Mechanisms of Quzhisanwei formula for non-alcoholic steatohepatitis based on a network pharmacology approach
Hai-Sheng Chai, Ting-Ting Shen, Yan-Hong Liu, Rong-Kun Yin, Zhi-Qin Wu, Yue-Lan Wu, Zhen-Yu Tan, Shang-Wei Hou*, Qin Zhang*
Mechanisms of Quzhisanwei formula for non-alcoholic steatohepatitis based on a network pharmacology approach
Hai-Sheng Chai1#, Ting-Ting Shen1#, Yan-Hong Liu1, Rong-Kun Yin1, Zhi-Qin Wu1, Yue-Lan Wu1, Zhen-Yu Tan1, Shang-Wei Hou1*, Qin Zhang1*
1Institution of Tongren Hospital, Shang Hai Jiao Tong University School of Medicine, ShangHai 200050, China.
The experienced prescription Quzhisanwei formula, a traditional Chinese medicine formula, has been demonstrated to be effective for the treatment of non-alcoholic steatohepatitis. However,the underlying pharmacological mechanisms remain unclear. This study aims to explore the potential action mechanisms of the experienced prescription Quzhisanwei formula in the treatment of non-alcoholic steatohepatitis.A network pharmacology approach including absorption, distribution, metabolism, and excretion evaluation, target prediction, known therapeutic targets collection, interacting pathways and network construction was used in this study. The active components and potential targets of the experienced prescription Quzhisanwei formula were screened by Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform, Swisspredict. Then, the targets of non-alcoholic steatohepatitis were collected in the GeneCards. Additionally, gene ontology and Kyoto Encyclopedia of Genes and Genomes pathway analyses were performed using WebGestalt. Drug-target-pathway networks were constructed using Cytoscape to give a visual view.It showed that 27 compounds and 89 potential targets were obtained in this work. These targets were further mapped to 109 gene ontology biological process terms. And the potential regulatory signaling pathway included non-alcoholic fatty liver disease, insulin, HIF-1 signaling pathway, PPAR signaling pathway, pathways in cancer, AGE-RAGE signaling pathway in diabetic complications, and so on.This study indicated that active compounds of the experienced prescription Quzhisanwei formula mainly had multi-target and holistic therapeutic effects on non-alcoholic steatohepatitis. However, in vivo and in vitro experiments are still needed.
Network pharmacology, Non-alcoholic steatohepatitis, Quzhisanwei formula
The experienced prescription Quzhisanwei formula has been demonstrated to be effective for non-alcoholic steatohepatitis. However, the pharmacological mechanisms remain unclear. This study investigated the potential mechanisms of the experienced prescription Quzhisanwei formula for non-alcoholic steatohepatitis. It indicated that active compounds of the experienced prescription Quzhisanwei formula mainly had multi-target and holistic therapeutic effects on non-alcoholic steatohepatitis. However, in vivo and in vitro experiments are still needed.
Background
Non-alcoholic steatohepatitis (NASH) is characterized by hepatic steatosis, inflammation, hepatocyte damage,and liver fibrosis [1]. The risk of cirrhosis,decompensated liver disease,and hepatocellular carcinoma is increasing in NASH patients, especially with advanced liver fibrosis [2].Recently, NASH has become the third most common sign for liver transplantation in the United States [3].
To date, the underlying mechanism of NASH is largely unknown.There is no effective pharmacotherapy for NASH except for lifestyle modification by diet and exercise. Lifestyle modification alone cannot be enough to reverse NASH-related liver fibrosis [4].Generally, treatments should control risk factors, such as obesity, insulin resistance, and hyperlipidemia. Drugs increasing insulin sensitivity, specifically the thiazolidinedione, are considered as therapeutic options for steatohepatitis. The experienced prescription Quzhisanwei formula (QZSWF) is composed of Huzhang (, PC), Shanzha (, CF), and Juemingzi (, CA), has been widely used for epilepsy in China. A clinical study has shown that QZSWF is safe and effective for the treatment of NASH, which can relieve NASH symptoms such as triglyceride, and controlled attenuation parameter of fibroscan [5]. QZSWF has been shown to exert a wide spectrum of pharmacological activities to improve liver steatosis, hepatic glucuronosyl transferases, and blood lipids in a high-fat diet-induced non-alcoholic fatty liver disease (NAFLD) models [6].
However, the underlying pharmacological mechanisms of QZSWF for NASH remain unclear. Previously,it is difficult to investigate the molecular mechanism of traditional Chinese medicine (TCM), especially TCM formula, due to the complex chemical composition and complex interactions among multiple components. Now, network pharmacology is applied accurate identification of therapeutic targets and biological mechanisms of ingredientsin herbs for treating disease [7].
Network pharmacology, including chemoinformatics, bioinformatics, network biology, and pharmacology, is a comprehensive method to reveal the bioactive components and potential mechanisms of TCM formulas from a systemic perspective [8].In this study, we aimed to investigate the mechanisms of QZSWF for treating NASH by a comprehensive network pharmacology-based approach.
