidine boxes are indicated in bold.
Multiple alignment of HpFAD2 against 16 other Δ12 desaturases is shown in Fig.2B. Three highly conservedhistidine boxes, such as HECGH (amino acids 95–99),HRRHH (amino acids 131–135), and HVCHH (aminoacids 305–309) were observed and scattered in the hydro- philic regions,., nontransmembrane regions. A com- parison of the histidine boxes of the ER desaturases with those of the plastid desaturases revealed the following com- mon characteristics. The first histidine box of ER desa- turases was HECXH, while that of plastid desaturases was HDCXH. However, the second histidine box (HRXHH/ HDXHH) and the third histidine box (HVXHH/HIXHH) were homogeneous. In the second motif, the sequence of HRRHH in the ER Δ12 desaturases was replaced with HDXHH in the plastid Δ12 desaturases. Additionally, the third histidine motif (HVXHH) found in ER Δ12 desaturases was substituted by HIPHH in the plastid Δ12 de-saturases.
3.2 Functional Analysis in Yeast
Heterologous expression in yeast was performed to validate the function of the putative HpFAD2. The heterologous hostINVSc1 was transformed with plasmid pYES2-HpFAD2, while pYES2 empty vector was employed as control. HpFAD2 was expressed under control of the galactose-inducible GAL promoter in the pYES2 expression vector. pYES2-transformed yeast showed the typical fatty acid composition ofwith C16:0, C16:1, C18:0, and C18:1 as the major fatty acids (Fig.3), which was identical to previous studies (Chodok., 2013; Kaye., 2015). However,expression resulted in two additional peaks, which were assigned C16:2Δ9, Δ12and C18:2Δ9, Δ12, respectively, based on the GC-MS analysis (Fig.3). This observation apparently confirmed the function of HpFAD2. Further- more, it was speculated thatHpFAD2 preferred C18:1 to C16:1, because the C18:2 to C18:1 ratio in the yeast trans- formant with pYES2-HpFAD2 was much higher than that of C16:2 to C16:1.

Fig.3 Fatty acid profile of the transformed yeast expressing pYES2-HpFAD2 or the pYES2 empty vector (control). Results are shown as mean±SD (n=3).
3.3 Growth Kinetics and Fatty Acid Profiles at Different Growth Stages
It took 30 days to monitorthegrowth pattern and obtain algal cells at different growth stages (Fig.4). Cell density increased slowly during the lag growth stage (days 1–5). The growth rate accelerated at the logarithmic growth stage (days 6–16), and then began to slow down on day 17 (stationary growth stage).Thecells were at the ‘green vegetative stage’ on days 5, 11, and 17 according to the microscopic observations (Fig.4).Light intensity was strengthened and lightduration was extended on day 21 to promote the conversion of green cells to red cells (Fig.4). Cell density and the dry biomass changed slightly thereafter.
The fatty acid profiles were determined based on cells collected on days 5, 11, 17, 22, 25, and 30 (Table 2). C16:0, C16:4, C18:1, C18:2, and C18:3n-3 were the major fatty acids in. Among them, C18:3n-3 was the most abundant, constituting 20%–30% of the total fatty acids during the whole course. No significant differences were observed in the contents of C14:0, C15:0, C17:0, C18:3n-6, C18:4, or C20:4n-6 whencul- tures were shifted to a light-stress condition. The concentrations of C16:0, C18:1, and C18:2 on day 30 increased by 22%, 77%, and 92%, respectively, compared to those on day 5, whereas the concentrations of C16:1, C16:2, C16:3, C16:4, and C18:3n-3 decreased by 74%, 72%, 27%, 43%, and 26%, respectively. The concentrations of PUFAs and unsaturated fatty acids (UFAs) decreased by 11% and 4% on day 30, while those of saturated fatty acids (SFAs), monounsaturated fatty acids (MUFAs), and double unsaturated fatty acids (DUFAs) increased by18%, 39%, and 60%, respectively, compared to day 5. In general, the C18:2 to C18:1 ratio in red cells was significantly higher than that in green cells, while there was no significant dif- ference in the C16:2 to C16:1 ratio between red cells and green cells.

