Transcriptome Responses of Hygromycin B Resistance Gene-Transformed, Hygromycin B-Adaptive and Wild Nannochloropsis oceanica Strains to Hygromycin B
LIU Hang, ZHANG Zhongyi, LIANG Sijie, GUO Li, , SUN Shiyang,PAN Kehou, and YANG Guanpin,
1) College of Marine Life Sciences, Ocean University of China, Qingdao 266003, China
2) Laboratory of Applied Microalgae, College of Fisheries, Ocean University of China, Qingdao 266003, China
3) Institutes of Evolution and Marine Biodiversity, Ocean University of China, Qingdao 266003, China
4) Key Laboratory of Marine Genetics and Breeding of Ministry of Education, Ocean University of China,Qingdao 266003, China
Abstract In order to decipher the hygromycin B tolerance and resistance mechanisms of Nannochloropsis oceanica, the transcriptome profiles of a transgenic strain carrying a randomly integrated hygromycin B resistant gene, a hygromycin B-adaptive strain and a wild type strain of N. oceanica were compared by transcriptome sequencing (RNA-seq) without referring to a high quality genome sequence. The results showed that the adaptive strain adapts to the hygromycin B existing environments mainly by intensifying the expressions of the efflux pump ABC and MFS superfamily transporter genes, thus reducing the intracellular concentration of hygromycin B. The transgenic strain obtains the hygromycin B resistance ability solely by expressing exogenous resistance gene. Accordingly, the screening and maintenance of N. oceanica transformants should be carried out at an antibiotics concentration higher than the adaptive threshold. Our findings can help the genetic modification of N. oceanica with the application of hygromycin B.
Key words Nannochloropsis oceanica; hygromycin B; tolerance; resistance; transcriptome
1 Introduction
Nannochloropsis are promising for industrial applications because they accumulate large quantities of polyunsaturated fatty acids, especially eicosapentaenoic acid (EPA)(Boussiba et al., 1987; Sukenik et al., 1989). Nannochloropsis belongs to class Eustigmatophyceae, order Eustigmatales, family Monodopsidaceae, and consists of six known species (Hibberd et al., 1981; Fawley et al., 2007) and one newly documented species (Fawley et al., 2015). At present, the genomes of two species of Nannochloropsis have been sequenced, which included N. gaditana (Radakovits et al., 2012; Corteggiani et al., 2014) and N. oceanica (Liang et al., 2012; Vieler et al., 2012). They are monoploidy and asexually reproducing, as revealed by mutation (Galloway, 1990), homologous recombination (Kilian et al., 2011)and preliminary genome sequencing (Pan et al., 2011). Various species of Nannochloropsis have been genetically modified through homologous recombination (Kilian et al.,2011) or genome editing (Wang et al., 2016; Verruto et al.,2018).
The screening and maintenance of transformants are critical steps for genetic modification. Hygromycin B is commonly used as a screening agent, and is an aminoglycoside (AGs) antibiotic produced by Streptomyces hygroscopicus. Hygromycin B contains a 2-deoxystreptamine(2-DOC) in which two or more amino-modified sugars are linked by a glycosidic bond (Pittenger et al., 1953).There are two functioning ways of hygromycin B in cells.It can bind to ribosomes to interfere ribosomal translocation and protein synthesis, or induce misinterpretation of mRNAs by ribosomes which can cause incorrect translation of proteins (Hermann et al., 2007). The aph7 gene(aph7, 1026 bp in length) from Streptomyces hygroscopicus encodes an aminoglycoside-4-phosphotransferase (Zalacain et al., 1987) which turns hygromycin B into a nonbiotoxic phosphate. In recent years, the combination of hygromycin B and aph7 has been widely used to screen the transgenic Nannochloropsis (Vieler et al., 2012; Ajjawi et al.,2017; Nobusawa et al., 2017).