Materials and methods
Screening of candidate compounds and identification of active compounds for the treatment of NASH with QZSWF
Compound information of QZSWF was collected from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform, which includes information of all 500 Chinese herbal medicines registered in thewith a total of 30,069 ingredients collected through literature mining and database integration [9]. Comprehensive data related to the pharmacokinetic properties of each chemical compound, including oral bioavailability (OB), intestinal epithelial permeability, drug-likeness (DL), blood-brain barrier, drug half-life, and Lipinski’s rule of five, are provided for the screening and evaluation of the compounds [9]. OB is one of the most important pharmacokinetic parameters in the absorption, distribution, metabolism, and excretion (ADME) processes [10].
Prior to target prediction, ADME was used to select bioactive components that conduce to its therapeutic effects, while those with poor pharmacological properties and bad drug ability compounds were removed [11]. For the purpose of obtaining compounds with higher oral absorption, utilization, and biological properties for further analysis, the active molecules were defined as those with OB indices ≥ 0.30 and DL index ≥ 0.18.
Identification of associated proteins and gene names
The protein targets of the compounds were retrieved from Swiss Target Prediction (http://www.swisstargetprediction.ch/), which is a database for target prediction of bioactive small molecules based on a combination of 2D and 3D similarity measures with known ligands [12]. The potential targets were carried out using the SMILES format of each constructed structure, and homo sapiens were chosen as the source of the target. The UniProt Knowledgebase (UniProtKB) is a protein database partially built by experts and contains 54,247,468 sequence entries. Furthermore, the gene names were extracted from UniProtKB (http://www.uniprot.org). Finally, the targets were mapped by theHuman Gene Database named GeneCards (http://www.genecards.org/) to obtain the targets related to their corresponding diseases.
Analysis by GeneMANIA
GeneMANIA (http://www.genemania.org) is a user-friendly and flexible web server for generating hypotheses regarding gene function, analyzing gene lists, and prioritizing genes for functional assays. Given a query list, GeneMANIA can list the genes that have shared properties, or function similar with the original query. It also shows a functional relationship network, expounding the association among the lists as well as curated genomics and proteomics data. The potential candidate target genes were entered into the search bar after selecting homo sapiens from the organism option, and the results were further collated.
Protein-protein interaction
The retrieved QZSWF active ingredient targets and their coexpressed protein are correlated with NASH targets by the Metascape [13](http://metascape.org).For each given gene list, protein-protein interaction (PPI) enrichment analysis has been carried out with the following databases: BioGrid, InWeb_IM, OmniPath. The resultant network contains the subset of proteins that form physical interactions with at least one other member in the list. If the network contains 3 to 500 proteins, the molecular complex detection (MCODE) algorithm [14] was applied to identify densely connected network components.
Gene function and pathway enrichment analysis
For the investigation of the underlying mechanism of 109 identified targets, genes of functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways analyses were performed to explore the above QZSWF function in the treatment of NASH. Webgestalt [15](http://www.webgestalt.org) which was updated in 2019, is a web-based tool that provides gene functional annotation and enrichment analysis. In the present study, gene ontology (GO) analysis of potential candidate targets and their interacting proteins was performed for the gene function annotation by Webgestalt (Overrepresentation Enrichment Analysis method), and another tool, Metascape [13](http://metascape.org) was applied for KEGG pathway enrichment analysis to illustrate which pathways may contribute to the treatment mechanism of QZSWF for NASH.Import the above common target into Metascape, set the species as human. Keep the results of protein interaction (BioGrid database) and module analysis, and analyze the results. Keep the results of GO biological process, Reactome pathway analysis, and KEGG pathway analysis, sort the results according to the number of targets, and retain the top-ranking results.
Network construction
Using candidate compounds and potential targets, a compound-tar acquisition network was constructed. The target pathway network is generated by linking potential targets to the signal pathways they participate in. The network is built using Cytoscape 3.6.1 software [16].
Results
Active compounds identification
A total of 187 compounds were collected,comprising 62 in PC, 57 in CF, and 68 in CA.Specifically,the chemical components with OB≥30% and DL≥0.18 were selected as candidate active ingredients.After eliminating those target-free chemical components,we obtained a total of 27 active ingredients,among which 10 components belong to PC, 4 belong to CF,and 13 belong to CA.Remarkably,among the 27 chemical components,some of which are present in a variety of TCMs. For example, rhein is the common compound of PC and CA(Table 1).
Identification of related proteins and genes of QZSWF
Based on the target fishing approach introduced above,a total of 2,067 target genes for the 27 compounds were obtained from the Swisspredict.Subsequently,the target genes were mapped to the GeneCards.Finally,89 target genes associated with NASH were remained (Table 2),and 26 compounds were finally obtained after removal of 1 candidate compound (MOL002280) without any relevant targets. These 89 target genes were considered to have effective therapeutic actions in the pathogenesis of QZSWF, and were used for further analysis.
GeneMANIA analysis
Among the 89 target genes and their interacting proteins,it was found that 26.15% had physical interactions,9.78% exerted co-localization,and 39.65% displayed similar co-expression characteristics.Other results of pathway,shared protein domains and genetic interactionsare shown in Figure 2.Finally, 109 targets were identified for further functional study.
