Bartonella Species lnvestigated among Rodents from Shaanxi Province of China*
AN Cui Hong, CHEN Bao Bao, LYU Wen, NIE Shou Min, LI Sheng Zhen, FAN Suo Ping,
SUN Yang Xin1,#, LIU Jin Ren3, LI Dong Mei4, and XU Ji Ru2,#
Bartonellaspp. are rod-shaped, Gram-negative,aerobic, fastidious, slow bacteria which cause diseases in humans and animals by parasitizing the endothelial and red blood cells of their hosts[1,2].Rodents are the most important natural reservoir hosts ofBartonella[3]. In 57 countries,epidemiological studies of rat-borneBartonellahave been carried out. Information on theBartonellainfection rate in rodents has been obtained from most of these countries. The infection rate of rodents in Portugal, Egypt, Japan, Canada, and the United States is more than 90%, the infection rate in Thailand and Russia is about 80%, and the infection rate in China is about 67%. To date, 22 species of rodent-borneBartonellahave been described. Of these,B. grahamii, B. vinsoniisubsp.arupensis,B.vinsoniisubsp.Berkhoffii, andB. elizabethaehave been found to be associated with human illness.People become infected with rodent-borneBartonellaincidentally, especially when they are exposed to the habitats of wild rodents harboring variousBartonellaspecies.Bartonellainfection can affect multiple organs and poses a risk to public health[4].
In China, 16 species ofBartonellahave been isolated from rodents. The investigation was carried out in various provinces including Heilongjiang,Fujian, Zhejiang, Yunnan, Hainan, Qinghai, Inner Mongolia, and Taiwan China[4]. No investigation has been reported in Shaanxi province.
From north to south in Shaanxi province, the North Mountains and Qinling Mountains divide Shaanxi province into three natural regions: Shanbei Plateau, Guanzhong Plain, and Qinba Mountain. The province spans the northern subtropics, warm temperate zone, and temperate zone. Because of the variety of landscape types, 55 species of rodents are found in Shaanxi province[5].
In order to investigateBartonellaspecies in rodents in Shaanxi Province, rodents were captured using mouse snap traps in 9 counties; three on the Shanbei Plateau (Dingbian, Yuyang, and Wuqi counties), two on the Guanzhong Plain (Dali and Chang’an counties), and four in the Qinba Mountains(Zhenping, Nanzheng, Ningshan, and Zhenba counties) (Table 1 and Supplementary Figure S1,available in www.besjournal.com), between 2014 and 2017. Following the capture of each animal, we recorded collection time, site, habitat, species,gender, weight, head-body length, and tail length,and collected spleen samples under sterile conditions, which were transported back to the laboratory in liquid nitrogen and stored at -80 °C until use.
Tweenty five mg spleen was homogenized within 200 μL sterilized trypsin soy broth, plated onto trypsin soy agar containing 5% (vol/vol) defibrinated sheep blood, incubated at 37 °C with 5% CO2, and later checked for growth ofBartonellaspecies on alternate days for up to 20 d. To obtain pure colonies, suspected colonies were selected and separately subcultured twice on fresh agar plates[6].Small, round, gray-white colonies were morphologically identified asBartonella, transferred onto fresh plates, and then stored in 30% glycerol in a freezer at -80 °C for further analysis.
DNA templates were prepared directly from bacterial colonies by the boiling method. The sample was added to 100 μL sterile deionized water, heated for 10 min at 100 °C and centrifuged at 6,000 rpm for 5 min at 4 °C. The supernatant was used as a source of DNA template for PCR to detect theBartonellacitrate synthase (gltA) gene. The primers used for the amplification of the 379-base pair fragment were forward BhCS781.p (5′-GGGGACCAGCTCATGGTGG-3′) and reverse BhCS1137.n (5′-AATGCAAAAAGAACAGTAAACA-3′)[7]. PCR was performed in 50 μL mixtures containing 25 μL 2X TaqPCR Master Mix,22 μL double-distilled H2O, 1 μL (10 mol/L) of each primer, and 1 μL of DNA template.

Table 1. Prevalence of Bartonella spp. in rodents
The PCR amplification conditions were as follows: an initial step of 94 °C for 2 min; 30 amplification cycles, each consisting of 94 °C for 30 s and 48 °C for 30 s; an elongation step of 72 °C for 1 min, and a final incubation at 72 °C for 5 min.Amplified products were electrophoretically analyzed on 1% agarose gels supplemented with 0.005% of GoldView and visualized under UV light.PCR products of the expected length were then purified and sequenced on both strands by Tsingke Biotechnology (Xi’an, China).
The nucleic acid sequence homology was blasted against reportedBartonellaspecies sequences in GenBank using the BLAST program at the National Center for Biotechnology Information Website(http://blast.ncbi.nlm.nih.gov/Blast.cgi). Individual gene sequences and concatenated sequences assembled forgltAwere aligned using ClustalW, and Neighbor-joining (NJ) phylogenetic tree analysis was performed using MEGA6.06[8]with the Kimura 2-parameter model and withBrucella abortusas the outlier group. Bootstrap calculations were carried out for 1,000 replicates[9]. A criterion of ≥ 96%homology togltAwas used to define phylogroups[10].