As we have observed in our transformation experiment,the resistance to hygromycin B of the transformants may lose very fast. We proposed that wild Nannochloropsis may tolerate hygromycin B to a threshold and below such threshold the transformants may lose their resistance gene easily. The tolerant threshold of wild Nannochloropsis to hygromycin B is not clear, and the mechanisms underlining the tolerance and resistance of Nannochloropsis to hygromycin B are unknown. We have acclimated a single cell derived wild type N. oceanica (sensitive to hygromycin B) and obtained a strain tolerating 350 μg mL-1hygromycin B. We have also genetically modified the wild strain by transferring aph7 into its genome, and obtained transformants resisting as high as 600 μg mL-1hygromycin B.In this study, we compared the transcriptomes of adaptive(tolerant), resistant and wild type strains of N. oceanica with the physiological mechanisms underlining hygromycin B tolerance and resistance deduced from the differential responses between tolerant and resistant strains.
2 Materials and Methods
2.1 N. oceanica and Growth Condition
N. oceanica was obtained from the Key Laboratory of Marine Aquaculture of Chinese Ministry of Education,Ocean University of China, and was purified by two rounds of colony picking. N. oceanica was continuously cultured at 26℃ and under 70 μmol photon m-2s-1following a rhythm of 12 h light:12 h dark. The medium was f/2 (Guillard,1975), which was prepared with natural seawater filtrated through a membrane (0.45 μm in pore size) and autoclaved at 121℃ for 30 min. The N. oceanica was batch cultured in a 250 mL flask containing 100 mL of the medium. The solid medium was prepared by adding 1% agar powder to the liquid.
2.2 Acclimatization
Wild-type N. oceanica was inoculated into fresh f/2 seawater medium containing 50 μg mL-1hygromycin B and cultured for 15 days, then a similar amount of cells (according to OD750value, transfer X volume in 100 mL) were inoculated to new medium containing 100 μg mL-1of hygromycin B. Such inoculation and cultivation were continued until the concentration of hygromycin B reached 350 μg mL-1. This concentration was believed to be the tolerance threshold of the wild strains, and N. oceanica stopped growing with higher antibiotic concentration. A colony was picked out and cultured into the tolerant (adaptive) strain.
2.3 Transformation
The plasmid pSELECT100 was donated by the Department of Biochemistry and Molecular Biology, Michigan State University (Vieler et al., 2012). The plasmid carries the hygromycin B resistance gene, aph7, and is controlled by endogenous lipid droplet surface protein gene (LDSP)promoter (Fig.1a) and nitric oxide synthase gene (NOS) terminator. Wild N. oceanica at the exponential growth phase was collected by centrifugation at 3000 × g, washed three times with sterilized 375 mmol sorbitol, and resuspended in fresh sorbitol. The algal density was adjusted to about 5×108cells mL-1. In total, 200 μL of microalgal cells were mixed with 3 μg Xmn I digested plasmid and 3–10 μg salmon sperm DNA and transferred into a 2 mm (thickness)electroporation cuvette. After ice bathing for 5 min, the cells were electroporated using a Bio-RAD electroporator at 2200 V, 500 Ω resistance and a capacitance of 50 μF. The electroporated cells were mixed with 1 mL of ice-cold f/2 slowly, suspended gently and transferred to a test tube containing 5 mL f/2. The cells were cultured at 25℃ under continuous light with shaking (150 r min-1) for 48 h. The cells were concentrated to 50 μL by centrifugation, and uniformly spread on a solid f/2 medium containing 400 μg mL-1hygromycin B, approximately 1×108cells each plate. The plate was placed at 26℃ and under 70μmol photon m-2s-1following a rhythm of 12 h light and 12 h dark. In 4 weeks,transformants were transferred into liquid f/2 medium containing 400 μg mL-1hygromycin B. The transformants were acclimated to resist 600 μg mL-1hygromycin B by increasing the antibiotics step by step, with 50 μg mL-1increment between every two steps. A colony was picked out and cultured into the resistant strain.