GO and pathway analysis of targets genes
GO and KEGG enrichment analyses were carried out using WebGestalt and Metascpe. The top ten functions were metabolic process (102/109), biological regulation (100/109), response to the stimulus (97/109), localization (83/109), cell comunication (82/109), multicellular organismal component processess (80/109), cellular component organization (73/109), developmental process (71/109), cell proliferation (56/109, Figure 1). These functional terms are highly associated with inflammatory activities, sugar, and lipids metabolic, especially in NAFLD or NASH.
After pathway analysis, the 109 associated protein participated in the top 20 KEGG pathways with significant false discovery rate-adjusted-value including HIF-1 signaling pathway, PPAR signaling pathway, pathways in cancer, AGE-RAGE signaling pathway in diabetic complications and so on (Figure 5).
Network analysis
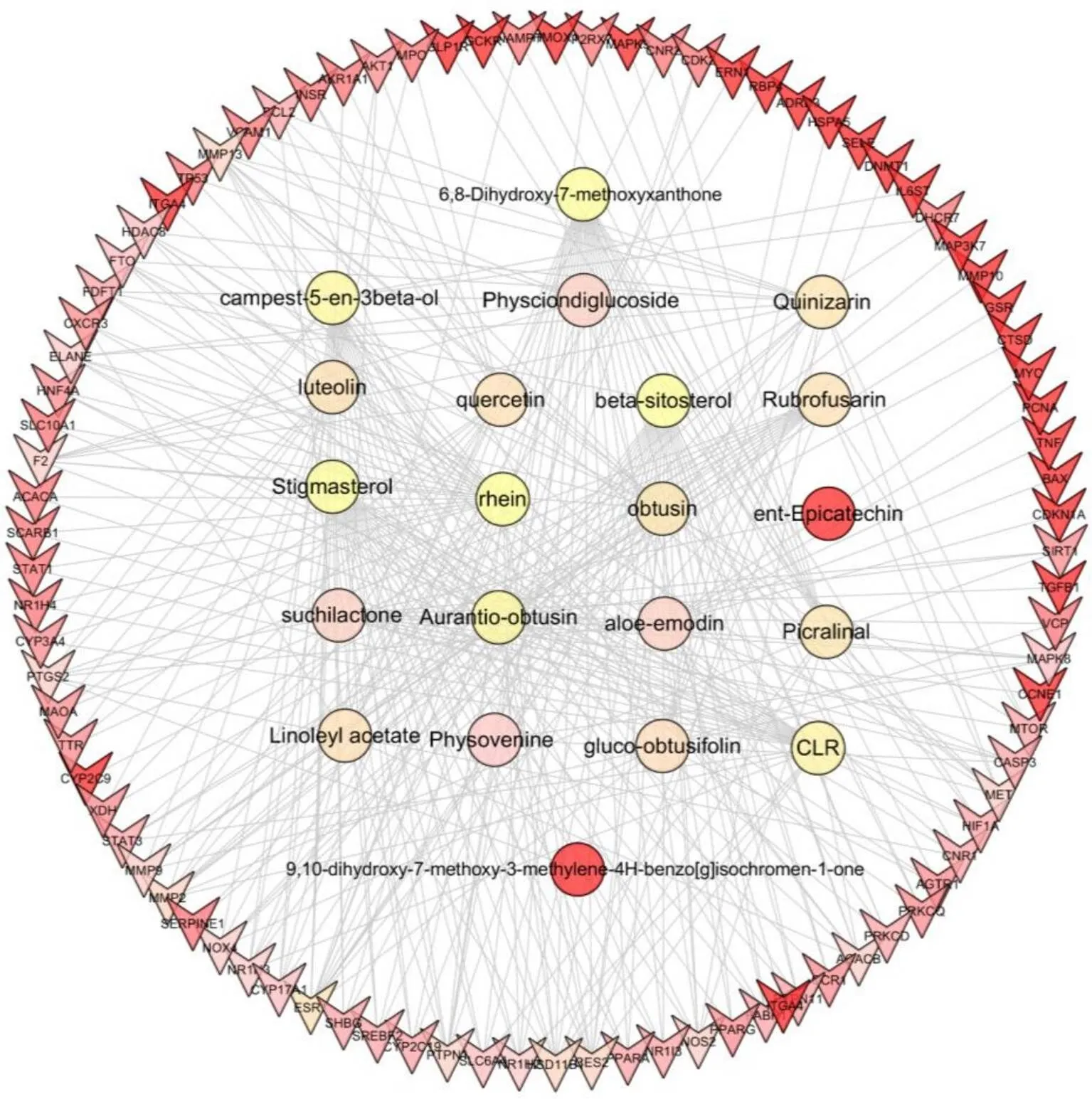
Based on target and pathway analyses, an entire compound and targets, targets and pathways network were constructed using Cytoscape3.6.1. This compound, targets, and diseases interaction network have 80 nodes and 129 edges. The red oblong, green inverted triangles, and blue circles correspond to geraniol, target genes, and pathways, respectively (Figure 3).

