Genetic Diversity, Antimicrobial Resistance, and Virulence Genes of Aeromonas lsolates from Clinical Patients, Tap Water Systems, and Food*
MENG Shuang, WANG Yong Lu, LIU Chen Geng, YANG Jing, YUAN Min, BAI Xiang Ning,JIN Dong, LIANG Jun Rong, CUI Zhi Gang,#, and LI Juan,#
1. State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing 10026, China; 2. Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Zhejiang 310058, Hangzhou, China;3. Ma’anshan Center for Disease Control and Prevention, Ma’anshan 243031, Anhui, China; 4. Clinical Laboratory of Xuanwu Hospital, Captital Medcial University, Beijing 100053, China
Abstract Objective This study aimed to evaluate the genetic diversity, virulence, and antimicrobial resistance of Aeromonas isolates from clinical patients, tap water systems, and food.Methods Ninety Aeromonas isolates were obtained from Ma’anshan, Anhui province, China, and subjected to multi-locus sequence typing (MLST) with six housekeeping genes. Their taxonomy was investigated using concatenated gyrB-cpn60 sequences, while their resistance to 12 antibiotics was evaluated. Ten putative virulence factors and several resistance genes were identified by PCR and sequencing.Results The 90 Aeromonas isolates were divided into 84 sequence types, 80 of which were novel,indicating high genetic diversity. The Aeromonas isolates were classified into eight different species. PCR assays identified virulence genes in the isolates, with the enterotoxin and hemolysin genes act, aerA, alt,and ast found in 47 (52.2%), 13 (14.4%), 22 (24.4%), and 12 (13.3%) of the isolates, respectively. The majority of the isolates (≥ 90%) were susceptible to aztreonam, imipenem, cefepime, chloramphenicol,gentamicin, tetracycline, and ciprofloxacin. However, several resistance genes were detected in the isolates, as well as a new mcr-3 variant.Conclusions Sequence type, virulence properties, and antibiotic resistance vary in Aeromonas isolates from clinical patients, tap water systems, and food.
Key words: Aeromonas; Multi-locus sequence typing; Multidrug resistance; Virulence gene;Antimicrobial resistance gene
INTRODUCTION
Aeromonasspp. are important human opportunistic pathogens that can cause intestinal and extra-intestinal diseases,particularly in immunocompromized individuals,including gastroenteritis, wound infections, and even life-threatening necrotizing fasciitis[1].Aeromonasspecies are often isolated from freshwater, seafood,and meat products[2-3]and are therefore a primary cause of food contamination and may act as intermediaries in transmitting disease to humans[4].
TheAeromonasgenus contains over 26 species and has a very complex taxonomy. Although great efforts have been made to correctly identify differentAeromonasspecies, particularly those related to human diseases, this has been difficult to achieve using traditional biochemical methods due to taxonomic complexity[1,5]. In addition,conventional biochemical methods such as matrix assisted laser desorption/ionization flight mass spectrometry (MALDI-TOF MS) are time consuming and tedious for routine use. Moreover, the 16S rDNA sequence used for bacterial identification has high between-species similarity and thus cannot adequately distinguish betweenAeromonasspecies[6-7]. Recently studies have shown that housekeeping gene sequencing (gyrBandrpoD) can be used for the phylogenetic analysis and identification ofAeromonasspecies[8-9]; for instance, Yano et al.[10]used housekeeping gene sequencing to identify 87Aeromonasstrains at the species level.
The pathogenesis ofAeromonasspp. involves a series of virulence factors[11]. These include hemolytic toxins such as aerolysin-related cytotoxic enterotoxin (Act)[12], heat-labile cytotonic enterotoxin (Alt), hemolysin (hlyA), heat-stable cytotonic toxins (Ast)[13], and aerolysin (aerA)[14]. In addition, the type III secretion system (TTSS), lateral flagella (laf), polar flagellum (fla)[15-16], elastase(ela)[17], and lipase (lip)[18]also contribute toward the pathogenicity ofAeromonas.
Aeromonasantibiotic resistance has increased globally in recent years; for example, some strains are resistance to aminoglycosides [aac(6’)-Ib], while others harbor plasmid-mediated quinolone resistance (PMQR) determinants[19]. InAeromonasisolates from South Africa and Korea, the prevalence ofaac(6’)-Ibwas found to be 29.23% and 29.00%,respectively[19-20]. The important PMQR determinantqnrShas been reported inAeromonas[21-22], with 73.85% ofAeromonasstrains in Korea found to harborqnrSgenes[19]. Conversely,qnrSwas found to be present in 21.00% ofAeromonasisolates from freshwater fish in South Africa[20]. The resistance ofAeromonasto several different classes of antibiotics poses a major problem for human health since the resistant bacteria can be transmitted from the aquatic environment to humansviathe food chain or direct contact[23]. Therefore, it is necessary to monitorAeromonasantimicrobial resistance to guide clinical treatment.
In this study, we evaluated the characteristics ofAeromonasstrains isolated from environmental sources, food, and clinical patients in Ma’anshan,Anhui province, China. In addition, we investigated the virulence-associated genes and antimicrobial resistance of theseAeromonasspp.
MATERIALS AND METHODS
Ethical Statement
Fecal samples and bodily fluids were acquired from patients who had provided informed consent.This study was reviewed and approved by the ethics committee of the National Institute for Communicable Disease Control and Prevention,Chinese Center for Disease Control and Prevention.