2.4 PCR and Southern Blotting Hybridization
Genomic DNA was extracted using the OMEGA E.Z.N.A HP Plant DNA Kit (Omega, USA). The aph7 was amplified with 5’-CGC GCT ACT TCG AGC GGA GG-3’ (HF)and 5’-GCG CTT CTG CGG GCG ATT TG-3’ (HR). The expected size of amplified aph7 fragment was 205 bp. For Southern blotting hybridization, the DNA was digested with Bam HI, separated through electrophoresis in 0.7%agarose gel, blotted and fixed onto a nylon membrane,hybridized with digoxin-labeled the amplified aph7 fragment, and visualized using the DIP High Prime DNA Labeling and Detection Starter Kit I (Roche, Germany) following manufacturer’s introductions.
2.5 Growth Curve Drawing
The growth curves of WT (wild type strain, sensitive to hygromycin B, in hygromycin B free f/2), WT350 (WT in f/2 containing 350 μg mL-1of hygromycin B), HA (hygromycin B adaptive strain in f/2 containing 350 μg mL-1of hygromycin B) and HBR (hygromycin B resistant strain,transgenic, in f/2 containing 400 μg mL-1hygromycin B)of N. oceanica were drawn. The algal strains were cultured to their exponential growth phase, diluted to 1×108cells mL-1, and inoculated 1 mL of diluted alga to 100 mL fresh f/2 medium containing desirable concentrations of hygromycin B, and cultured at 26℃ under 70 μmol photon m-2s-1following a rhythm of 12 h light and 12 h dark. Three milliliters of algal culture was taken every day to measure the absorbance at 750 nm. The growth curves were drawn using the data collected from 4 replicates.
2.6 RNA Extraction
N. oceanica were collected on day 9 by centrifuging the cells at 6000 r min-1for 5 min, then transferred to the fresh f/2 seawater medium containing 350 μg mL-1hygromycin B and incubated for 24 h. The cells were collected again by centrifuging at 6000 r min-1for 5 min with total RNA extracted with TRIZOL Reagent (Sigma, USA) following manufacturer’s instructions. The integrity, concentration and ratios of OD260/OD230and OD260/OD280of the RNA were determined through agarose gel electrophoresis and on Nannodrop 2000 spectrophotometer and Agilent 2100, respectively.
2.7 Transcriptome Sequencing
A total amount of 1.5 μg RNA was used to construct the sequencing library using NEBNext®Ultra™ RNA Library Prep Kit for Illumina®(NEB, USA) following manufacturer’s instructions during which the index code was added to distinguish algal materials. In brief, mRNA was purified from total RNA using polyT coated magnetic beads.First strand cDNA was synthesized using random hexamers as primers and M-MuLV reverse transcriptase. Second strand cDNA was subsequently synthesized using DNA polymerase I and RNase H. The ends of DNA fragments were polished, adenylated and ligated to NEBNext Adaptor. The fragments were purified with AMPure XP system(Beckman Coulter, Beverly, USA) with cDNA (250-300 bp in length) enriched. The cDNA fragments in desirable lengths were mixed with 3 μL of USER Enzyme (NEB,USA) at 37℃ for 15 min and then at 95℃ for 5 min. These assemblage of cDNA fragments were amplified with Phusion High-Fidelity DNA polymerase, universal PCR primers and index (X) Primer and purified with AMPure XP system as the sequencing library. The quality of libraries was assessed on the Agilent Bioanalyzer 2100 System.
Raw reads were processed through in-house perl scripts.Clean reads were obtained by removing those containing adapter, ploy-N or at low-quality and calculated for Q20,Q30 and GC-content and duplication level simultaneously.Differential expression analysis was performed using the DESeq R package (Young et al., 2010). A gene was believed to express differentially between two compared samples if log2|foldchange| > 1 and q-value < 0.005. The differentially expressed genes (DEGs) were enriched into Gene Ontology (GO) function groups with GOseq R package(Young et al., 2010), and KEGG pathway (http://www.genome. jp/kegg/) with KOBAS (Mao et al., 2005) software.
3 Results and Discussion
3.1 Acclimation of N. oceanica and Isolation of an Adaptive Strain
The single cell derived wild type (WT) N. oceanica was acclimated to grow in liquid medium containing 350 μg mL-1by continuously increasing the concentration of hygromycin B with 50 μg mL-1in each batch culture cycle (1/10 volume of inoculation, at the plateau growth phase). By streaking on the solid medium at the last batch culture and colony picking, an adaptive strain tolerating 350 μg mL-1of hygromycin B was obtained. The WT failed to grow in medium containing 350 μg mL-1hygromycin B while the adaptive strain grew well in such medium (Fig.1).
