β-catenin in intranuclear inclusions of hepatocellular carcinoma
Suzan Schwertheim, Holger Jastrow, Julia Kälsch,3, Thomas Herold, Sarah Theurer, Saskia Ting, Kurt Werner Schmid,4, Hideo Andreas Baba
1Institute of Pathology, University Hospital of Essen, University of Duisburg-Essen, Essen 45147, Germany.
2Institute of Anatomy and Electron Microscopy Unit of Imaging Center Essen, University Hospital of Essen, University of Duisburg-Essen, Essen 45147, Germany.
3Department of Gastroenterology and Hepatology, University Hospital of Essen, University of Duisburg-Essen, Essen 45147,Germany.
4Member of the West German Cancer Centre Essen (WTZE), University Hospital of Essen, University of Duisburg-Essen, Essen 45147, Germany.
Abstract
Keywords: Wnt/β-catenin pathway, KDM2A, intranuclear inclusions, hepatocellular carcinoma
INTRODUCTION
The Wnt/β-catenin signaling pathway is regarded as playing the most important role in hepatocellular regeneration[1-4]. Speeet al.[4]reported that the Wnt/β-catenin signaling pathway is active in the ductular reaction of acute liver failure patients[4,5]. Apteet al.[1]demonstrated a significant correlation between nuclear/cytoplasmic-β-catenin staining and hepatocellular regeneration. Recently, we published a detailed study on intranuclear inclusions (NI) in hepatocellular carcinoma (HCC). In general, NI are considered to be a morphological feature, without any notable function or influence on disease. On the contrary, our earlier studies revealed that these NI are surrounded by a completely closed nuclear membrane and contain degenerated organelles, autophagy-associated proteins and lysosomes, suggesting a biological function of NI.The presence of NI was also significantly correlated with survival in HCC[6]. However, the factors that play a role in the occurrence of NI are mostly still unknown. Thus, we questioned whether proteins of the Wnt/β-catenin pathway are involved in the functioning of NI.
We performed immnuohistochemical analyses to investigate the presence of β-catenin, glutamine synthetase and KDM2A in NI. Glutamine synthetase, an enzyme involved in the removal of ammonia in the liver[7],nitrogen balance and pH regulation[8]is also a target of the Wnt/b-catenin pathway in the liver[9]. Longet al.[8]showed increased levels of expression of glutamine synthetase in HCC and in liver tissue with cirrhosis and chronic hepatitis B. KDM2A (FBXL11) is known to demethylate histone H3K36[10], which contains an F-box,a JmjC domain, a CxxC zinc finger, a PHD domain, and three leucine-rich repeat elements[10-13]. KDM2A is involved in various signaling pathways including NF-κB signaling, p53 activity and WNT/β-catenin pathway; demethylation of β-catenin with consequent degradation has been reported[14]. Thus, to investigate a possible correlation of KDM2A expression levels on survival in HCC patients, we performed Kaplan-Meier survival studies. Further, we examined if positive immunoreactivity for β-catenin within NI is associated simultaneously with the presence of the autophagy-associated proteins p62/sequestosome1, ubiquitin, LC3B,cathepsin B and cathepsin D in NI.
The goal of the current study was to investigate whether proteins of the Wnt/β-catenin pathway are associated with NI morphology and disease survival.
METHODS
Patients
Seventy two patients with HCC diagnosed at the Institute of Pathology, University Hospital of Essen,Germany between 1999 and 2005 were included in this study. Formalin-fixed and paraffin-embedded (FFPE)material from untreated patients were provided in all cases, prepared according to institutional standards and stained with H&E as described before[6]. The current WHO criteria[15]were used for the diagnosis of all tumors and the classification of tumors was based on the TNM system (8th edition). Table 1 provides an overview of patient data and tumor characteristics. In 70 HCC cases, complete clinical records and follow-up data were available.
Informed consent was obtained from each patient. The study complied with the Helsinki Declaration of 1975 and the Ethics Committee (Institutional Review Board) of the University Hospital Essen (reference number:16-6917-BO).