Aeromonas Isolates
In 2018, 90Aeromonasisolates were obtained from 33 stool samples from patients with diarrhea,36 tap water systems, and 21 foods in Ma’anshan Anhui Province, China (Figure 1). The isolated strains were identified using an automatic bacteriologic analyzer (Vitek 2 Compact, BioMerieuX). Bacteria were cultured on Luria-Bertani (LB) broth or brain heart infusion (BHI) agar plates overnight at 37 °C.
Multi-locus Sequence Typing (MLST) and Subtyping of Aeromonas Isolates
To analyze the subtype of theAeromonasisolates, we used theAeromonasMLST scheme(http://pubmlst.org/Aeromonas/) with six housekeeping genes:gyrB,groL,gltA,metG,ppsA,and recA. PCR was carried out using previously described primers and protocols[5]. The sequences of the six loci were compared to those published in theAeromonasMLST database, as well as the STs. New alleles and STs were submitted to theAeromonasMLST database for name assignment.
In this study, 90Aeromonasstrains were identified at the species level by analyzing the housekeeping genesgyrBandcpn60[8,24]. The reference nucleotide sequences of these genes were taken from the GenBank database and included the 28 representative species listed in Supplementary Table S1 (available in www.besjournal.com). A phylogenetic tree was constructed using the neighbor-joining method in Clustal-W[25]. All primers were synthesized by Beijing Tsingke Biological Technology Company (Beijing, China).
Detection of Virulence-associated Genes
To detect virulence-associated genes in theAeromonasisolates, we performed PCR using previously describedalt,ast,hlyA,aerA,act,ascV,aexT,laf,lip,fla,andelaprimers. PCR amplification was performed in a 50 μL reaction volume containing 25 μL of Taq PCR MasterMix (Takara Bio,Inc., Japan), 1 μL of 10 μmol/L primer, 21 μL of ddH2O, and 2 μL of DNA template under the following cycling conditions: pre-denaturation at 95 °C for 5 min, 30 cycles of denaturation at 95 °C for 30 s, annealing at 55-60 °C for 30 s, and extension at 72 °C for 1 min, followed by a final cycle at 72 °C for 5 min. Positive PCR products were confirmed by sequencing, detecting a total of 11 virulenceassociated genes.
Antimicrobial Susceptibility Test
Antimicrobial susceptibility tests were carried out using the broth microdilution method according to CLSI guidelines (Clinical and Laboratory Standards Institute, 2018). The minimum inhibitory concentrations (MICs) of the following 13 antibiotics were measured: amoxicillin/clavulanate (AMC),ampicillin (AMP), cefepime (FEP), ceftriaxone (CRO),ceftazidime (CAZ), imipenem (IPM), aztreonam(ATM), gentamycin (GEN), tetracycline (TCY),ciprofloxacin (CIP), trimethoprim/sulfamethoxazole(SXT), chloramphenicol (CHL), and colistin (CT).E. coliATCC 25922 was used as the quality-control strain for susceptibility testing.
Detection of Resistance Genes
To detect antimicrobial resistance genes, we performed PCR amplification on tetracycline resistance (tetA,tetB, andtetE), extended-spectrum β-lactamase (ESBL) (blaTEM,blaSHV, andblaCTX)[19],aminoglycoside resistance [armA,aphAI-IAB,aac(6’)-Ib, andaac(3)-IIa][26], sulphonamide resistance(sul1andsul2)[27], and mobile colistin resistance (mcr-1,mcr-2,mcr-3,andmcr-4) genes, as well as PMQR(qnrA,qnrB, andqnrS) genes[19]using previously described primers and protocols (Table 1)[19,28-32].Positive PCR products were confirmed by sequencing.
RESULTS
MLST of Aeromonas Isolates
The 90Aeromonasisolates were divided into 84 STs of which 80 were novel (ST569-ST644 and ST649-ST652), indicating high genetic diversity. No STs were predominant.
Diversity and Distribution of Aeromonas Species
We evaluated the phylogeny of the 90Aeromonasisolates based on theirgyrBandcpn60sequences (Figure 2). Sequencing analysis classified 82 (91.1%) of the strains into eight different species, of which the three most common wereA.jandaei(32.2%),A. veronii(25.5%), andA. caviae(13.3%). Notably, eight strains did not belong to any of the 28 known species and may be regarded as new species. In addition, the distribution ofAeromonasspecies isolated from clinical patients,food, and tap water samples varied (Table 2).A.caviae(36.4%) was the most prevalent species in clinical isolates,A. veronii(18.1%) was the most common in food isolates, andA. jandaei(58.3%)was the most prevalent in environmental isolates,with the of these three species differing significantly between patient-, food-, and environment-derived isolates (P< 0.05,χ2test).
Distribution of Virulence-associated Genes in Aeromonas Strains
We detected 11 virulence-associated genes in theAeromonasisolates (Table 3), of which 77.8%carriedfla, 52.2% carriedact, 44.4% carriedela,and 43.3% carriedascV. Two additional genes,lafandast, were present in 8.9 and 13.3% of the isolates, respectively. The prevalence ofast,lip,andeladiffered significantly in the patient-, food-,and environment- derived isolates (P< 0.05,Fisher's exact test), while onlylipandaexTwere found to be more prevalent in patient-derived isolates than food-derived or environmental isolates. As shown in Table 4, the 11 virulenceassociated genes differed significantly among the most common species. The hemolytic geneactwas prevalent inA. hydrophilaandA. veronii, whereas the enterotoxin genealtwas prevalent inA.aquariorumandA. hydrophila. The enterotoxin geneast, hemolytic geneaerA,and hemolytic genehlyAwere more prevalent inA. hydrophila;however, both extracellular protease geneselaand lip were rare in A. jandaei and A. veronii but very common in other species.