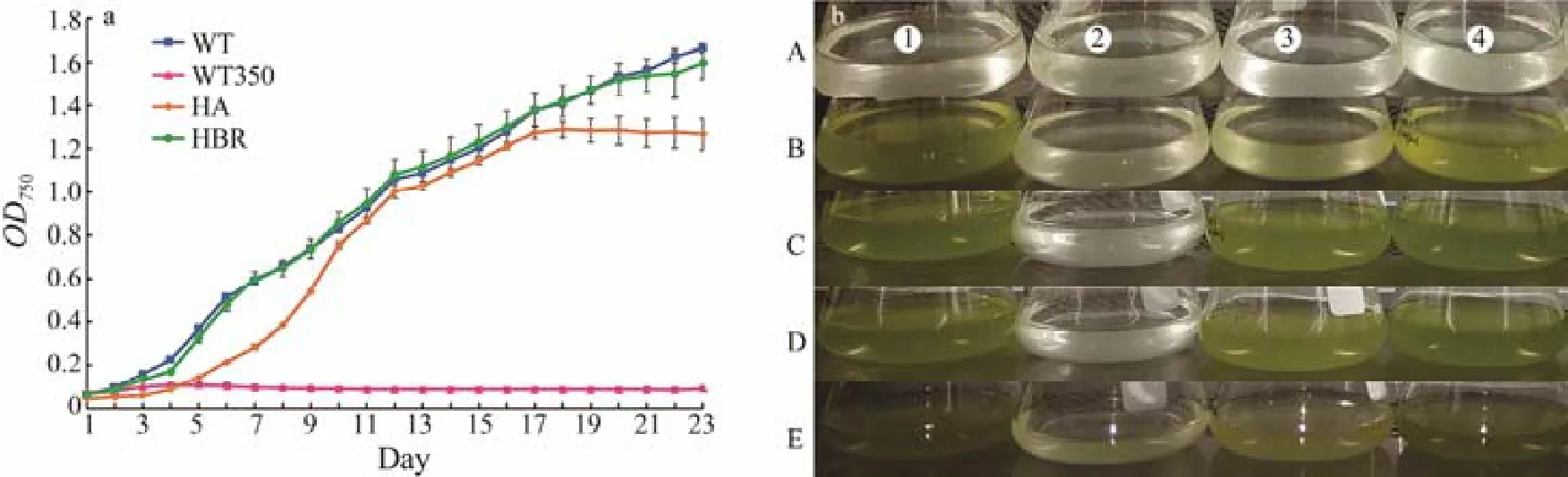
Fig.1 The growth performance of WT (wild type strain in f/2 without hygromycin B), WT350 (WT strain in f/2 containing 350 μg mL-1 of hygromycin B), HA (hygromycin adaptive strain in f/2 containing 350 μg mL-1 of hygromycin B) and HBR(hygromycin resistant strain, transgenic, in f/2 containing 350 μg mL-1 of hygromycin B) of N. oceanica (a). The growth characteristics of WT in f/2 without hygromycin B (column 1) and WT350 (column 2), HA (column 3) and HBR (column 4) in f/2 containing 350 μg mL-1 of hygromycin B on day 0 (A), day 5 (B), day 10 (C), day 15 (D) and day 20 (E) (b).
3.2 Development of Transgenic (Hygromycin B Resistant) Strains
Six transgenic colonies were randomly selected, and were verified by amplifying a fragment of aph7 and through Southern blotting hybridization. As expected, an aph7 fragment (Fig.2a) with 205 bp in length was amplified with primers HF and HR from the genomic DNA of 6 colonies(Fig.2b). Different numbers of hybridization bands (1–4)were detected among 3 strains derived from 3 transgenic colonies through Southern blotting hybridization (Fig.2c),which were longer than the plasmid corresponding bands.These results indicated that N. oceanica has been successfully transformed with aph7 gene. A strain showing a single hybridization band (lane 3 in Fig.2c) was used to decipher the resistance mechanism. The transgenic (or resistant) strains were acclimated to resist 600 μg mL-1hygromycin B and maintained at this concentration.