Fig.4 The growth curve, biomass accumulation, and light microscopic images of Haematococcus pluvialis. Black diamond represents the time point that light intensity was changed. Results are shown as mean±SD (n=3). Scale bar: 10μm.
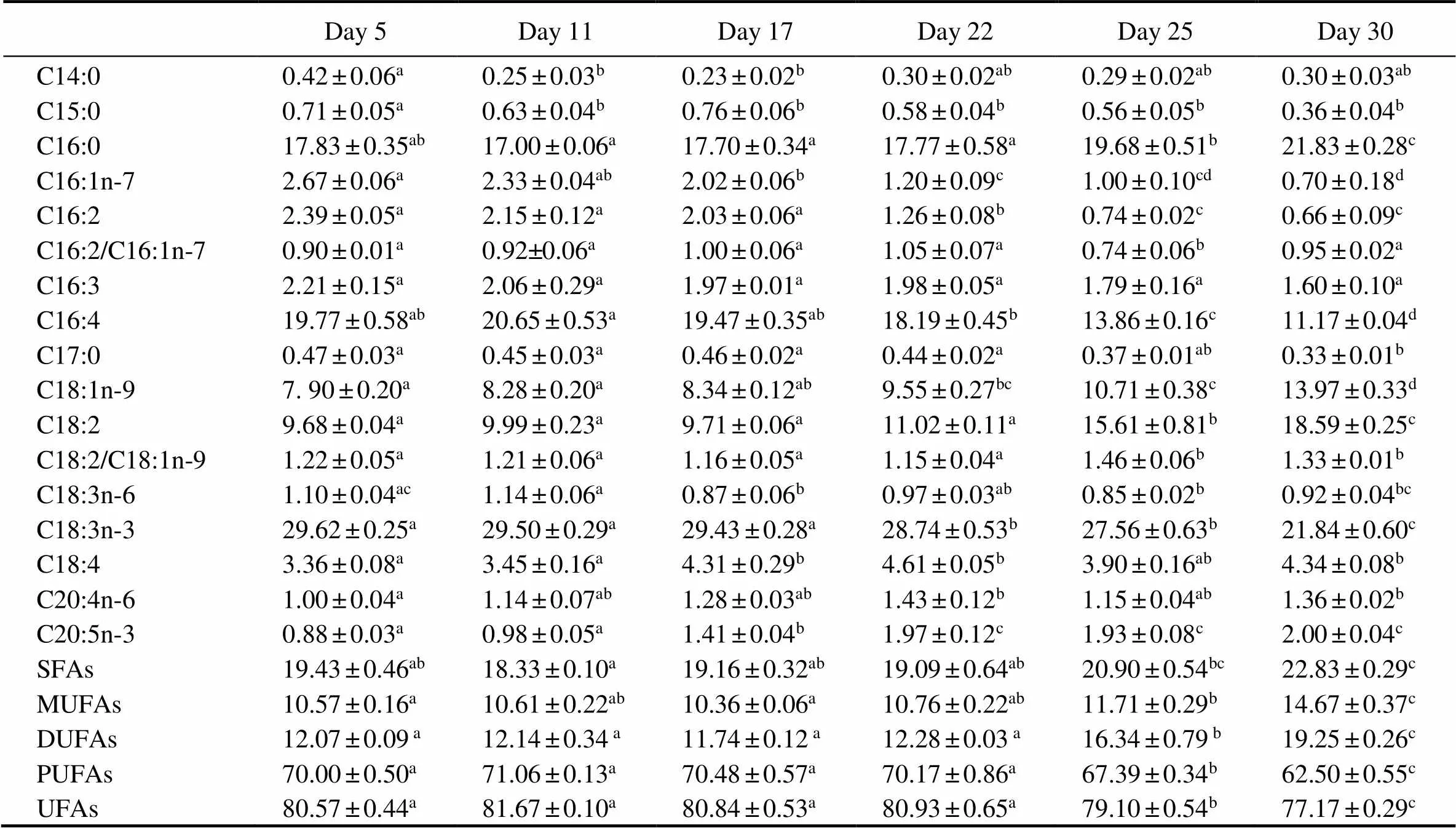
Table 2 Fatty acid compositions (% of total fatty acids) of Haematococcus pluvialis at different growth stages
Notes: SFAs, saturated fatty acids; MUFAs, monounsaturated fatty acids; DUFAs, double unsaturated fatty acids; PUFAs, polyunsaturated fatty acids; UFAs, unsaturated fatty acids. C16:2/C16:1n-7 represents the C16:2 to C16:1n-7 ratio. C18:2/C18:1n-9 represents the C18:2 to C18:1n-9 ratio. Data are presented as mean±SD (=3). One-way ANOVA was performed and different letters (a, b, c, and d) represent a significant difference between different groups at the 95% confidence level (<0.05).