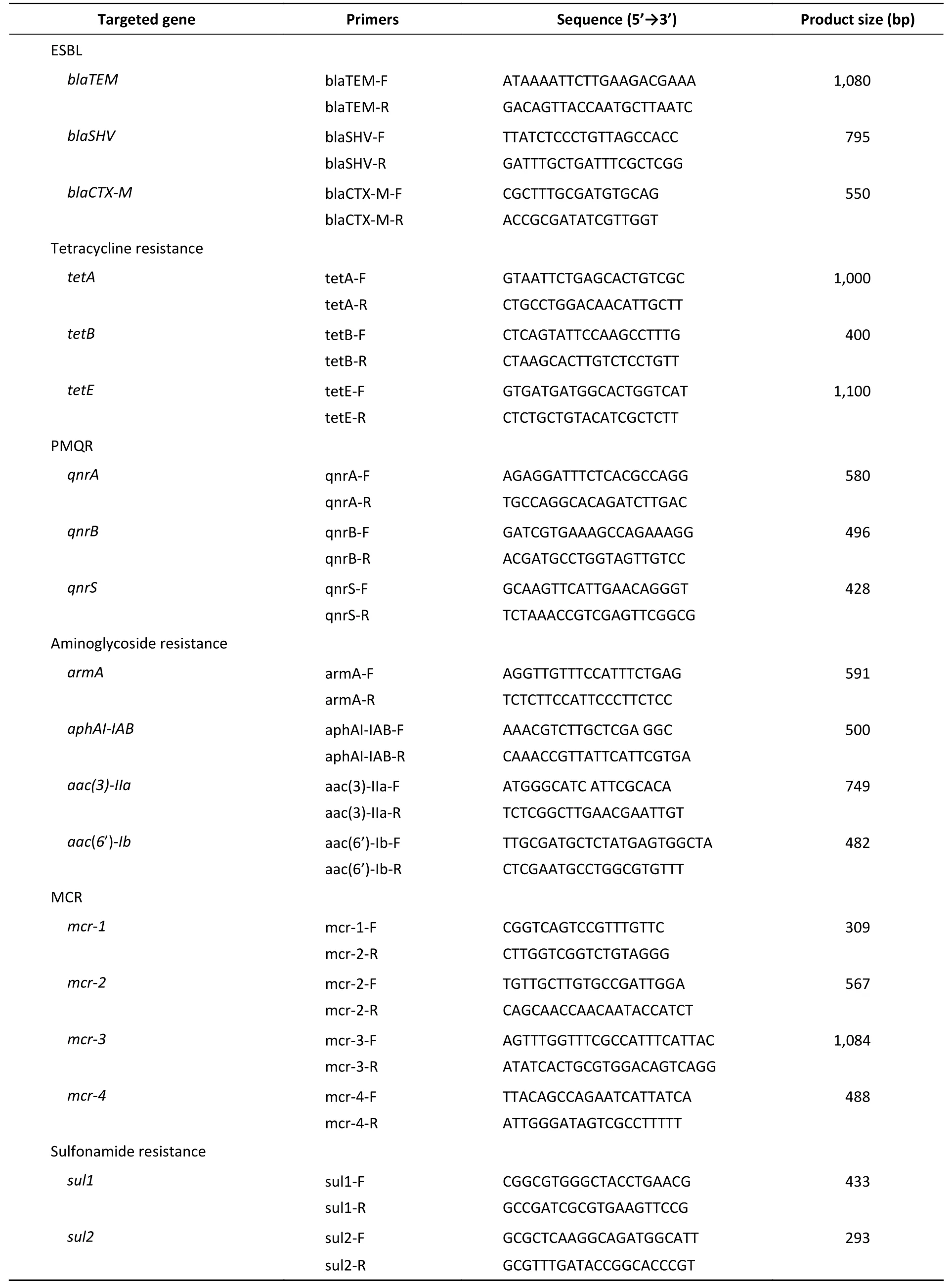
Table 1. Primer sequences used to amplify antimicrobial resistance genes
Prevalence of Antimicrobial Resistance
Next, we evaluated the susceptibility of the 90Aeromonasisolates to 13 antibiotics belonging to ten classes of antibiotic using the broth microdilution method according to CLSI recommendations (Table 5). High ampicillin (100%)and amoxicillin/ clavulanic acid (86.7%) resistance was observed in theAeromonasstrains; however,the majority of the isolates (≥ 90%) were susceptible to aztreonam, imipenem, cefepime, CHL, gentamicin,tetracycline, and ciprofloxacin. Notably, cefepime and ciprofloxacin resistance were significantly higher in patient isolates than in food or environmental isolates (P< 0.05, Fisher's exact test), whereas only one antibiotic (colistin) displayed significantly higher resistance rates in environmental isolates (Table 5).Nineteen isolates (21.1%) were found to be multidrug-resistant (MDR), displaying resistance to at least three of the antibiotics tested in this study.Of these 19 MDR isolates, 10 (52.6%) were isolated from patients, 7 (36.8%) were isolated from the environment, and 2 (10.5%) were isolated from food(Figure 1).