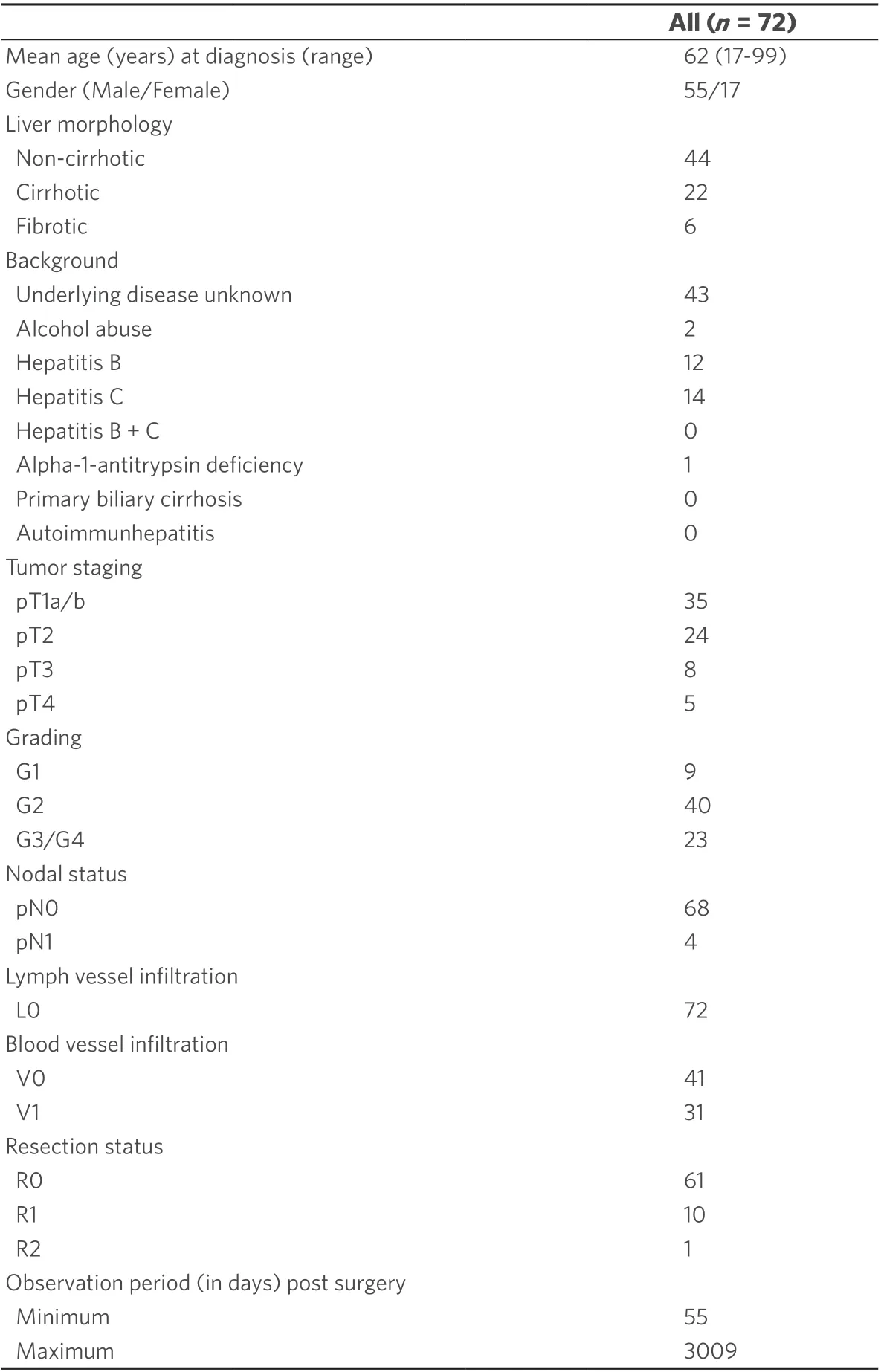
Table 1. Clinical and pathological parameters of the study group with 72 HCC cases
Tissue microarray construction and immunohistochemistry
We investigated the expression of selected candidate proteins immunohistochemically using tissue microarrays (TMAs). Regions of tumors were selected with matching H&E stained slides and marked on the donor block. Construction of the TMAs was performed by using a manual tissue-array instrument (Beecher Instruments, Silver Spring, MD, USA) as described before[6]. Briefly, we took three 1-mm-thick tissue cores from each specimen. Each TMA contained three corresponding tumor-free liver tissue cores as controls and cores with myocardial tissue for TMA orientation and 10 sections of about 3 µm each were cut from each TMA. Immunohistochemistry (IHC) of the paraffin sections for the antibodies β-catenin, KDM2A,glutamine synthethase, ubiquitin, p62, LC3B, cathepsin B and cathepsin D was conducted as described before[6]by using an automated staining device (Dako Autostainer, Dako, Glostrup, Denmark). Further, we stained one section from each TMA with H&E; Supplementary Table 1 provides detailed information on the antibodies used and staining protocols. Additionally, we included negative controls in every run: slides were incubated with non-immune immunoglobulin at the same concentration as the primary antibody instead of the primary antibody as described before[6]. We counted the membrane-bound intranuclear inclusions (NI)in all 10 serial sections of each case; cases lacking evaluable material on all 10 serial sections were excluded as described before[6]. We defined an inclusion only as positive if it was delimited by an intact membrane and completely closed. In a standardized area, inclusions were counted and calculated by: 10 × (3 × 1 mm tissue cores) = 10 × (3 × 0.78 mm2) = 23.5 mm2. The qualitative detection of membrane-bound nuclear inclusions was recorded as positive (1) or negative (0). We defined cases containing at least one membrane-bound nuclear inclusion as positive; haematoxylin counterstaining enabled the enumeration of the inclusions. We analyzed the immunostainings within NI. Quantitative analysis of immunopositive NI was performed for β-catenin, KDM2A, ubiquitin, p62, LC3B, cathepsin B and cathepsin D. Due to the small size of the NI and staining surface, we did not quantify immunostaining intensity. Cases with positively stained membranebound nuclear inclusions were classified as positive (1) or negative (0).
Next generation sequencing
FFPE tumor tissue of all cases was analyzed by next generation sequencing (NGS) with the Illumina MiSeq sequencer (Illumina, San Diego, CA, USA) following the manufacturer’s instructions as described before[16].Briefly, 45 ng of DNA was used to perform multiplex-PCR and Biomedical Genomics Workbench (CLC Bio,Qiagen, USA) was used for analysis. We designed a customized HCC-panel containing regions of interest: 23 genes of the Wnt pathway including theCTNNB1gene.