Hygromycin B resistant (transgenic) (HBR) strain grew well in medium containing 350 μg mL-1hygromycin B. Its growth curve was highly similar with that of WT in medium without hygromycin B. Hygromycin B adaptive (HA)grew in in medium containing 350 μg mL-1hygromycin B;however its growth curve was different from that of HBR in medium containing 350 μg mL-1hygromycin B and WT in medium without hygromycin B (Fig.1a). The color of HA and HBR strains in antibiotics containing medium was similar to that of WT in medium without hygromycin B, indicating that HA was tolerant and HBR was resistant to hygromycin B (Fig.1b).
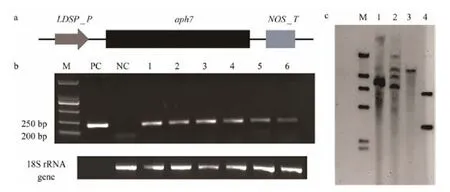
Fig.2 Resistance gene construct (a), amplification of aph7 gene fragment (b) and Southern blotting verification of resistance gene integration into chromosomes (c). In a, LDSP_P, LDSP promoter; NOS_T, NOS terminator; In b, M, DNA Mark2000; PC, positive control, pSELECT100 as template; NC, negative control, DNA of wild type N. oceanica as template; Lanes 1–6, DNA of 6 colony derived strains as template; In c, M, DNA length marker; lanes 1–3, 3 strains corresponding to those in B, lane 4, plasmid pSELECT100. aph7 is 1026 bp in length. The amplified fragment is 205 bp in length (partial). The multiple bands represent multiple copies of aph7 gene in the genome, and different band patterns of aph7 gene indicate the different position in genome where the gene is integrated. The PCR product is labeled and used as the probe of hybridization.
3.3 Transcriptomic Analysis
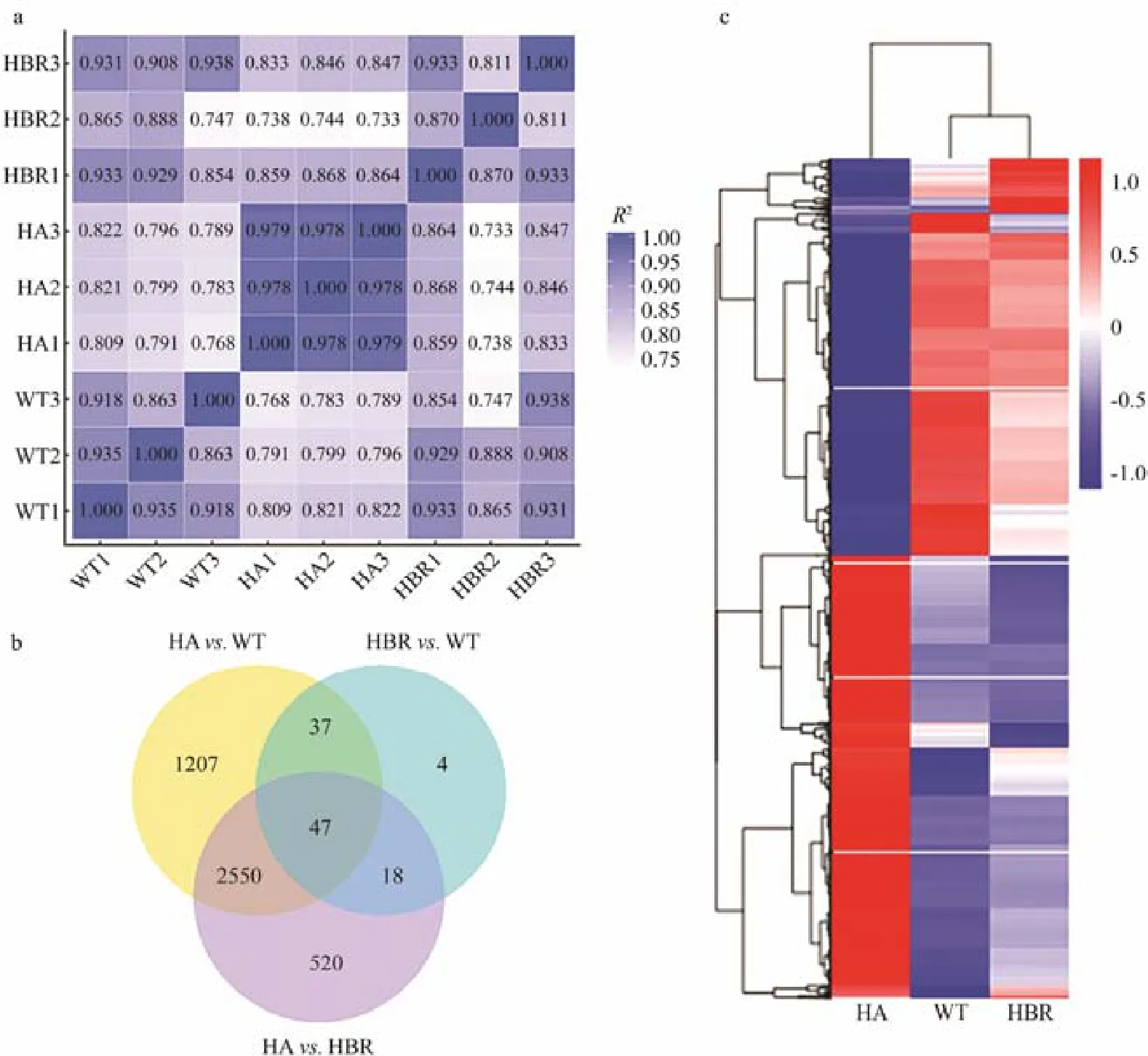
Fig.3 The differences among WT, HA and HBR N. oceanica transcriptomes. (a) The heat map of Pearson correlation between biological replicates and strains. (b) Cluster analysis of differentially expressed unigenes. WT, wild-type N. oceanica; HA, hygromycin B adaptive N. oceanica; HBR, hygromycin B resistant (transgenic) strains of N. oceanica. (c)Venn diagram of differentially expressed unigenes.