Table 1 Information on candidate active compounds from PC,CA, and CF herbs
PC, Huzhang (); CF, Shanzha (); CA, Juemingzi ();OB, oral bioavailability; DL, drug-likeness.
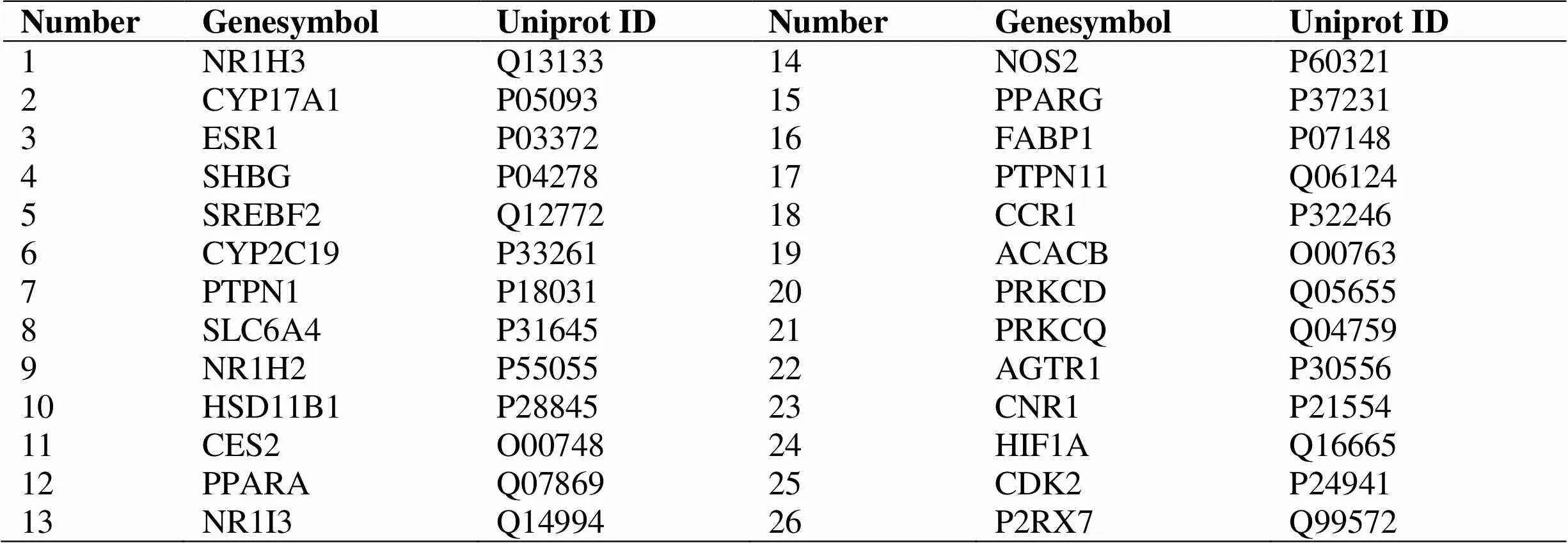
Table 2 The proteins and genes QSWF treatment of NASH
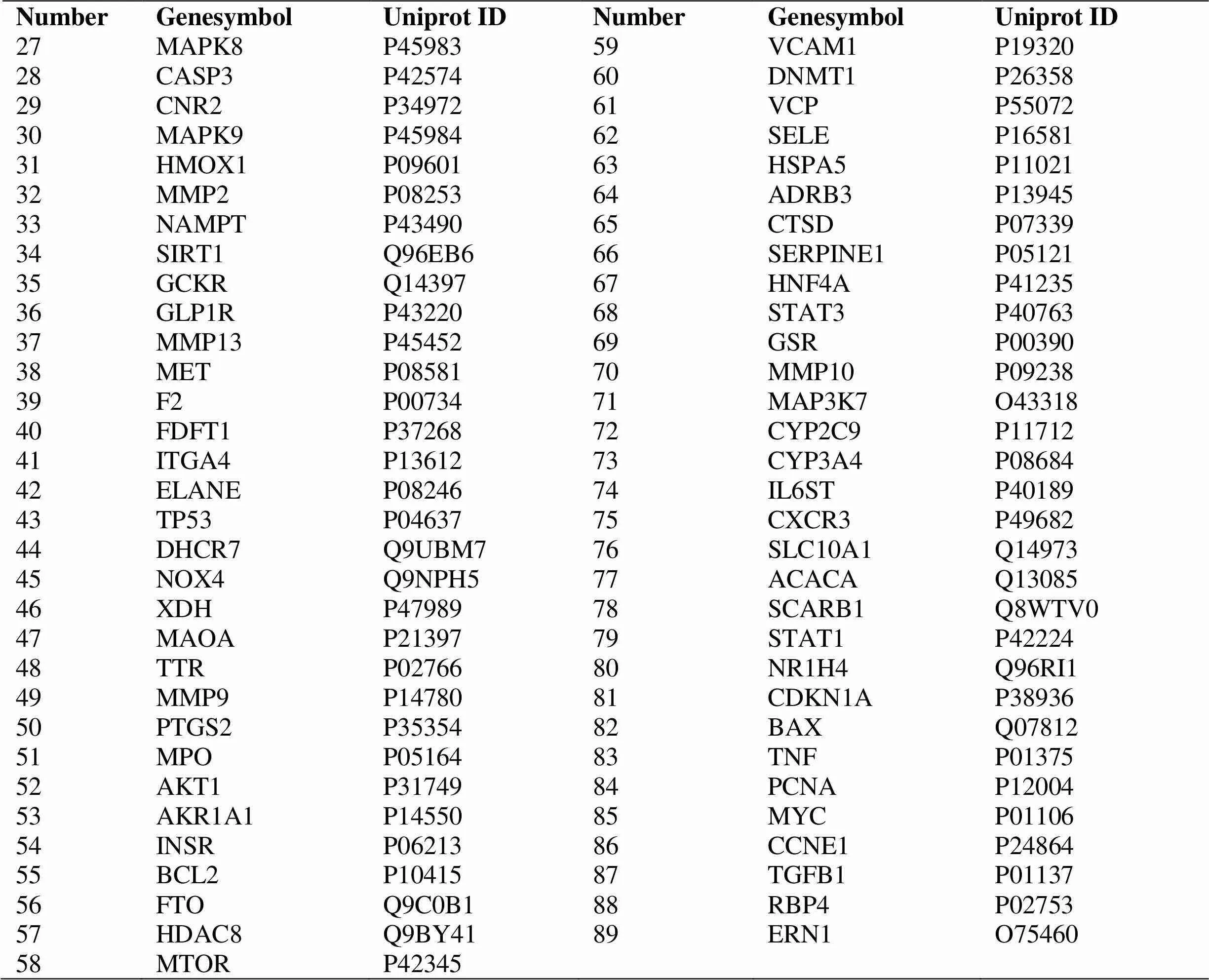
Table 2 The proteins and genes QSWF treatment of NASH (continued)
QZSWF, the experienced prescriptionQuzhisanwei formula; NASH, non-alcoholic steatohepatitis.
PPI analysis of targets genes
The MCODE algorithm has been applied to identify densely connected network components of the proteins network (Figure 4). The key target genes were found by MCODE algorithm using Metascape. Combine protein interaction data with pathway and biological process enrichment data to add biological significance to the substructures identified in the protein interaction network. The three most significantly related GO terms in each MOCDE were kept in this PPI network. There are 7 NASH-related targets in those clusters.Cluster1 includes BCL2,BID,BAX,MCL1,PCNA,MAPK9,BCL2L11,SIRT1,HNF4A,DNMT1,PTPN11,STAT5B,CDKN1B and JAK2.Cluster 2 gets TP53,VCP,MYC,MAPK8,STAT1,CDK2,CDKN1A,HSPA5,ESR1,MDM2,JUN, and HIF1A.TNF,CASP3,MAP3K7,AKT1,INSR,PTPN1 and STAT3 were involved in cluster3.CNR1,CNR2,CCR1 and CXCR3 belong to cluster4.CYP3A4,CYP17A1,CYP2C9 and CYP2C19 belong to cluster5.PPARG,NR1H2 and NR1H3 belong to cluster 6.PPARA,RXRA and FABP1 are the member of cluster 7.
Discussion
NASH is a severe disease of liver that occurs in 3%–7% of population. It can cause fibrosis, cirrhosis and hepatocellular carcinoma, negatively impacting the patient’s quality of life and resulting in significant financial burden. The aim of treatments in those patients is to improve inflammation, fibrosis and to positively improve the patient’s quality of life. Currently, the therapies for these patients only are based on the control of associated conditions such as: obesity, diabetes mellitus and hypertriglyceridemia, as well as discontinuation of hepatotoxic drugs. The recommendation of the loss of excess weight and physical exercises should always be recommended, regardless of the histopathological grade of the patient’s liver biopsy, which can effectively improve insulin sensitivity and may decrease hepatic steatosis and the degree of inflammation [17, 18]. Despite the high prevalence of hepatic steatosis and the potential to disease progress to fibrosis and cirrhosis, there was no approved drug treatments for NASH [19]. Therefore, better understandings of the possible mechanisms might contribute to more specificity and effectiveness of the therapeutic regimens. The 10-year application of QZSWF has confirmed its therapeutic effect on NASH. However, the pharmacological mechanisms are still not clear completely.