Transmission electron microscopy
Ultrastructural analysis of NI was carried out as described previously[6,16]. Briefly, for transmission electron microscopy (TEM), fresh tissue from liver biopsy of a representative HCC patient was fixed in 2%glutaraldehyde in 0.1M cacodylate buffer (cb), pH 7.3, for 4 h at room temperature. Afterwards it was washed in cb, post-fixed with 1% osmium tetroxide in cb, dehydrated in a graded series of alcohol and embedded in epoxy resin. In order to determine blocks of adequate quality, semi-thin sections were stained with basic fuchsin and methylene blue. Ultrathin sections of selected blocks were mounted on copper grids, and then treated with uranyl acetate (1%) and lead citrate (0.4%) for contrast enhancement. Digital TEM images were acquired on the Zeiss EM 902A (Zeiss, Oberkochen, Germany) equipped with a Morada slow-scan CCD camera using ITEM 5.2 software (both Olympus Soft-Imaging-Systems, Münster, Germany).
Double immunofluorescence of tissue sections
Spatial localization of β-catenin and p62 was investigated by double immunofluorescence staining.One-µm-thick FFPE tissue sections (HCC) were cut, dewaxed, rehydrated and pretreated with Target Retrieval Solution (Agilent Technologies, Ratingen, Germany) at pH 9.0 for 20 min at 97 °C. For double labeling immunofluorescence, the primary antibodies anti-β-catenin (Transduction) and anti-p62 (Enzo)were used. Anti-p62 antibody was labeled with Donkey Anti Rabbit AF555. For labeling of anti-β-catenin antibody Goat Anti Mouse HRP and Goat Anti HRP AF488 were used. Details are provided in Supplementary Table 2. DNA was stained with DAPI; image analysis and microscopy was performed by using an Olympus BX43 (Olympus Deutschland, Hamburg, Germany).
Statistical analysis
Statistical analysis was performed using the Statistical Package for Social Sciences (SPSS 24.0, Chicago,IL, USA) program. Relationships between categorical parameters were investigated using the two-sided Fisher’s exact test. Overall survival (OS) curves were performed using the Kaplan-Meier method, and differences in survival curves were compared by the log-rank test. Mann-WhitneyU-test was used to assess whether positive KDM2A immunostaining in NI correlates with the number of NI;P≤ 0.05 was defined as statistically significant.
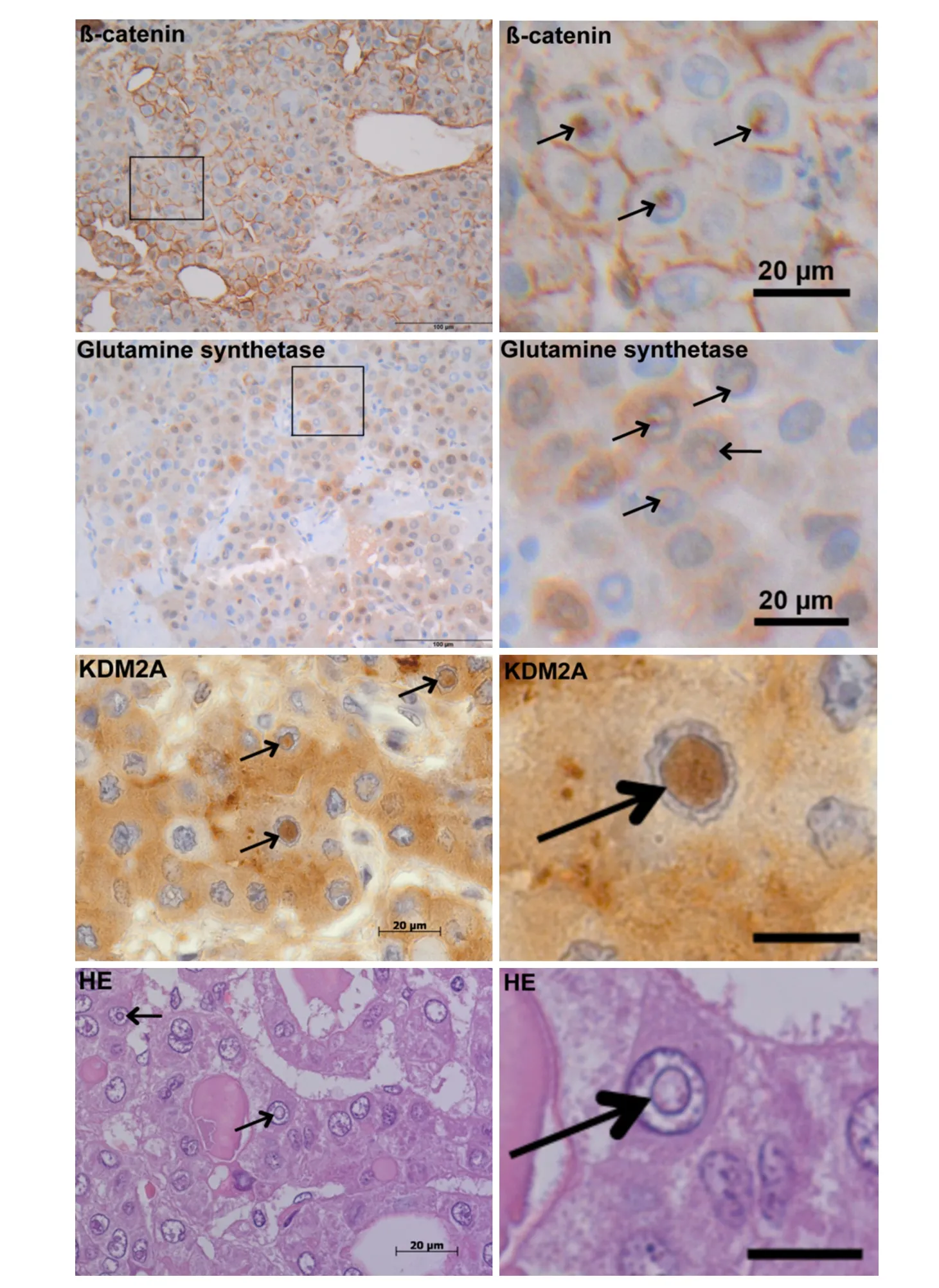