Detection of Aimicrobial Resistance Genes
The PMQR geneqnrSwas detected in 4 (4.4%)isolates. The ESBL geneblaCTX-Mwas detected in 2(2.22%) isolates. The aminoglycoside resistance genesaac(6’)-IbandarmAwere detected in 2(2.22%) and 1 (1.11%) isolates, respectively. The sulfonamide genessul1andsul2were found in 3(3.33%) and 9 (10%) isolates, respectively (Figure 1).The mobile colistin resistance genemcr-3was detected in 3 (3.33%) isolates. Sequence analysis revealed that one isolate (E1006) harboredmcr-3.25(GenBank accession no. KM985469.1) while two (P92 and F1015) harbored a newmcr-3variant which differed from themcr-3.8gene by three amino acid changes according to sequence alignment(unpublished data). The ESBL genesblaTEMandblaSHV, aminoglycoside resistance genesaphAI-IABandaac(3)-IIa, tetracycline resistance genestetA,tetB,andtetE, colistin resistance genesmcr-1,mcr-2,andmcr-4genes, and PMQR genesqnrAandqnrBwere not detected in any isolates.
DISCUSSION
Aeromonasis a genus of bacteria that are ubiquitously present in aquatic environments and have been linked to infections in both humans and animals[1]. In this study, we evaluated 90Aeromonasisolates from patients, tap water, and food, and assessed the genetic diversity, putative virulence genes, and antimicrobial resistance of these isolates.
The 90 isolates were separated into 84 STs of which just four were found to match those published in theAeromonasMLST database, suggesting that 80 were novel (ST569-ST644 and ST649-ST652) and indicating high genetic diversity. We also evaluated the phylogeny of the 90Aeromonasisolates based on the concatenatedgyrB-cpn60gene sequences(Figure 2), revealing that the isolates were closely related and includedA. jandaei(29 isolates),A.veronii(23 isolates),A. caviae(12 isolates),A.aquariorum(7 isolates),A. hydrophila(6 isolates),A.salmonicida(2 isolates),A. enteropelogenes(2 isolates),A. media(1 isolate), and new species (8 isolates). The most common species wasA. jandaei,which comprised 32% of all isolates and was mostly isolated from tap water systems. The second most prevalent species wasA. veronii, which is distributed in various environments such as water and fish[1];indeed, someA. veroniistrains have been isolated from snail lion, suggesting that this species may be related to invertebrates in aquatic environments.The third most prevalent species wasA. caviae,which has previously been shown to have clinical relevance. Zhou et al.[33]reported that the four most prevalent species ofAeromonasin clinical isolates wereA. caviae(41.7%),A. veronii(31.3%),A.dhakensis(13.9%), andA. hydrophila(5.2%). Another report[34]aboutAeromonasrecovered from Patients Suffering from Diarrhea in Israel were evaluated forAeromonasspecies, and the most prevalent species wereA. caviae(65%) andA. veronii(29%). In this study, the most prevalent species in clinical isolates wasA. caviae, accounting for 36.4% of the isolates,followed byA. veronii(18.1%). In brief, the prevalent species ofAeromonasin clinical settings reported in this study is correspond with that reported by other scholars.
The pathogenic mechanism ofAeromonasis complex and multifactorial, which may be related to some of its virulence-associated genes; therefore,we evaluated the virulence-associated genes present in these isolates (Table 3). The enterotoxin and hemolysin genesact,aerA,alt, andastwere present in 47 (52.2%), 13 (14.4%), 22 (24.4%), and 12 (13.3%)of the 90 isolates, respectively. Theactgene was detected in 91.3% ofA. veroniiisolates and 83.3% ofA. hydrophilaisolates,while theaerAgene was detected in 83.3% ofA. hydrophilaisolates and 42.9% ofA. aquariorumisolates. Thealtgene was detected in 100% ofA. hydrophilaandA. aquariorumstrains and 16.7% ofA. caviae, whereas theastgene was present in 28.6% ofA. aquariorumstrains and allA. hydrophila. Thefla,ela, andlipgenes were present in 70 (77.8%), 40 (44.4%), and 34 (37.7%) of the 90 isolates, withflaharbored in the majority of species andelaandlipboth prevalent inA.aquariorum,A. caviae, andA. hydrophilaisolates.The TTSS genesascVandaexTwere detected in 39(43.3%) and 20 (22.2%) of the 90 isolates,respectively:ascVwas present in 62.1% ofA.jandaei, 52.2% ofA. veronii, and 28.6% ofA.aquariorum; whileaexTwas present in 56.5% ofA.veroniiand 16.7% ofA. caviae. Enterotoxins and hemolysins are very important virulence factors inAeromonas spp.[35]and many studies have shown a positive correlation between the number of toxin genes harbored by an isolate and its potential virulence[35,13]. The virulence genes detected in this study indicate the potential pathogenicity of the isolates from clinical, food, and environmental sources, as well as their possible risk to human health.