3.4 Relative Transcriptional Levels at Different Growth Stages
The relativetranscriptional levels were determined by qPCR with the 2−ΔΔCtmethod (Fig.5). Thetranscriptional levels during days 5–11 rose slightly and were significantly lower than those on day 17. After being exposed to the light-stress condition, thetranscriptional level in red cells increased dramatically. Therefore, the values on days 22, 25, and 30, which were close to each other, were significantly higher than those on days 5, 11, and 17 (<0.05). The transcriptional levels on days 22, 25, and 30 were 9.14, 10.58, and 9.55 times of those on day 5, respectively.
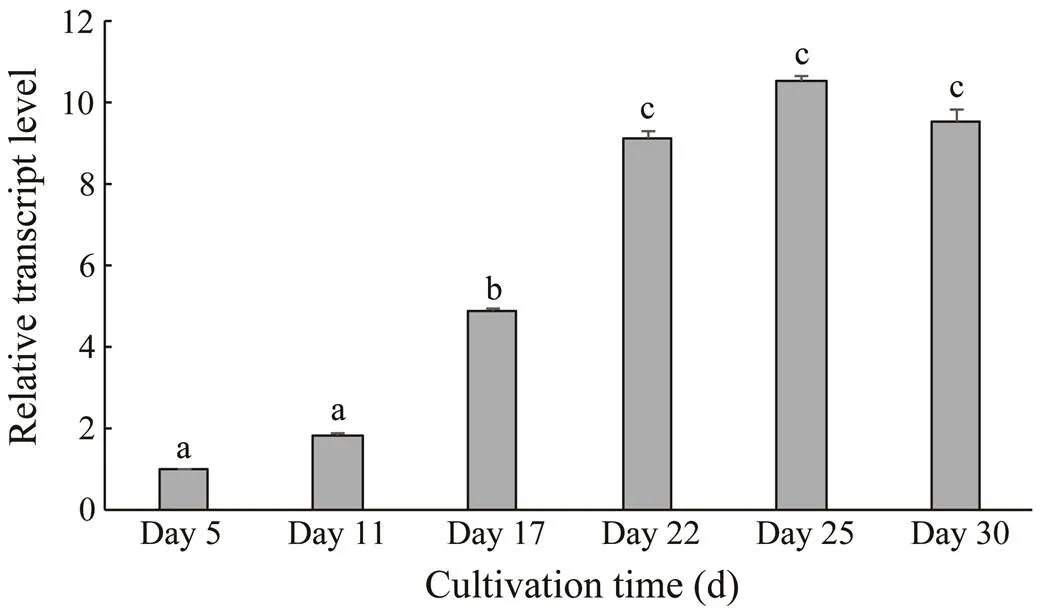
Fig.5 HpFAD2 transcriptional levels in Haematococcus pluvialis at different growth stages. Values are mean±SD of triplicates. One-way ANOVA was performed and different letters (a, b, and c) represent a significant difference between different groups at the 95% confidence level (P<0.05).