WT, HA and HBR were exposed to 350 μg mL-1of hygromycin B, 3 replicates each, with their transcriptomes sequenced and analyzed. It was found that 78.84% of 12442 unigenes can be allocated to at least one of NR, NT, KO,SwissProt, PFAM, GO and KEG databases. The biological replicates were highly similar each other, indicating that the transcriptomes were profiled appropriately. Pearson correlation coefficients between HA and WT, and between HA and HBR were low, while those between WT and HBR were high (Fig.3a). These similarities were also revealed by differentially expressed gene clustering (Fig.3b). HA was obviously different from WT and HBR while WT and HBR were highly similar with each other, indicating that that the mechanisms underlining hygromycin B tolerance of HA and hygromycin resistance of HBR are different.
3.4 Deduced Physiological Mechanisms Under Tolerance and Resistance
In total, 3841 DEGs were identified between HA and WT (Fig.3c), of which 2079 genes were up-regulated and 1762 genes were down-regulated. Four KEGG pathways were significantly enriched (corrected P-value < 0.05; Table 1), which included proteasome (ko03050), DNA replication (ko03030), mismatch repair (ko03430) and ribosome biogenesis (ko03008). It is clear that N. oceanica adapts to hygromycin B by degrading degraded and wrong folded proteins, less synthesizing proteins thus reducing growth, speeding up DNA synthesis and intensifying error correcting of DNA replication (Table 1). In addition,among the 2079 up-regulated genes, 8 were major facilitator superfamily transporter genes and 28 were ABC transporter genes (Table 2). This finding indicates that N. oceanica intensifies its ion transportation to respond the antibiotic stress. In contrast, in HBR strain only N-Glycan biosynthesis (ko00510) was barely enriched (2 out of 22 members in the pathway). No transporter gene was up-regulated in expression in HBR except for the foreign resistance gene. These findings indicate that HBR may resist hygromycin B solely by degrading the hygromycin B with the enzyme encoded by aph7.