Chi-Squaretests of difference in the prevalence ofBartonellastrains in rodents were conducted withP< 0.05 considered the threshold for statistical significance using the Statistical Package for the Social Sciences (SPSS 19, Chicago, IL).
Rodents (487 individuals) were sampled from nine counties within three regions of Shaanxi Province to detect the presence ofBartonellainfection. The tested rodents represented 27 species belonging to four families (Table 1). The overall prevalence ofBartonellaspecies in rodents was observed to be 26.08% (127/487). Statistical analysis showed that the prevalence ofBartonellainfection varied significantly among regions (χ2= 15.056,P=0.001), counties (χ2= 36.964,P= 0.000), and species(χ2= 42.758,P= 0.000).
Bartonellainfection was found in 14 species of rodents in eight counties. The highestBartonellaprevalence were inMeriones meridianus(40.00%,2/5) andMus musculus(40.00%, 2/5), followed by
Niviventer confucianus(39.06%, 25/64) andMeriones unguiculatus(34.72%, 75/216) (Table 1). A highBartonellaprevalence was found in rodents collected from Zhenping county (40.38%, 21/52) and Dingbian county (34.98%, 78/223) (Table 2).
Sequencing and phylogenetic analysis of thegltAfragment was performed among representative isolates (Figure 1 and Supplementary Table S1,available in www.besjournal.com). A criterion of≥ 96% homology togltAwas used to define phylogroups, and the sequences were divided into 10 phylogroups. The homology of GP1, Gp4, Gp5,Gp8, and Gp10 was above 97% withB.queenslandensis,B. sylvatica,B. taylorii,B. jaculi,andB. japonica, respectively. GP2, GP3, and Gp9 were above 97% withB. elizabethae,B. grahami, andB. washoensis, respectively. The latter strains are pathogenic to humans. We did not find sequences of ≥ 96% homology clustered with Gp6 and Gp7 in the GenBank. We used the NJ and maximum likelihood (ML) methods to construct phylogenetic trees and obtained the same results. Thus, NJ method was used for further analysis.
In this study,B. elizabethaewas detected fromMeriones unguiculatusin Dingbian county in the Shanbei Plateau region. In this county,Meriones unguiculatusis the dominant species, accounting for 90% of rodent communities, and thrives in the desert and semi-desert steppe, as well as sandy or salinized farmland, ridges, wasteland, thickets and so on. In China,Meriones unguiculatusis distributed in Inner Mongolia, Jilin, Liaoning, Hebei, Shanxi,Shaanxi, Gansu, and Ningxia provinces.
B. washoensiswas detected fromSpermophilus alashanicusin Dali county in the Guanzhong Plain region.Spermophilus alashanicusis very common in this area, mainly living in dry grassland and desert steppe. It is a solitary species, each individual inhabiting a single hole in a habitat of low and sparse plants, roadsides, ridges, fallow land and so on. This animal is distributed in Inner Mongolia, Shanxi,Shaanxi, Gansu, Ningxia provinces.
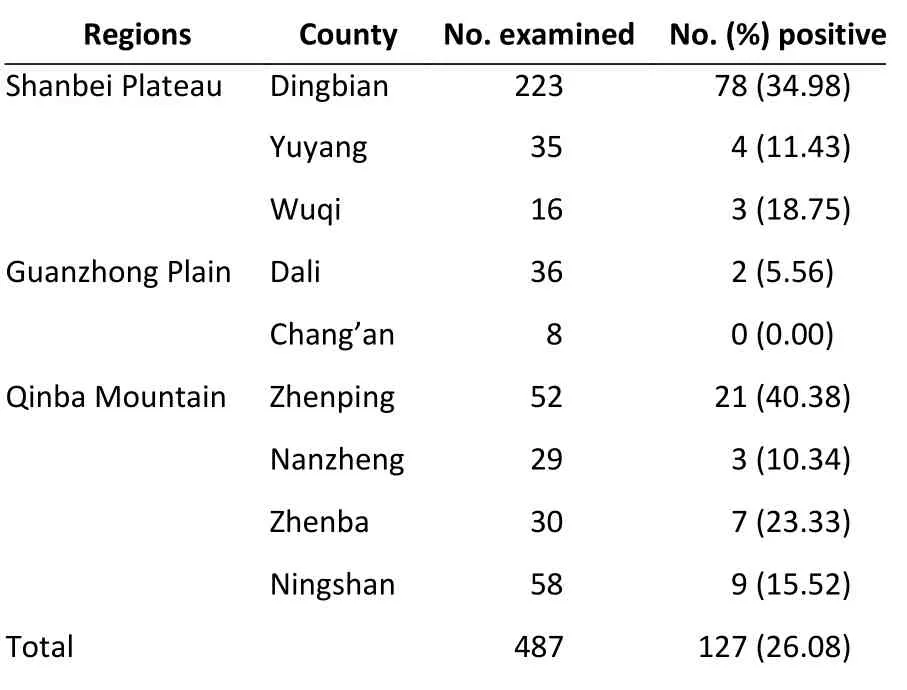
Table 2. Prevalence of Bartonella spp. in counties
B. grahamiiwas detected in Ningshan county fromRattus norvegicusand in Zhenba county from
Rattus confucianusin Qinba Mountain.Rattus norvegicusis distributed all over the world. It is very adaptable to different environments and can inhabit and breed in cities and villages in every season.Rattus confucianusis a common species in Qinba Mountain and it is widely distributed especially in the provinces located south of the Yangtze River.This species lives in vegetation-rich mountain forests, thickets, thatched grass, brook grass, weeds near farmland, rock seams, wasteland, and vegetable gardens, etc.