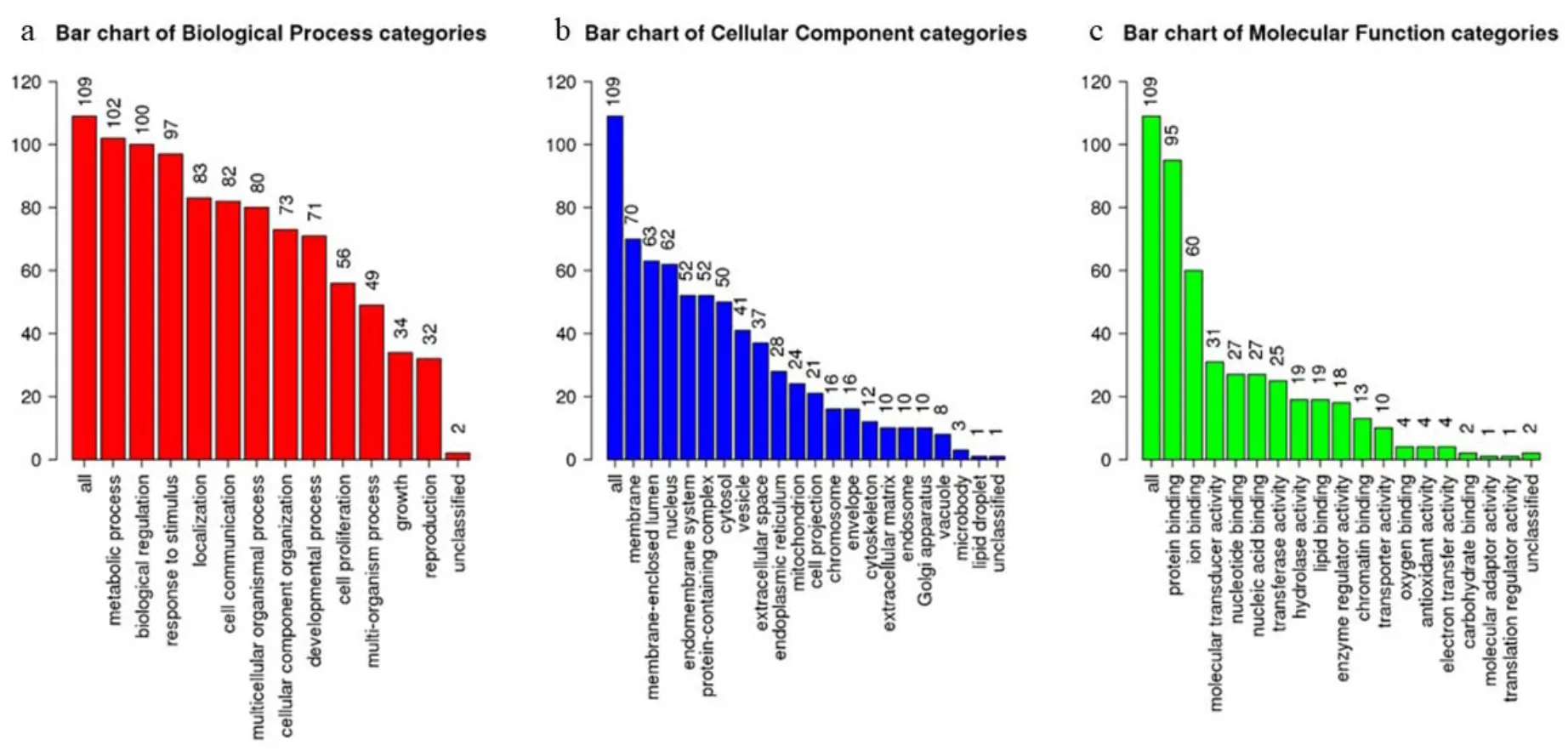
Figure 1 GO map of putative target genes. (a) Biological process categories. (b) Cellular component categories. (c) Molecular function categories.GO, gene ontology.
Figure 2 Protein network of QZSWF on NASH. Black nodes represent target proteins, and connecting colors indicate different correlations. Functional associations between targets were investigated using GeneMANIA. Different colors of the network edge indicate the bioinformatics methods applied: co-expression, website prediction, pathway, physical interactions and co-localization. The different colors for the network nodes indicate the biological functions of the set of enrichment genes. QZSWF, the experienced prescriptionQuzhisanwei formula; NASH, non-alcoholic steatohepatitis.

Figure 3 QZSWF compounds-target network. The rectangle, inverted triangle and circle correspond to compoud, target gene and pathway, respectively. Different colors indicate different weights in the network. QZSWF, the experienced prescriptionQuzhisanwei formula.
Figure 4 QZSWF for NASH target protein interaction network and module analysis.Different colors represent different lists, indicating the sharing and association between different list genes in the protein interaction module. QZSWF, the experienced prescriptionQuzhisanwei formula; NASH, non-alcoholic steatohepatitis.