4 Discussion
As an essential component of cell membranes, LA is indispensable for growth and reproduction in eukaryotes. Moreover, LA is also the precursor of other-6 and-3 long-chain PUFAs. Therefore, the OA to LA reaction, ca- talyzed by Δ12 desaturase, is the committing step of PUFA biosynthesis.accumulates large amounts of fatty acids under adverse conditions. However, knowle- dge of genes related to fatty acids biosynthesis inis insufficient, and no study has been performed on Δ12 desaturases. In the current study, we successfully clone and functionally characterize a Δ12 fatty acid de- saturase gene from. These results will be help- ful forfurther analysis of the fatty acid biosynthesis path- way in.
This gene, designated, possesses approximate- ly 70% identity to other ER Δ12 desaturases but only 44% identity to plastid Δ12 desaturases. Moreover, the results of the PSORT algorithm and phylogenetic analyses also confirm thatbelong to ER Δ12 desaturases.
Many transmembrane ER proteins contain consensus motifs in their cytoplasmically exposed tails, which serve as the retrieval signal to bring proteins back from the sorting compartment adjacent to the ER (Jackson, 1990). The ER retention motif generally features two lysine residues. One is immutably located at the −3 position from the C-terminus, while the other one is usually positioned four or five residues from the C-terminus. Site- directed mutagenesis analyses have demonstrated that these two lysine residues are essential for the function of the enzyme and cannot be substituted by other amino acid re- sidues (Jackson, 1990). The presence of two lysines at the −3 and −5 positions of HpFAD2 proves the hypo- thesis that HpFAD2 belongs to ER Δ12 desaturases.
Introns are widespread and variable in eukaryotic genomes and are always related to gene duplication and the evolution of the species (Han, 2018). Several types of introns have been identified and one is the spliceo- somal intron (Han., 2018). Spliceosomal introns are typical of GT/AG motifs, which separately act as donor and acceptor splice sites (Zhang., 2011; Rogozin.,2012). The length, number, and organization of introns vary considerably among different taxa. For example, the introns of nitrate reductase genes have been well studied in several algae (Bhadury, 2011). The results show that there are 15, 18, 10, and 2 intronsin the nitrate reductasegenes of,,, and, respectively (Da- wson., 1996; Gruber., 1996; Zhou and Klein- hofs, 1996; Song and Ward, 2004). Lu. (2009) characterized a Δ12 desaturase gene in Antarctic. It contains eight introns, seven of which were spliceoso- mal introns, with lengths ranging from 54 to 520bp. Only one intron (259bp) follows the GT/AG rule in theΔ12 desaturase gene (Kaye., 2015). In the present study, the nine introns identified inare longer than those of Antarctic,and all possess canonical GT/AG splicing signals.
, as the most representative yeast, can pro- duce MUFAs (C16:1 and C18:1) but not PUFAs. Hence, it has been regarded as a good host for functional charac- terization of desaturases and elongases related to fatty acid biosynthesis (Niu., 2007; Chodok., 2013; Cui., 2016).has been employed in many studies on the functions of genes from microalgae (Lu., 2009; Kaye., 2015). Heterologous expression was performed withINVSc1 in this study to verify the desaturation activity and substrate specificity of HpFAD2. The GC-MS results indicated that HpFAD2 led to two new fatty acids (C16:2 and C18:2), confirming its Δ12 desaturation activity and substrate preference for C18:1 rather than for C16:1. This is consistent with a previous study that Δ12 desaturases from(Domer- gue., 2003) also recognize C16:1 and C18:1 as sub- strates. Nevertheless, the reported Δ12 desaturases from(Lu., 2009) and(Kaye., 2015) only show specificity for C18:1 as a yeast substrate.
The green cells began to transform into red cells with an increase of astaxanthin after the light treatment was en- hanced. The quantitative results indicated that thetranscriptional levels in red cells were significantly higher than those in green cells.The trend in the C18:2 to C18:1 ratio was similar to thetranscriptional level. When light intensity was strengthened at day 22, the C18:2 to C18:1 ratio did not markedly increase, although thetranscriptional level was significantly enhanced, which may has been due to the response delay of transcription enhancement to the increase of fatty acids. The C18:2 to C18:1 ratio in red cells became significantly higher than that in green cells as cultivation time was prolonged. Little correlation was detected between the C16:2 to C16:1 ratio with thetranscriptional levels. This observation was in line with the results of yeast expression,., HpFAD2 preferred C18:1 to C16:1.