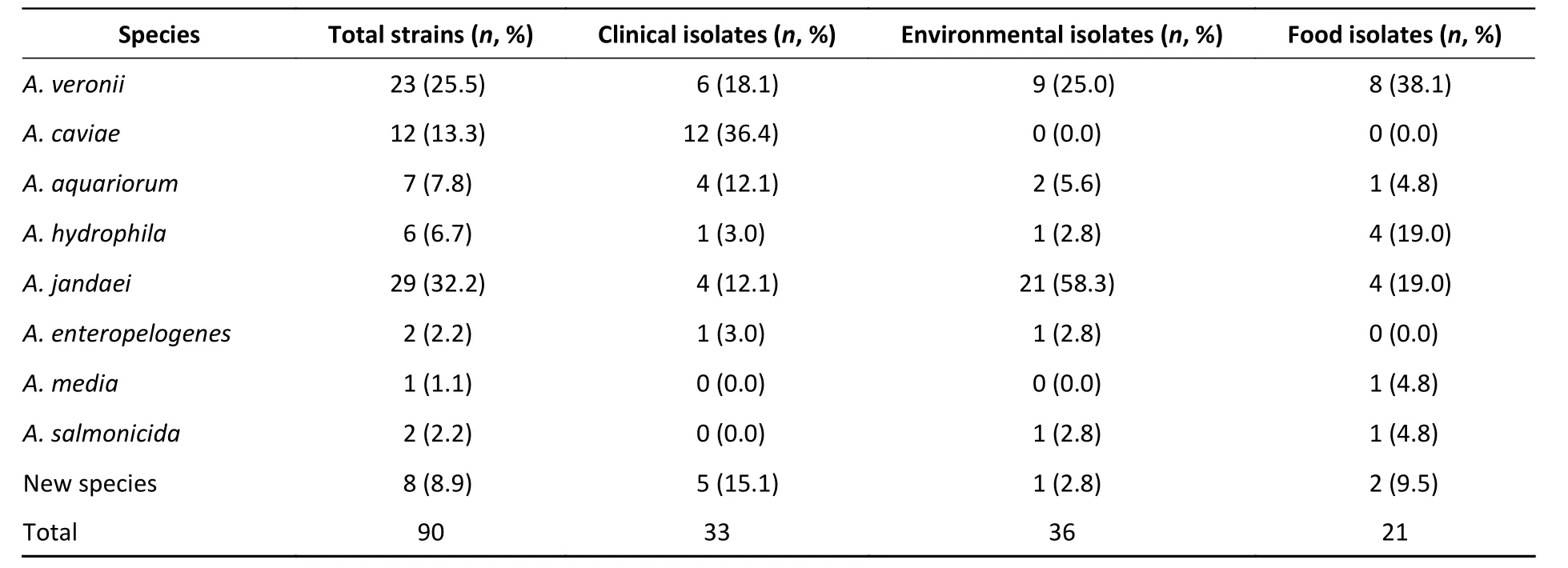
Table 2. Distribution of Aeromonas spp. in isolates collected from clinical patients, food,and tap water samples

Table 3. Distribution of virulence-associated genes in Aeromonas strains isolated clinical patients, food,and tap water samples
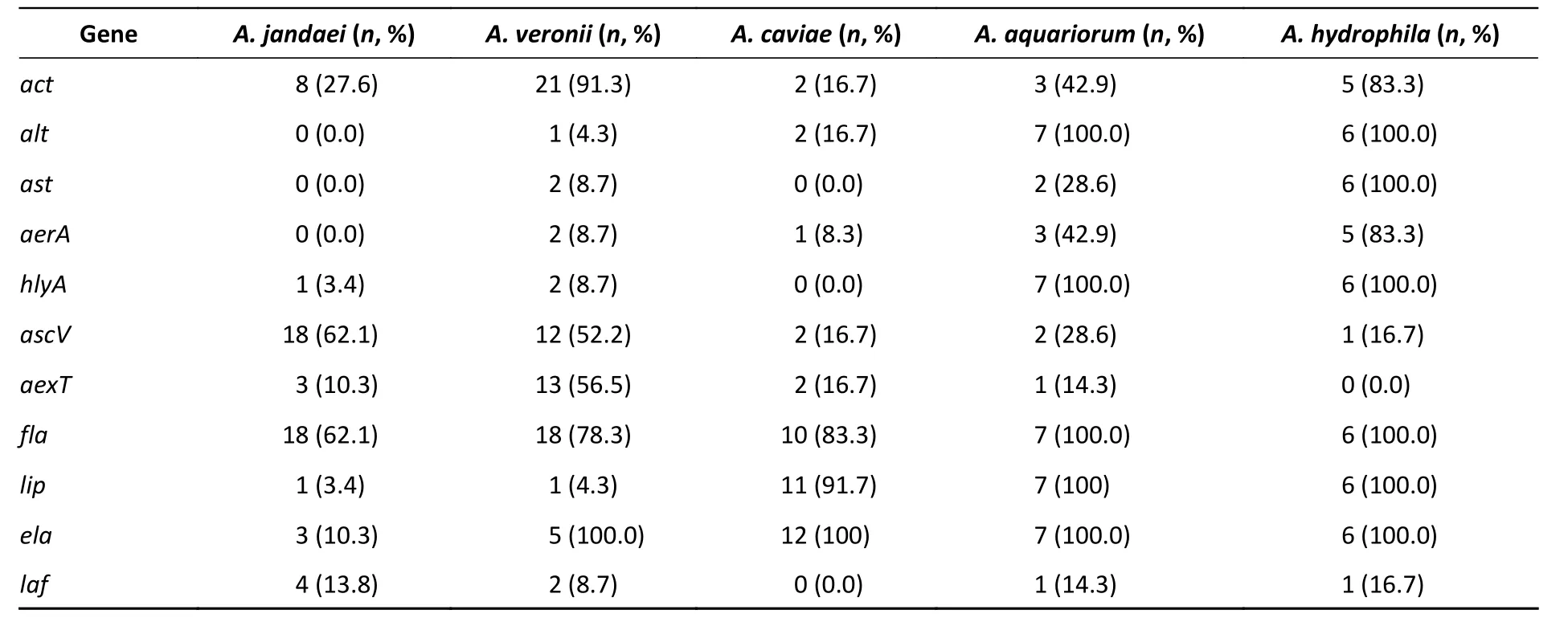
Table 4. Distribution of virulence genes in the five most common Aeromonas spp.