B. elizabethaewas detected fromMeriones unguiculatus;B. washoensiswas detected fromSpermophilus alashanicus; B. grahamiiwas detected fromRattus norvegicusandRattus confucianusand so on. This indicates thatBartonellahas host specificity.
Merionesunguiculatus,Spermophilus alashanicus,Rattus norvegicus, andRattus confucianusare widely distributed in the abovementioned habitats. People working or living in these places who come into close contact with rodents are vulnerable to pathogenicBartonellainfection. Ningshan county is located in the Qinling Mountain area and Zhenba county is located in the Bashan area. Qinling Mountain and Bashan Mountain are the main tourist areas of Shaanxi province, and visitors to this area are vulnerable to infection by pathogenic bacteria if they do not pay attention to personal hygiene. Therefore, the risk ofBartonellainfection should be assessed.
Small mammals, as the largest host group ofBartonella, have great regional differences.Bartonellahas a wide adaptability to different geographical environments and its geographical distribution characteristics are associated with its host animals[4]. In this study, the highest infection rate ofBartonellawas found in Zhenping county(40.38%). Zhenping county is located in the southernmost end of Shaanxi province, at the northern foot of the Daba Mountain range, and at the junction of Sichuan, Shaanxi, and Hubei provinces. The second highest infection rate was found in Dingbian county (34.98%), which is located in the northwest corner of Shaanxi province, at the transition zone between the Loess Plateau and the desert steppe, and at the junction of Shaanxi, Gansu,Ningxia, and Inner Mongolia provinces. The sampling area of Zhenba county was located at the junction of Shaanxi, Sichuan, and Chongqing provinces, and Yuyang area is located at the border between Shaanxi and Inner Mongolia. The epidemiological significance of the detection ofBartonellain these four districts is also useful for reference to neighboring provinces. Of the nine counties surveyed,Bartonellawas detected in all except Chang’an county, which indicates thatBartonellais widely distributed in Shaanxi province.Bartonellawas isolated from 14 rodent species belonging to 12 genera, indicating thatBartonellais distributed in many host species. In the Chang’an area, noBartonellawas detected in 15 rat species, which may be related to the sample size and requires further investigation.
In conclusion, our study identified eight genotypes ofBartonellain Shaanxi province. Three of these genotypes,B. elizabethae,B. grahamii, andB. washoensis, are associated with human illness.There is therefore a risk infection of the human population, so population monitoring and assessment of the infection risk should be carried out. However, two genotypes could not be determined, and further studies are required to identify these genotypes through detection of related genes such asrpoBandftsZ.
#Correspondence should be addressed to SUN Yang Xin, Tel/Fax: 86-29-68655600, E-mail: sxpco@126.com; XU Ji Ru, 86-29-82657814, E-mail: xujiru@mail.xjtu.edu.cn
Biographical note of the first author: AN Cui Hong,female, born in 1972, Deputy Director Technician,majoring in plague, brucellosis, and etiologic biological vector control.

Supplementary Table S1. The alignment of gltA gene sequences with sequences retrieved from GenBank database
Received: June 17, 2019;Accepted: January 13, 2020
 Biomedical and Environmental Sciences2020年3期
Biomedical and Environmental Sciences2020年3期
- Biomedical and Environmental Sciences的其它文章
- Macular Perfusion Changes and Ganglion Cell Complex Loss in Patients with Silicone Oil-related Visual Loss*
- lnhibitory Effects of Cypermethrin on lnteractions of the Androgen Receptor with Coactivators ARA70 and ARA55*
- Comparative Ligandomic Analysis of Human Lung Epithelial Cells Exposed to PM2.5*
- A New Method for Ultra-sensitive P24 Antigen Assay Based on Near-infrared Fluorescent Microspherelmmunochromatography*
- The Rise and Fall of the Secular Trend in Body Height in Sardinia: An Age-Period-Cohort Analysis
- Spatial and Temporal Variations of Pneumoconiosis in the Pearl River Delta Region in 2006-2015*