Figure 1. Intranuclear inclusions in hepatocellular carcinoma. Positive immunoreactivity for β-catenin, glutamine synthethase and KDM2A in intranuclear inclusions (NI). The images depict NI (arrows) containing accumulations of β-catenin, glutamine synthethase and KDM2A. For β-catenin and glutamine synthethase IHC increased magnification of the marked area shows positive immunostainings within NI. Further, HE-staining (bottom-left image) reveals that NI (arrows) are completely closed with no contact to the cytoplasm on this plane. Additionally increased magnification of the intranuclear inclusion in the center of the HE and KDM2A-images reveal the bordering membrane (right images; arrow). The black bars at the right (third and forth row) equal 10 µm. First and second row images are original magnifications: 400 ×; third and fourth row images are original magnifications: 1,000 ×
RESULTS
Accumulation of β-catenin and proteins associated with Wnt/β-catenin signalling in NI
Our IHC analysis of NI in HCC showed positive immunoreativity for β-catenin within NI and in the surrounding cytoplasma [Figure 1]. Further, we observed an accumulation of glutamine synthetase, a target gene of β-catenin in NI. Additionally, we detected immunopositivity for the lysine demethylase KDM2A,which is involved in β-catenin degradation. We performed serial tissue sections for β-catenin, glutamine synthetase and KDM2A IHC staining [Supplementary Figure 1]. The number of cases containing at least oneintranuclear inclusion with positive immunoreactivity was analyzed for β-catenin and KDM2A. We detected in 19 of 72 (26.4%) HCCs at least one membrane-bounded intranuclear inclusion with positive β-catenin immunostaining; for KDM2A 19 of 71 (26.8%) valid HCC cases showed at least one membrane-bounded intranuclear inclusion with positive immunostaining. β-catenin immunohistochemistry was available for all cases (72/72) while for KDM2A, suitable material for immunohistochemistry was available in 71 of 72 cases.
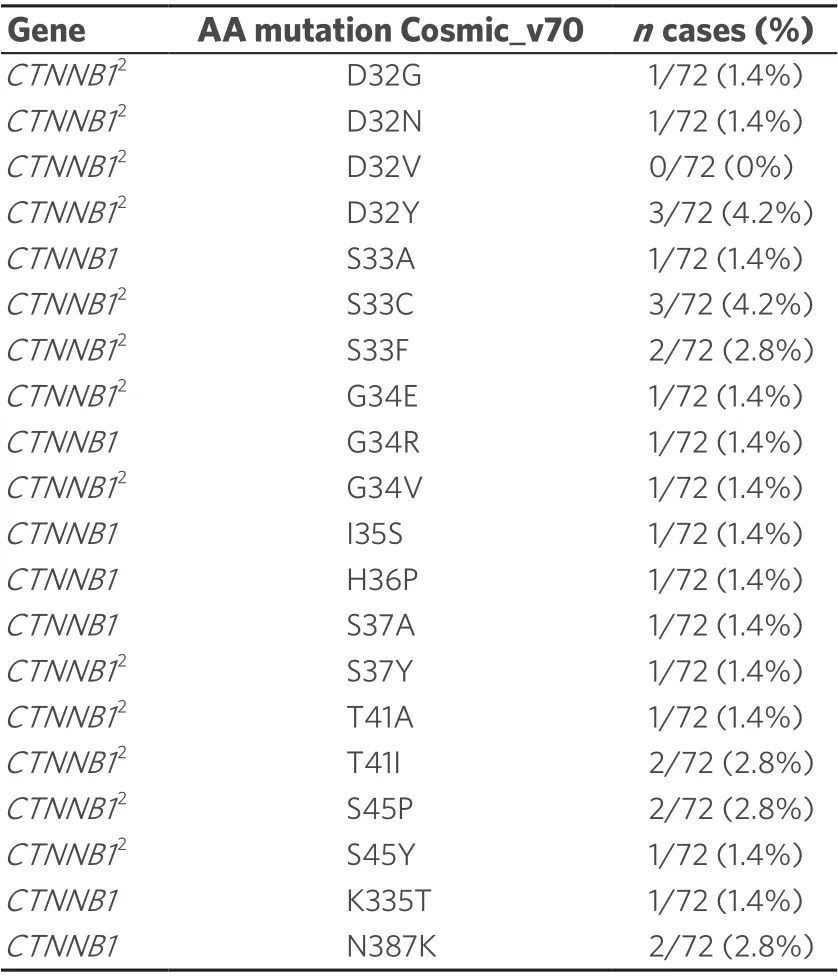
Table 2. NGS study: CTNNB1 mutations in the HCC cohort1
NGS study of CTNNB1 mutations
To clarify the reason for the high number of NI containing β-catenin, we investigated HCCs with possible mutations of the β-catenin geneCTNNB1by NGS. We found up to twenty differentCTNNB1mutations in our cases. We have listed all mutations found in COSMIC with a prevalence of at least 5%. The mutations that are also recorded in the ClinVar database are marked [Table 2]. Chi-square cross table analysis of the HCC cases demonstrated that a positiveCTNNB1mutation status was significantly associated with the occurrence of positive nuclear β-catenin immunostaining (P≤ 0.001). Additionally, we performed cross table analysis to examine a possible association between the occurrence of NI and the presence ofCTNNB1mutations; no significant association was found.