Intriguingly, we found that both MUFAs and DUFAs contents increased considerably but content of PUFAs de- creased significantly during cell transformation, which might be caused by the astaxanthin storage. In, about 95% of astaxanthin is stored in cytosolic lipid bodies in the form of fatty acyl mono- or diesters (Holtin., 2009; Chen., 2015). In other words, the storage of astaxanthin requires MUFAs and DUFAs rather than PUFAs.
5 Conclusions
In this study, we reported the cloning and functional characterization of a Δ12 fatty acid desaturase gene innamed.HpFAD2 showed typical struc- tural homology with other ER Δ12 fatty acid desaturases. Its functional characterization was performed by hetero- logous expression in, confirming its Δ12 fattyacid desaturase activity and substrate preference for C18:1 over C16:1. Further analysis indicated that thetranscriptional levels of red cells were significantly higher than those of green cells. This report provides insight into the biosynthesis of fatty acids in.
Acknowledgements
This study was supported by the Zhejiang Provincial Na- tural Science Foundation of China (No. LQ16D060001), the National Natural Science Foundation of China (No. 41606163), the Natural Science Foundation of the Ningbo Government (No. 2017A610288), the Ningbo Science and Technology Research Projects, China (No. 2019B10006), the Zhejiang Major Science Project, China (No. 2019C0 2057), the Earmarked Fund for Modern Agro-Industry Technology Research System, China (No. CARS-49) and partly sponsored by K. C. Wong Magna Fund at Ningbo University.
Avelange-Macherel, M. H., Macherel, D., Wada, H., and Murata, N., 1995. Site-directed mutagenesis of histidine residues in the Δ12 acyl-lipid desaturase of., 361 (1): 111-114.
Bhadury, P., Song, B., and Ward, B. B., 2011. Intron features of key functional genes mediating nitrogen metabolism in marine phytoplankton., 4 (3): 207-213.
Chen, G., Wang, B., Han, D., Sommerfeld, M., Lu, Y., Chen, F., and Hu, Q., 2015. Molecular mechanisms of the coordination between astaxanthin and fatty acid biosynthesis in(Chlorophyceae)., 81 (1): 95- 107.
Chen, G., Yang, J., Eggersdorfer, M., Zhang, W., and Qin, L., 2016. N-3 long-chain polyunsaturated fatty acids and risk of all-cause mortality among general populations: A meta-ana- lysis., 6: 28165.
Chodok, P., Eiamsa-ard, P., Cove, D. J., Quatrano, R. S., and Kaewsuwan, S., 2013. Identification and functional characterization of two Δ12-fatty acid desaturases associated with essential linoleic acid biosynthesis in., 40 (8): 901-913.
Cui, J., He, S., Ji, X., Lin, L., Wei, Y., and Zhang, Q., 2016. Identification and characterization of a novel bifunctional Δ12/ Δ15-fatty acid desaturase gene from., 38: 1155-1164.
Dawson, H. N., Pendleton, L. C., Solomonson, L. P., and Cannons, A. C., 1996. Cloning and characterization of the NR- encoding gene from: Structure and identi- fication of transcription start points and initiator sequences., 171 (2): 139-145.
Domergue, F., Spiekermann, P., Lerchl, J., Beckmann, C., Kilian, O., Kroth, P. G., Boland, W., Zahringer, U., and Heinz, E., 2003. New insight intofatty acid metabolism. Cloning and functional characterization of plastidial and microsomal Δ12-fatty acid desaturases., 131 (4): 1648-1660.
Gao, G., Wu, M., Fu, Q., Li, X., and Xu, J., 2019. A two-stage model with nitrogen and silicon limitation enhances lipid pro- ductivity and biodiesel features of the marine bloom-forming diatom., 289:121717.
Gruber, H., Kirzinger, S. H., and Schmitt, R., 1996. Expression of the Volvox gene encoding nitrate reductase: Mutation-de- pendent activation of cryptic splice sites and intron-enhanced gene expression from a cDNA., 31 (1): 1-12.