In this study, the majority ofAeromonasstrains displayed MDR phenotypes, with 100.0% resistance against amoxicillin and 86.7% resistance against amoxicillin/ clavulanic acid, consistent with previous studies[36,37]. Due to their chromosomal β-lactamase expression,Aeromonas spp. are naturally resistant to β-lactams. As such, the resistance rate ofAeromonasstrains derived from patients was significantly higher than those from tap water systems or food, with the exception of colistin. Moreover, the drug resistance rate of strains isolated from tap water systems was significantly higher than that of clinical and food strains. Colistin is a last-resort antibacterial used to treat clinically serious infections caused by MDR gram-negative bacteria[38]. A new mobile colistin resistance gene,mcr-3, has been detected in MDR bacteria isolated from severely ill patients in many countries. It is particularly important to determine the presence of these strains in meat products and drinking water due to their direct impact on public health[31]. In this study, three of theAeromonasstrains that were resistant to colistin harboredmcr-3genes and were derived from the feces of patients with diarrhea, tap water, and fresh pork from the supermarket.
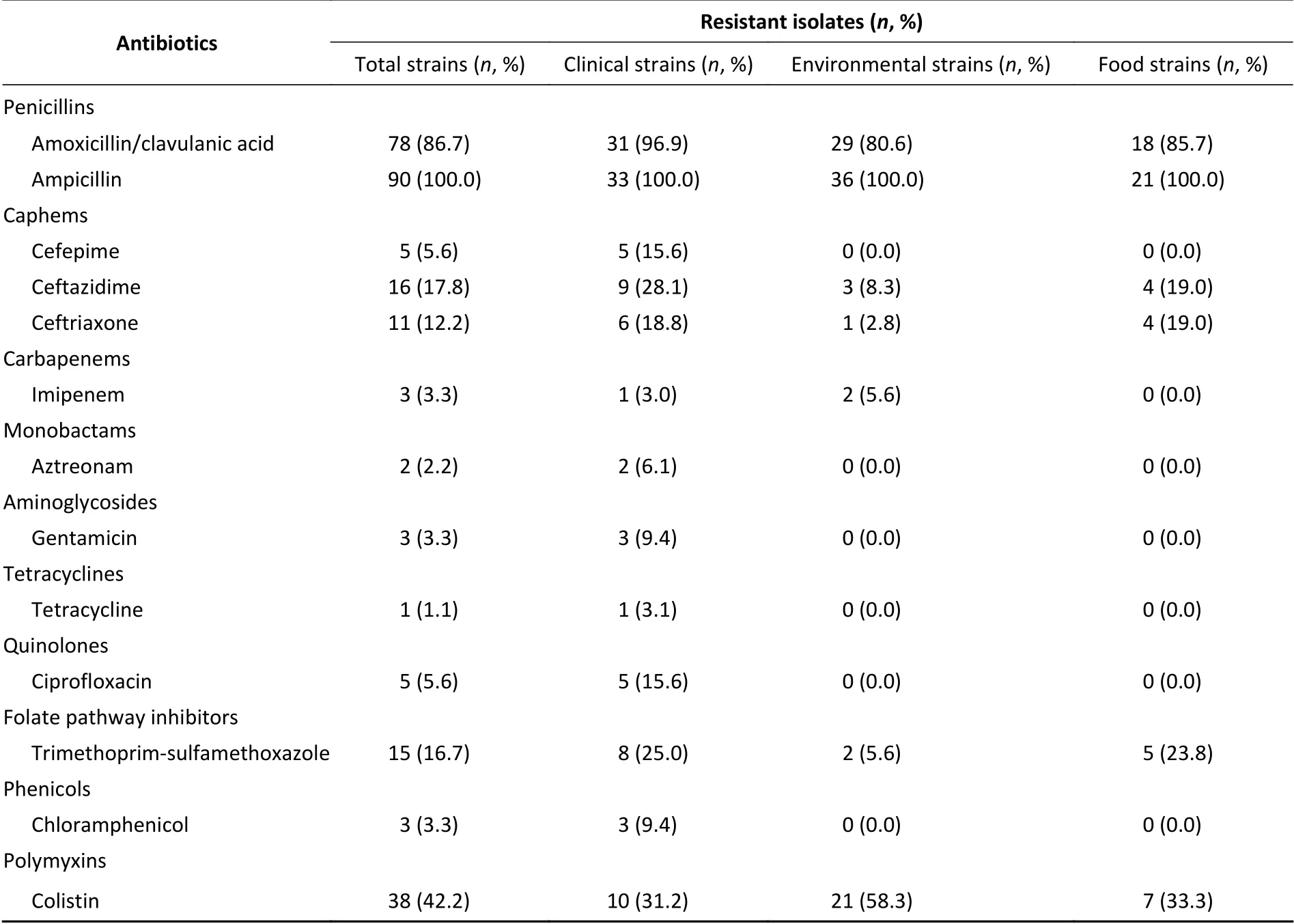
Table 5. Prevalence of resistance to different antibiotics
The existence of thesemcr-3genes is of great importance to global public health because obtaining anmcr-3gene may lead to high levels of colistin resistance inAeromonas, particularly since it is ubiquitous in soil and water systems and has the opportunity to interact with bacteria from a variety of different sources.Aeromonasspecies may therefore be a reservoir formcr-3and contribute toward its potential spread. In China,mcrgenes have not only been detected in a large number of human pathogens, but also have a high positive test rate in animals (livestock, pets, and even wildlife) and the environment (soil and water)[39]. Since colistin is being used at increasingly high frequencies in veterinary and human medicine, it is essential to continuously monitormcrgenes in both clinical and environmental settings.