TEM analysis of NI
We performed ultrastructural analysis in order to investigate the content and shape of NI in more detail[Figure 2]. Our TEM studies showed that NI in HCC in most cases was completely enclosed by the nuclear membrane. NI contained degenerated cell material, lysosomes and heterolysosomes. In general, the content of the inclusions had a higher electron density than the cytoplasm, suggesting that NI are not simply passive invaginations of the cytoplasm into the nucleus [Figure 2].
Immunohistochemical analysis of NI
Association ofβ-catenin with autophagy-associated proteins in NI

Figure 2. Ultrastructural analysis of intranuclear inclusions (NI) in hepatocellular carcinoma. A: The image depicts two nuclei, one of which contains a large NI, which we analyzed in more detail at higher magnification; B: NI is bordered by the two (inner and outer) nuclear membranes (arrowhead) with attached heterochromatin (h); degraded cellular material resembling fine filamentous material (asterisk), a small vesicle with homogenous content of high electron-density, most probably a lysosome (black arrow) and heterolysosomes (white arrows) are seen. In addition, the NI shows fine granular, ribosome-like material (triangles) and a larger round area with no clearly detectable membrane (#) and hardly electron-dense content, which might be a lipid droplet
We analysed NI by immunohistochemistry to clarify whether there was an association between the occurrence of intranuclear inclusions with β-catenin immunopositivity and autophagy [Figure 3]. Recently,we have examined the autophagy-associated proteins p62, ubiquitin, LC3B, cathepsin B and cathepsin D in HCC demonstrating positive immunoreactivity for all investigated proteins in NI[6]. In the current study[Figure 3], we used chi-square cross table analysis to investigate if positive β-catenin immunostaining in NI is associated with immunoreactivity for the autophagy-associated proteins p62, ubiquitin, LC3B, cathepsin B and cathepsin D. We defined cases as positive (1) if at least one membrane-bound inclusion showed positive immunostaining, whereas cases lacking stained inclusions were classified as negative (0) according to the same system as described before[6]. Analysis of 19 cases with positive β-catenin immunostaining revealed that 15 also had nuclear inclusions with positive ubiquitin immunostaining (P< 0.001), 16 showed p62 and 17 had cathepsin B immunoreactivity (P≤ 0.001). We also revealed a significant relationship for LC3B (P=0.041) and cathepsin D (P= 0.005); details are listed in Table 3.
Correlation of β-catenin immunopositivity within NI with β-catenin immunopositivity in the surrounding cytoplasm
Further chi-square cross table analysis was used to investigate the relationships between β-catenin immunoreactivity within NI and in the surrounding cytoplasm. We found that of the 19 cases that contained positive β-catenin immunostaining within NI, 11 also showed β-catenin immunopositivity in the surrounding cytoplasm (P= 0.002; Table 3).
Spatial co-localization of β-catenin with p62 in nuclear inclusions
To analyse if these autophagy-associated proteins are located within the same inclusion, we performed double immunofluorescence labeling. Figure 4 shows a DAPI-stained nucleus with NI. Co-localization of β-catenin with p62 in the same inclusion is proven by the formation of the merged color yellow [Figure 4,arrows].
The occurrence of NI with KDM2A immunopositivity correlates with recurrence-free and overall survival
We performed Kaplan-Meier survival curves to check if disease specific overall survival (OS) and recurrencefree survival depends on the accumulation of KDM2A protein within the inclusions.