Guo, M., Chen, G., Chen, J., and Zheng, M. J., 2019. Identification of a long-chain fatty acid elongase fromsp. involved in the biosynthesis of fatty acids by heterologous expression in., 18 (5): 1199-1206.
Haas, S., Bauer, J. L., Adakli, A., Meyer, S., Lippemeier, S., Schwarz, K., and Schulz, C., 2016. Marine microalgaeandsp. as n-3 PUFA source in diets for juvenile european sea bass (L.)., 28: 1011-1021.
Han, D. X., Li, Y. T., and Hu, Q., 2013. Astaxanthin in microalgae: Pathways, functions and biotechnological implications., 28 (2): 131-147.
Han, J., Zhang, L., Wang, P., Yang, G., Wang, S., Li, Y., and Pan, K., 2018. Heterogeneity of intron presence/absence insp. (Bacillariophyta) contributes to the understanding of intron loss., 54 (1): 105-113.
Hashimoto, K., Yoshizawa, A. C., Okuda, S., Keiichi, K., Goto, S., and Kanehisa, M., 2008. The repertoire of desaturases and elongases reveals fatty acid variations in 56 eukaryotic genomes., 49 (1): 183-191.
Holtin, K., Kuehnle, M., Rehbein, J., Schuler, P., Nicholson, G., and Albert, K., 2009. Determination of astaxanthin and asta- xanthin esters in the microalgaeby LC-(APCI)MS and characterization of predominant carotenoid isomers by NMR spectroscopy., 395 (6): 1613-1622.
Iskandarov, U., Khozin-Goldberg, I., and Cohen, Z., 2010. Identification and characterization of Δ12, Δ6, and Δ5 desaturases from the green microalga., 45 (6): 519-530.
Jackson, M. R., Nilsson, T., and Peterson, P. A., 1990. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum., 9 (10): 3153-3162.
Jiang, X. M., Han, Q. X., Gao, X. Z., and Gao, G., 2016. Conditions optimising on the yield of biomass, total lipid, and va- luable fatty acids in two strains of., 194: 723-732.
Kamat, S. G., and Roy, R., 2016. Evaluation of the effect of n-3 PUFA-rich dietary fish oils on lipid profile and membrane fluidity in alloxan-induced diabetic mice ()., 416: 117-129.
Kaye, Y., Grundman, O., Leu, S., Zarka, A., Zorin, B., Didi- Cohen, S., Khozin-Goldberg, I., and Boussiba, S., 2015. Me- tabolic engineering toward enhanced LC-PUFA biosynthesis in: Overexpression of endogenous Δ12 desaturase driven by stress-inducible promoter leads to enhanced deposition of polyunsaturated fatty acids in TAG., 11: 387-398.
Kotajima, T., Shiraiwa, Y., and Suzuki, I., 2014. Functional screening of a novel Δ15 fatty acid desaturase from the coccolithophorid., 1841 (10): 1451-1458.
Kyte, J., and Doolittle, R. F., 1982. A simple method for displaying the hydropathic character of a protein., 157: 105-132.
Lepage, G., and Roy, C. C., 1984. Improved recovery of fatty acid through direct transesterification without prior extraction or purification., 25 (12): 1391-1396.
Los, D. A., and Murata, N., 1998. Structure and expression of fatty acid desaturases., 1394: 3- 15.
Lu, Q., Li, J., Wang, J., Li, K., Li, J., Han, P., Chen, P., and Zhou, W., 2017. Exploration of a mechanism for the production of highly unsaturated fatty acids insp. at low temperature grown on oil crop residue based medium., 244: 542-551.
Lu, Y., Chi, X., Yang, Q., Li, Z., Liu, S., Gan, Q., and Qin, S., 2009. Molecular cloning and stress-dependent expression of a gene encoding Δ12-fatty acid desaturase in the Antarctic microalgaNJ-7., 13: 875-884.