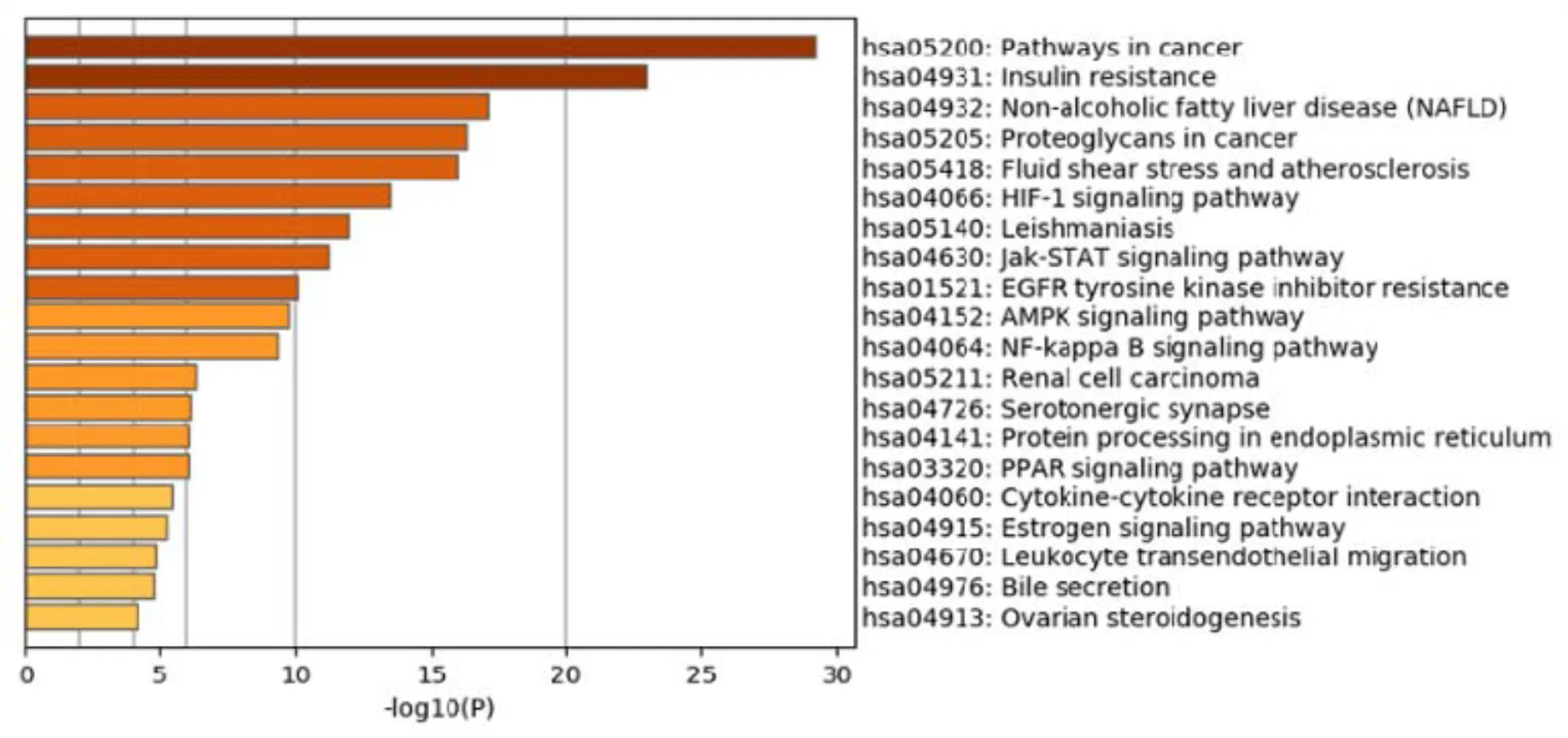
Fig 5 KEGG pathway analysis of putative target genes. The 89 targets participate in 10 KEGG pathways with significant FDR-adjusted-value including pathways in cancer, insulin resistance, NAFLD, and so on.KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate; NAFLD, non-alcoholic fatty liver disease.
In our study, we analyzed the molecular targets and involved pathway mechanisms of QZSWF based on network pharmacology. For this reason, we mainly focused on three aspects. Firstly, common targets of QZSWF for NASH were enriched in pathological changes of insulin resistance (INSR, PTPN1, AKT1, STAT3), cancer and hypoxic stress (HIF-1 signaling pathway, EGFR), apoptosis (BCL2, MCL1, BCL2L11, BAX, BID, MAPK9) and PPAR signaling pathway (FABP1, PPARA, RXRA). Secondly, enrichment analysis suggested the regulatory effects of QZSWF on the insulin resistance and that the apoptosis may be the most important mechanism of NASH treatment. Thirdly, HIF regulation was also considered as the critical section of QZSWF for NASH. To sum up, the therapeutic effect of QZSWF for NASH was mediated by controlling the levels of its targets, which has been shown to be critically related to NASH progression in a multicomponent, multitarget, and multilink pattern.
We cautiously speculate that the main mechanism of the QZSWF formula in treating NASH may be with targeting multiple goals, exerting an anti-inflammatory, anti-hypoxia-oxidative stress injury, improvement of insulin resistance and even anti-fibrosis, anti-tumor multi-directional pharmacological treatment. Indeed, this has been proved by accumulating evidence, which polygonum cuspidatum and its active ingredients have systematic pharmacological effects on liver lipid accumulation [20], NAFLD [21] and cancer [22] by targeting multiple proteins and pathways, as well as CA [23, 24] and CF [25]. Multiple target therapeutic medicaments are more effective for the treatment of complex diseases, such as NASH [26].
Conclusion
In summary, this study provides novel insight into the perspectives and challenges for QZSWF research and its application in future clinical investigation. However, in vivo and in vitro experiments are needed to provide more information on the mechanism of QZSWF formula.
1. Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012, 55: 2005–2023.
2. Younossi ZM, Blissett D, Blissett R, et al. The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 2016, 64: 1577–1586.
3. Takahashi Y, Sugimoto K, Inui H, et al. Current pharmacological therapies for nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J Gastroenterol 2015, 21: 3777–3785.