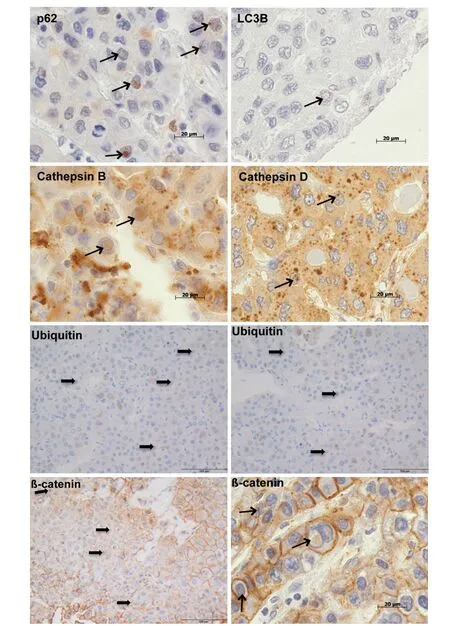
Figure 3. Immunohistochemical analysis of β-catenin and autophagy-associated proteins in hepatocellular carcinoma. The images depict intranuclear inclusions (arrows) containing positive immunoreactivity for β-catenin, p62, LC3B, ubiquitin, cathepsin B and cathepsin D. Original magnifications for ubiquitin and β-catenin (left image bottom) images: 400 × and for p62, LC3B, ubiquitin,cathepsin B, cathepsin D, β-catenin (right image bottom) images: 1,000 ×
We analyzed 10 serial sections from each TMA. Data on OS were available for 69 valid HCC cases. 19 of these 69 patients had at least one membrane-bound intranuclear inclusion showing immunoreactivity for KDM2A. We observed that most of them (16/19, 84%) survived whereas 26 (52%) of 50 patients who contained no KDMN2A protein within NI died during the observation period. Kaplan-Meier survival analysis depicted that patients with NI containing KDM2A protein accumulations displayed a lower risk for death compared to patients with lower number of KDM2A immunopositive NI (P= 0.014; Figure 5A). We also found that patients with KDM2A immunopositivity in the cytoplasm show a significant survival benefit compared to patients with KDMN2A negative cytoplasm (P= 0.009; Figure 5B). Briefly, 42 of 69 patients showed immunopositive KDM2A cytoplasm and 30 of 42 (71.4%) survived compared to 17 of 27 patients with no immunoreactivity for KDM2A in the cytoplasm who died. Further, we examined recurrencefree survival in dependence on KDMN2A immunoreactivity in the cytoplasm. Data on recurrence-free survival was available for 47 valid HCC cases. We found that significantly more patients with KDM2A immunopositive cytoplasm showed recurrence-free survival than patients with a lack of KDM2A in thecytoplasm (P= 0.009; Figure 5C); briefly 27 (84.4%) of 32 KDM2A immunopositive patients showed no local recurrence compared to 7 (46.7%) of 15 patients with lack of KDM2A in the cytoplasm and developed recurrence. Additionally, we studied the adjacent normal tissue sections (NTS) on all serial sections of the HCC patient cohort by Kaplan-Meier survival curves. We also detected in the adjacent normal tissue that the occurrence of KDM2A in the cytoplasm was associated with a significant benefit in recurrence-free survival(P= 0.027; Figure 5D).
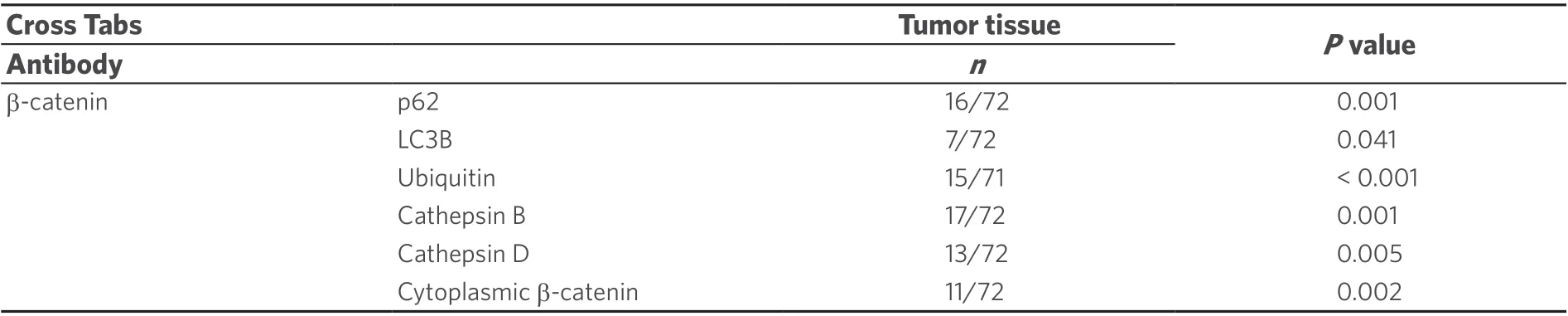
Table 3. Correlation of β-catenin immunopositivity within NI with autophagy associated proteins in NI and with cytoplasmic β-catenin
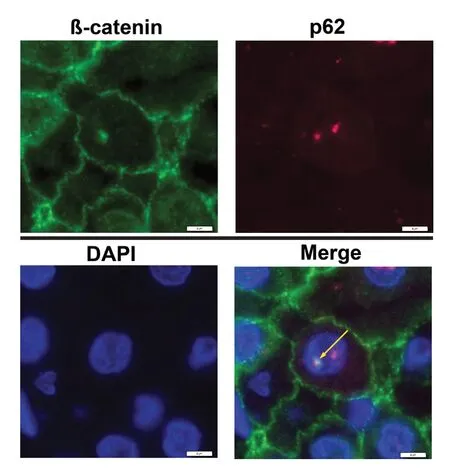
Figure 4. Double-IF studies demonstrate co-localization of β-catenin with p62 in intranuclear inclusions (NI) of hepatocellular carcinoma sections. Nuclei were stained with DAPI (blue) to detect NI. The merged images show superimposition of the signals for β-catenin (green staining) with p62 (red staining) in the same inclusion; the merged color yellow (arrow) proves co-localizations
Increased occurrence of NI in HCC cases with KDM2A immunopositivity
Mann-WhitneyUTest analysis showed a significant positive correlation between the number of NI and positive KDM2A immunostaining in HCC (P≤ 0.001; Figure 6). Our results revealed that cases with at least one KDM2A immunopositive inclusion had, at the same time, a significantly higher number of NI than HCCs lacking KDM2A immunoreactivity within NI.