Marventano, S., Kolacz, P., Castellano, S., Galvano, F., Buscemi, S., Mistretta, A., and Grosso, G., 2015. A review of recent evidence in human studies of n-3 and n-6 PUFA intake on cardiovascular disease, cancer, and depressive disorders: Does the ratio really matter?, 66 (6): 611-622.
Niu, B., Ye, H., Xu, Y., Wang, S., Chen, P., Peng, S., Ou, Y., Tang, L., and Chen, F., 2007. Cloning and characterization of a novel Δ12-fatty acid desaturase gene from the tree., 29: 959-964.
Otero, P., Saha, S. K., Gushin, J. M., Moane, S., Barron, J., and Murray, P., 2017. Identification of optimum fatty acid extraction methods for two different microalgaeandfor food and biodie- sel applications., 409 (19): 4659-4667.
Petrie, J. R., Mackenzie, A. M., Shrestha, P., Liu, Q., Frampton, D. F., Robert, S. S., and Singh, S. P., 2010. Isolation of three novel long-chain polyunsaturated fatty acid delta 9-elongases and the transgenic assembly of the entiredo- cosahexaenoic acid pathway in., 46 (5): 917-925.
Porebski, S., Bailey, G., and Baum, B. R., 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components., 15: 8-15.
Rogozin, I. B., Carmel, L., Csuros, M., and Koonin, E. V., 2012. Origin and evolution of spliceosomal introns., 7 (1): 11.
Saha, S. K., McHugh, E., Hayes, J., Moane, S., Walsh, D., and Murray, P., 2013. Effect of various stress-regulatory factors on biomass and lipid production in microalga., 128: 118-124.
Shanklin, J., and Cahoon, E. B., 1998. Desaturation and related modifications of fatty acids., 49: 611-641.
Song, B., and Ward, B. B., 2004. Molecular characterization of the assimilatory nitrate reductase gene and its expression in the marine green alga(Chlorophyceae)., 40: 721-731.
Thiyagarajan, S., Arumugam, M., Senthil, N., Vellaikumar, S., and Kathiresan, S., 2018. Functional characterization and sub- strate specificity analysis of D6-desaturase from marine mi- croalgasp., 40: 577-584.
Xue, W. B., Liu, F., Sun, Z., and Zhou, Z. G., 2016. A Δ-9 fatty acid desaturase gene in the microalgaReisigl:Cloning and functional analysis., 17 (7): 1143.
Zhang, X., Tolzmann, C. A., Melcher, M., Haas, B. J., Gardner, M. J., Smith, J. D., and Feagin, J. E., 2011. Branch point iden- tification and sequence requirements for intron splicing in., 10 (11): 1422-1428.
Zhang, Y., Min, J., and Zhang, L., 2019. Anti-inflammatory and immunomodulatory effects of marine n-3 polyunsaturated fatty acids on human health and diseases., 18 (2): 481-492.
Zhou, J., and Kleinhofs, A., 1996. Molecular evolution of nitrate reductase genes., 42 (4): 432- 442.
. Tel: 0086-574-87609570
E-mail: xujilinnbu@163.com
December 25, 2019;
March 30, 2020;
June 19, 2020
(Edited by Qiu Yantao)
 Journal of Ocean University of China2020年6期
Journal of Ocean University of China2020年6期
- Journal of Ocean University of China的其它文章
- Numerical Simulation and Analysis of Electromagnetic Fields Induced by a Moving Ship Based on a Three-Layer Geoelectric Model
- Applying Both Chemical Liquefaction and Enzymatic Catalysis Can Increase Production of Agaro-Oligosaccharides from Agarose
- In situ Assemblies of Bacteria and Nutrient Dynamics in Response to an Ecosystem Engineer, Marine Clam Scapharca subcrenata, in the Sediment of an Aquaculture Bioremediation System
- Harvesting Microalgae Biomass Using Sulfonated Polyethersulfone (SPES)/PES Porous Membranes in Forward Osmosis Processes
- Simulation of the Internal Wave of a Subsurface Vehicle in a Two-Layer Stratified Fluid
- Effects of Different Weaning Protocols on Survival,Growth and Nutritional Composition of Pharaoh Cuttlefish (Sepia pharaonis) Juvenile