4. Chitturi S, Wong VW, Chan WK, et al. The Asia-Pacific working party on non-alcoholic fatty liver disease guidelines 2017-part 2: management and special groups. J Gastroenterol Hepatol 2018, 33: 86–98.
5. Chai HS, Zhang Q. Clinical efficacy and safety analysis of Quzhi Sanwei recipe for non-alcoholic steatohepatitis. Chin J Hepatol 2018, 23: 778–781.
6. Shen TT, Fan L, Zheng C, et al. Effects of Quzhi Sanwei Fang on liver function and serum lipid in mice with nonalcoholic fatty liver disease. J Tradit Chin Med 2017, 6: 175–182.
7. Su M, Guo C, Liu M, et al. Therapeutic targets of vitamin C on liver injury and associated biological mechanisms: a study of network pharmacology. Int Immunopharmacol 2019, 66: 383–387.
8. Song W, Ni S, Fu Y, et al. Uncovering the mechanism of Maxing Ganshi decoction on asthma from a systematic perspective: a network pharmacology study. Sci Rep 2018, 8: 1–11.
9. Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014, 6: 1–6.
10. Xu X, Zhang W, Huang C, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012, 13: 6964–6982.
11. Li B, Rui J, Ding X, et al. Exploring the multicomponent synergy mechanism of Banxia Xiexin decoction on irritable bowel syndrome by a systems pharmacology strategy. J Ethnopharmacol 2019, 233: 158–168.
12. Gfeller D, Grosdidier A, Wirth M, et al. SwissTargetPrediction: a web server for target prediction of bioactive small molecules. Nucleic Acids Res 2014, 42: 32–38.
13. Tripathi S, Pohl MO, Zhou Y, et al. Meta-and orthogonal integration of influenza "OMICs" data defines a role for UBR4 in virus budding. Cell Host Microbe 2015, 18: 723–735.
14. Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003, 4: 1–27.
15. Liao Y, Wang J, Jaehnig EJ, et al. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res 2019, 47: 199–205.
16. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003, 13: 2498–2504.
17. Younossi ZM, Loomba R, Rinella ME, et al. Current and future therapeutic regimens for nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatology 2018, 68: 361–371.
18. European Association for the Study of the Liver (EASL), European Association for the Study of Diabetes (EASD), European Association for the Study of Obesity (EASO). EASL-EASD-EASO clinical practice guidelines for the management of non-alcoholic fatty liver disease.J Hepatol 2016, 64: 1388–1402.
19. Kleiner DE, Makhlouf HR. Histology of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis in adults and children. Clin Liver Dis 2016, 20: 293–312.
20. Zhao XJ, Chen L, Zhao Y, et al.extractattenuates fructose-induced liver lipid accumulation through inhibiting Keap1 and activating Nrf2 antioxidant pathway. Phytomedicine 2019, 63: 1–9.
21. Pan B, Shi X, Ding T, et al. Unraveling the action mechanism of polygonum cuspidatum by a network pharmacology approach. Am J Transl Res 2019, 11: 6790–6811.
22. Wu X, Li Q, Feng Y, et al. Antitumor research of the active ingredients from traditional Chinese medical plant. Evid Based Complement Alternat Med 2018, 2018: 2–10.
23. Meng Y, Liu Y, Fang N, et al. Hepatoprotective effects ofethanol extract on non-alcoholic fatty liver disease in experimental rat. Pharm Biol 2019, 57: 98–104.
24. Tzeng TF, Lu HJ, Liou SS, et al.(leguminosae) seed extract alleviates high-fat diet-induced nonalcoholic fatty liver. Food Chem Toxicol 2013, 51: 194–201.
25. Liu ZL, Xie LZ, Zhu J, et al. Herbal medicines for fatty liver diseases. Cochrane Database Syst Rev 2013, 8: 1–216.
26. Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology 2010, 52: 1836–1846.
10.12032/MDM2020063005
:
QZSWF, the experienced prescription Quzhisanwei formula; NASH, non-alcoholic steatohepatitis; ADME, absorption, distribution, metabolism, and excretion; GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; NAFLD, non-alcoholic fatty liver disease; TCM, traditional Chinese medicine; OB, oral bioavailability; DL, drug-likeness; PPI, protein-protein interaction; MCODE, molecular complex detection; FDR, false discovery rate; PC, Huzhang (); CF, Shanzha (); CA, Juemingzi ().
:
The authors declare that they have no conflict of interest.
:
Hai-Sheng Chai, Ting-Ting Shen, Yan-Hong Liu, et al. Mechanisms of Quzhisanwei formula for non-alcoholic steatohepatitis based on a network pharmacology approach. Medical Data Mining 2020, 3 (2): 92–101.
:Yu-Ping Shi.
: 01 March 2020,
04 June 2020,
:25 June 2020
contributed equally to this article.
Shang-Wei Hou, Email: housw@sjtu.edu.cn; Qin Zhang, Email: zq1980@shtrhospital.com.Institution of Tongren Hospital, Shang Hai Jiao Tong University School of Medicine, No.1111 Xianxia Road, Changning District, ShangHai 200050, China.