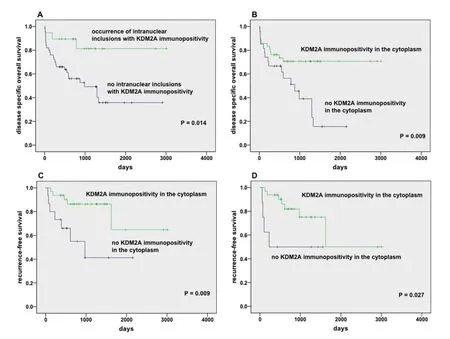
Figure 5. Kaplan-Meier survival studies. Disease specific OS was analyzed in 69 valid hepatocellular carcinoma (HCC) samples in relation to the presence of KDM2A immunopositivity in intranuclear inclusions (A) and in the cytoplasm (B). Additionally, recurrencefree survival was examined in 47 valid HCC samples in relation to the presence of KDM2A immunopositivity in the cytoplasm of tumor(A) and in adjacent normal tissue (B). We counted inclusions in a standardized area of 23.5 mm2 (ten serial sections of each case). Only cases with evaluable material on all ten serial sections were included. Cases containing at least one membrane-bound nuclear inclusion were considered positive; P M 0.05 was defined as statistically significant
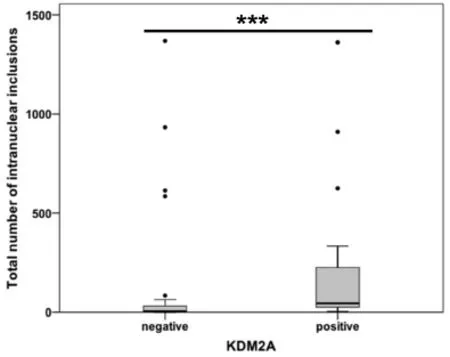
Figure 6. Association between KDM2A immunopositivity and intranuclear inclusions (NI) in hepatocellular carcinoma. The diagram depicts a significant association between positive KDM2A immunoreactivity within NI and the occurrence of NI. ⋆⋆⋆P ≤ 0.001
DISCUSSION
In the present study, we have demonstrated accumulation of β-catenin and proteins associated with the Wnt/beta-Catenin pathway in NI, partly together with autophagy-associated proteins in the same inclusion. The presence of inclusions with KDM2A immunopositivity correlated significantly with overall survival in HCC.We found positive immunostaining for β-catenin, glutamin syntethase and KDM2A within NI [Figure 1]. In addition, other immunohistochemical studies have reported strong immunopositivity for β-catenin within NI in thyroid carcinomas and pulmonary neuroendocrine tumors[17,18]. Rezket al.[18]reported that some cases had β-catenin positive inclusions although they had almost negative cytoplasmic/nuclear staining. This is in line with our results. Though we found in 11 of 19 cases a correlation of β-catenin immunopositivity within NI with immunopositivity in the surrounding cytoplasm, there were 8 cases nearly lacking in cytoplasmic staining. Rezket al.[18]suggested that β-catenin might play a role in the development of NI as it is involved in organizing actin and microtubule polymerization[18,19]. Additionally, Gamachi et al. also observed NI with immunopositivity for biotin, biotin-binding enzymes and β-catenin in pregnancy-related endometrium and morule-associated neoplastic lesions[20]; they revealed that though biotin and biotin-binding enzymes were present in neoplastic and non-neoplastic tissues, β-catenin was lacking in non-neoplastic endometrial lesions. Further, NI with immunoreactivity for PAX8 protein were observed in conventional clear cell renal cell carcinoma[21]. The immunopositivity for PAX8 is interesting as it is a transcription factor that is also involved in the proliferation of tumor cells via the Wnt/β-catenin pathway; overexpression of PAX8 has been reported in various carcinomas[21]. These studies did not clarify why NI contained these proteins. In our current study, to clarify the reason for β-catenin immunopositivity in NI, we examined our HCC cases on CTNNB1 exon 3 mutations. We differentiated between immunopositivity within the nucleus and in the intranuclear inclusion and found up to 20 different CTNNB1 mutations, all of which are listed in the COSMIC catalog and most, reported in the NCBI ClinVar database. A positive mutation status for CTNNB1 correlated significantly with nuclear immunopositivity, which is supported by the literature as CTNNB1 mutations are known to be associated with the translocation of β-catenin proteins from the membrane to the nucleus and activation of Wnt/β-catenin signalling[22]. However, we found no association between CTNNB1 mutations and positive β-catenin-immunostaining within the inclusions. Recently, we have shown that NI in HCC contained autophagy-associated proteins[6]. Since we could simultaneously detect degraded cell material and lysosomes in NI by TEM studies, we assumed that biological processes similar to autophagy are taking place in NI[6]. These autophagy-associated proteins were partly co-localized within the same NI.In the current study, we demonstrate that cases with β-catenin immunopostive NI also harbored NI with immunopositivity for autophagy-associated proteins; this association was significant for p62, cathepsinB/D, ubiquitin and LC3B. We found that p62 immunoreactivity was almost exclusively located in NI whereby cytoplasmic staining was diffuse and very weak. We suggest that high p62 positivity in NI could be caused by several reasons including alterations in p62 nucleocytoplasmic shuttling[23]as previously reported in our HCC study[6]. Intriguingly, we also detected co-localization of β-catenin with p62. The latter strongly suggests that biological processes with involvement of β-catenin are taking place within NI. We found in 11 of 19 cases a significant correlation between β-catenin immunopositivity within NI and β-catenin immunoreactivity in the surrounding cytoplasm, and we suppose that some NI with positive β-catenin immunostaining have developed by invagination of the cytoplasm into the NI with further closure of the invagination.