Resistance to SXT and quinolone, which are antimicrobials used to treatAeromonasinfection,has been widely documented. Deng et al.[40]reported thatAeromonasisolates collected from cultured freshwater animals have the 5% resistance for Ciprofloxacin and 18.86% resistance for SXT, at the same time, the detection rate ofsul1gene was 18.86% and that ofqnrSgene was 4.7%. The researchers noted[33]thatAeromonasisolated from clinical patients have the 6.1% resistance rates of Ciprofloxacin, as well as 5.2% resistance rates of SXT.A total of 186Aeromonas, collected from commercially reared fish and ornamental fish, were evaluated for their antimicrobial susceptibilities. The researchers[28]found that the resistance rate of SXT was 9.4%, and the detection rate ofsul1was 9.4%. In our study, the resistance rate of SXT was 16.7% and the detection rate ofsul1was 3.3%. As reported, the SXT resistance and its determinants are highly prevalent inAeromonas[28,41,42], most likely due to the overuse of sulfonamide drugs in animal farms and fish ponds. PMQR genes have recently been characterized inAeromonasstrains[22,43]; Chenia[20]reported thatqnrSwas found to be present in 21%ofAeromonasisolates from freshwater fish in South Africa. In the present study, the detection rate ofqnrSwas 4.4%. however, when we screened the 90Aeromonasisolates for the three PMQR genesqnrA,qnrB,andqnrS, onlyqnrSwas detected in strains isolated from clinical specimens. It is thought thatAeromonasmay act as a carrier of these resistance genes via horizontal transfer[44]; therefore, the prevalence of MDR inAeromonasspecies could be considered a threat to public health.
CONCLUSIONS