To clarify the factors contributing to β-catenin accumulation in NI, we also studied the immunoreactivity of proteins associated with the Wnt/β-catenin pathway. We detected immunopositivity within NI for glutamine synthetase, which is a target of β-catenin signalling implicated in the development of HCC[7,24,25]and for KDM2A having a role in β-catenin degradation[14,26,27]. Luet al.[14]reported that lysine demethylase KDM2A demethylases nuclear β-catenin, which then induces the degradation of β-catenin and consequently, the downregulation of Wnt/β-catenin target genes. In our study, the presence of NI containing KDM2A protein was significantly associated with longer survival in HCC patients; this is in line with our previous work in HCC demonstrating by Kaplan-Meier survival curves that the presence of NI significantly correlated with survival[6]. Additionally, HCC patients with KDM2A immunopositivity in the cytoplasm showed a significant benefit in both, disease-specific OS and recurrence-free survival. Further, we found that even the presence of immunopositive KDM2A in the cytoplasm of normal tissues adjacent to tumor was associated significantly with recurrence-free survival. It has been documented by immunohistochemical studies that KDM2A positive staining, though predominantly found in the nucleus, can also be detected in the cytoplasm[28].Luet al.[14]reported methylation by KDM2A in both non-phosphorylated and phosphorylated β-catenin,and in both the cytosol and the nucleus; they suppose that methylation/demethylation plays a role in modulating β-catenin activity. Henderson et al. described that β-catenin can be exported from the nucleus to the cytoplasm where its levels are regulated by degradation[29]. Our results suggest that methylation of β-catenin by KDM2A and consequently, repressing of Wnt/β-catenin signaling might play a role in the beneficial survival of patients with immunopositive inclusions and cytoplasm. However, mechanistically driven experiments are necessary to prove this hypothesis. The role of KDM2A in disease is contradictory and seems to be context dependent[26]. On one hand, KDM2A is upregulated in ovarian[30], breast and lung cancer[13,30,31]and associated with a poor prognosis; on the other, it is downregulated in prostate cancer[32]and in the liver, KDM2A regulates hepatic gluconeogenesis whereby its exogenous expression reduces blood glucose levels[33]. Reasons for the dual effects of KDM2A may be due to the involvement of KDM2A in various biological signaling pathways, including interacting with the p53-binding protein and NFkappaB activity by demethylation of the p65 subunit of NF-kappaB[34]. Further, we found that the increased occurrence of NI was significantly associated with KDM2A immunopositivity in NI. KDM2A is described as a heterochromatin-associated and HP1-interacting protein[32]and KDM2A is required to sustain centromeric integrity and genomic stability, particularly during mitosis[32]. A change in chromatin stability has been discussed as a factor contributing to the development of NI[6,16,35,36]. Thus, we suppose that KDM2A might play a role in the formation of inclusions. The mechanisms that participate in the formation of inclusions are still poorly understood, in spite of several investigations[35,37]. However, we believe that various factors can induce the formation of inclusions and that KDM2A may be one.
In this study, we demonstrated accumulations of β-catenin and proteins associated with the Wnt/β-catenin pathway in NI. The simultaneous presence of β-catenin with autophagy-associated proteins, partly colocalized to the same inclusion suggests that biological processes similar to autophagy might take place in NI. Further, we found that the presence of KDM2A immnuopositive NI provides a survival benefit to HCC patients. To what extent this is related to possible degradation of β-catenin by KDM2A, and whether NI play a role in these biological processes needs to be analyzed by further experiments.
DECLARATIONS
Acknowledgments
We thank Dorothe Möllmann (Institute of Pathology, University Hospital of Essen, University of Duisburg-Essen) for her excellent technical assistance. Additionally, the authors thank Laura Malkus (Institute of Pathology, University Hospital of Essen, University of Duisburg-Essen) for technical assistance in the preparation of the samples for TEM.
Authors’ contributions
Made substantial contributions to conception and design of the study and performed data analysis and interpretation: Schwertheim S, Baba HA, Jastrow H, Herold T
Conceptualization: Schwertheim S, Baba HA, Schmid KW
Data acquisition: Herold T, Jastrow H, Baba HA, Schmid KW
Investigation: Schwertheim S, Theurer S, Jastrow H, Herold T, Ting S, Kälsch J, Baba HA, Schmid KW
Methodology: Schwertheim S, Theurer S, Jastrow H, Herold T, Ting S, Kälsch J, Baba HA
Supervision: Schwertheim S, Baba HA, Schmid KW
Writing - original draft preparation: Schwertheim S, Baba HA
Writing - review and editing: Schwertheim S, Theurer S, Jastrow H, Herold T, Ting S, Kälsch J, Baba HA,Schmid KW
Availability of data and materials
The source of the data came from the Institute of Pathology, University Hospital of Essen.
Financial support and sponsorship
None.
Conflicts of interest
All authors declared that there are no conflicts of interest.
Ethical approval and consent to participate
Informed consent was obtained from each patient. The study complied with the Helsinki Declaration of 1975 and the Ethics Committee (Institutional Review Board) of the University Hospital Essen (reference number:16-6917-BO).
Consent for publication
Not applicable.
Copyright
© The Author(s) 2020.
- Hepatoma Research的其它文章
- Risk factors of portal vein thrombosis after splenectomy in patients with liver cirrhosis
- Stereotactic body radiation therapy for the management of HCC
- Genetics of alcohol-related hepatocellular carcinoma - its role in risk prediction
- Comprehending the therapeutic effects of stereotactic body radiation therapy for small hepatocellular carcinomas based on imagings
- Imaging assessment after SBRT for hepatocellular carcinoma