We obtained 90Aeromonasisolates from clinical patients, tap water systems, and food in Ma’anshan,Anhui Province, China. High genetic diversity was observed in these isolates, which belonged to 80 novel STs. ConcatenatedgyrB-cpn60gene sequences classified 82 (91.1%) of theAeromonasisolates into eight different species as well as several new species. Virulence genes were examined by PCR,indicating that the isolates may be pathogenic and pose a risk to human health. When measuring antibiotic resistance to ten distinct antibiotic classes,21.1% of the strains were found to be MDR (≥ 3). The PMQR, ESBL, aminoglycoside resistance,sulphonamide, andmcr-3genes were detected in the isolates, as well as a newmcr-3gene variant.Thus, this study sheds light on the genetic diversity,antibiotic resistance, and pathogenicity ofAeromonasspecies identified from a variety of sources.
CONFLICT OF INTEREST
The authors have declared that no competing interests exist.
Received: April 6, 2020;
Accepted: May 13, 2020
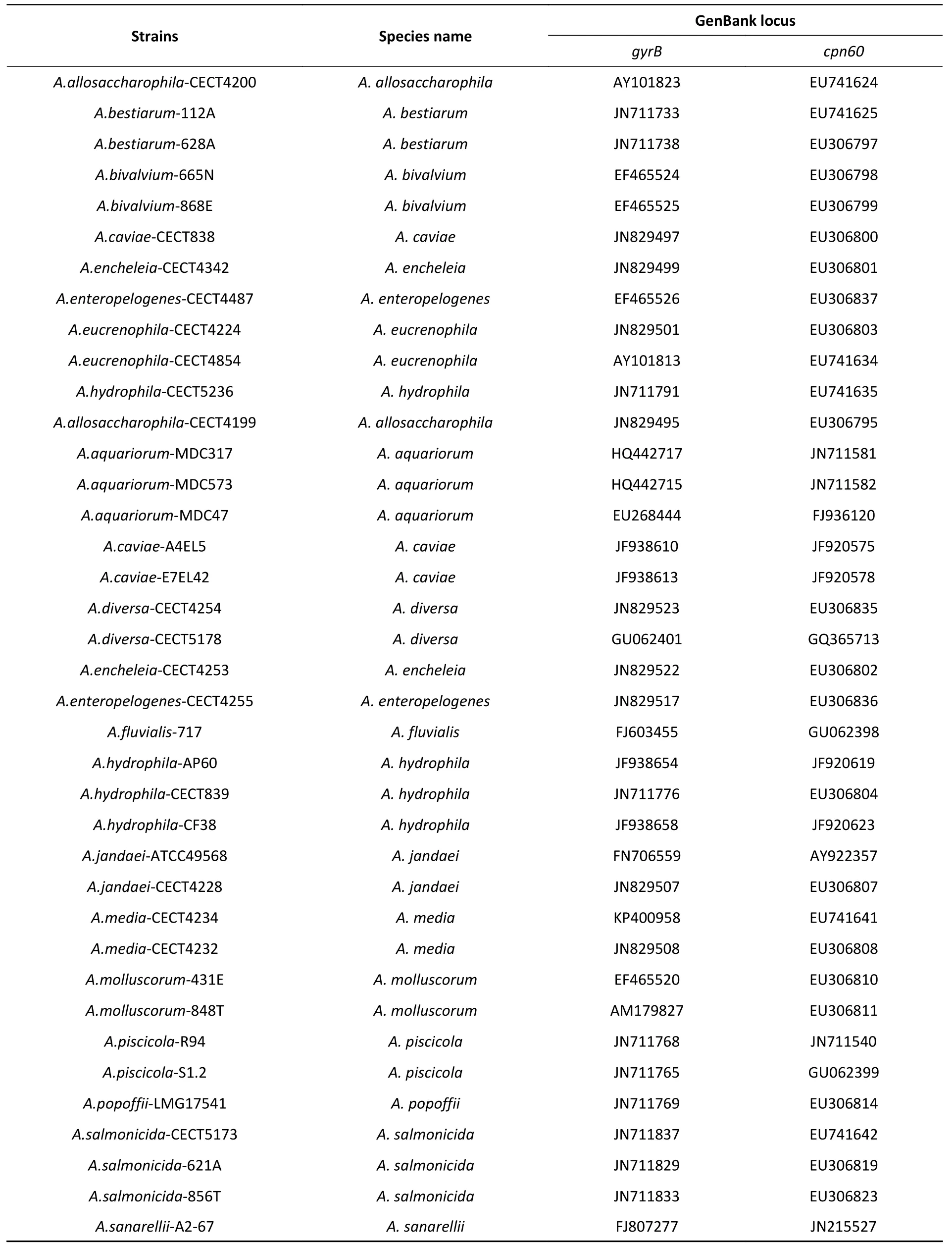
Supplementary Table S1. The gyrB and cpn60 genes of twenty-eight representative Aeromonas species available in GenBank
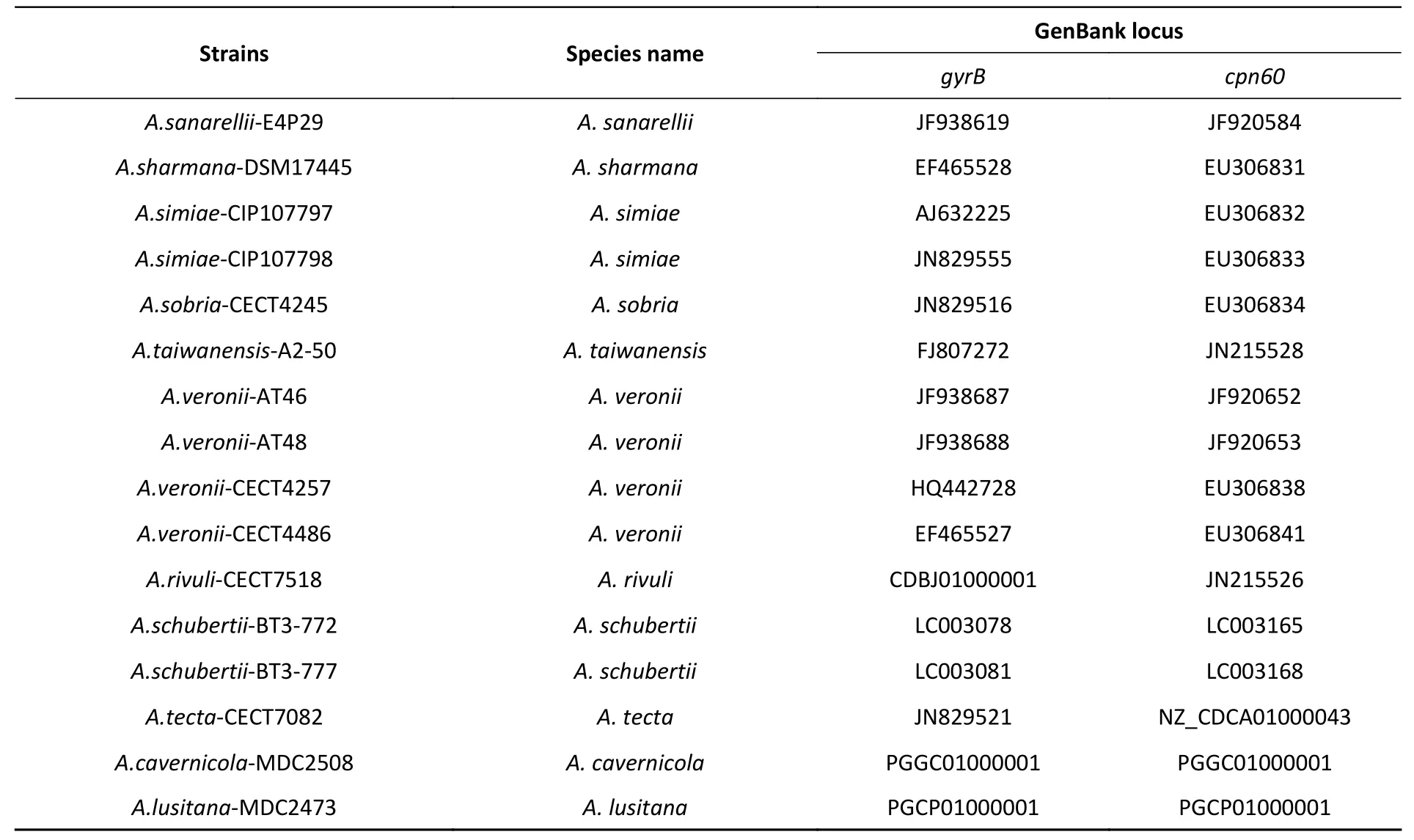
Continued
 Biomedical and Environmental Sciences2020年6期
Biomedical and Environmental Sciences2020年6期
- Biomedical and Environmental Sciences的其它文章
- Gender Differences in the Relationships between Pubertal Stages and the Perpetration of Self-inflicted and lnterpersonal Violence among Middle SchoolStudents in China*
- Capacities and Functionalities Assessment of Veterinary Laboratories in South-west Nigeria Using the FAO Laboratory Mapping Tool
- Grape Seed Procyanidin Extract Attenuate Sodium Fluoride-induced Oxidative Damage and Apoptosis in Rat Kidneys*
- Prevalence of Hyperuricemia and Associated Factors in the Yi Farmers and Migrants of Southwestern China: A Cross-sectional Study
- Detection of Pseudorabies Virus Antibodies in Human Encephalitis Cases*
- Risk or Beneficial Factors Associated with Unplanned Revascularization Risk Following Percutaneous Coronary lntervention: A Large Single-Center Data*