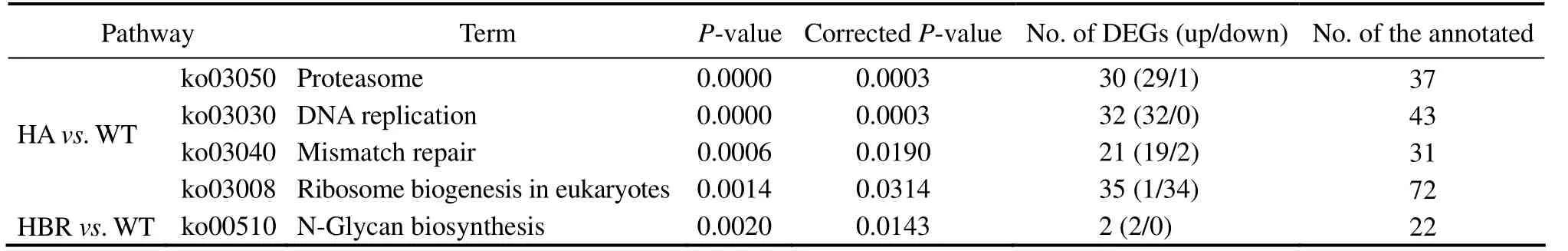
Table 1 KEGG pathways significantly enriched by DEGs between HA and WT and between HBR and WT
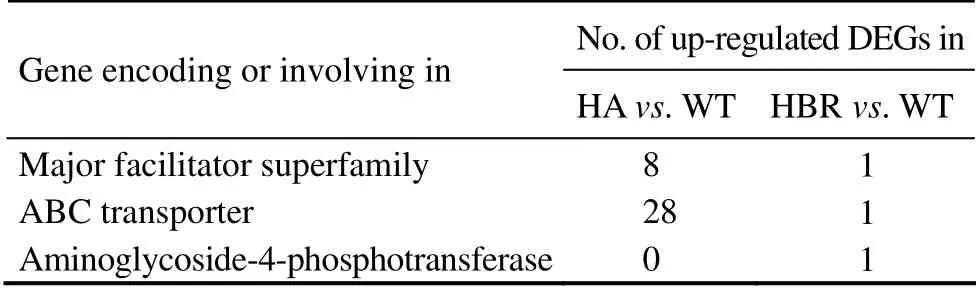
Table 2 Differentially expressed transporter and hygromycin inactivating enzyme genes in HA and HBR in comparison with WT
N. oceanica obtained the tolerance to hygromycin B by up-regulating the expression of its diverse transporter genes,as well as the misfolded protein degrading and DNA repairing associated genes, and down-regulating the expression of protein synthesis related genes. The microalga obtained its resistance to the antibiotics by expressing solely the foreign resistance gene. The tolerance of N. oceanica to hygromycin B was as high as 350 μg mL-1hygromycin B while its resistance to this antibiotics reached 600 μg mL-1hygromycin B. Its tolerance to hygromycin was obtained by acclimating to the antibiotics. The resistant transformants should be screened at a concentration higher than the threshold of tolerance, for example, 400 μg mL-1hygromycin B. The transformants should be maintained at even a higher concentration that is 600 μg mL-1hygromycin B in this study. In summary, it was extremely important that the resistance to 600 μg mL-1hygromycin B should be obtained through acclimation after transformation.
Acknowledgements
This study was financially supported by the National Key R&D Program of China (Nos. 2018YFD0900305 and 2018YFD0901506), the Marine S & T Fund of Shandong Province for Pilot National Laboratory for Marine Science and Technology (Qingdao) (No. 2018SDKJ0406-3),and the Fundamental Research Funds for the Central Universities (No. 201762017).
 Journal of Ocean University of China2020年2期
Journal of Ocean University of China2020年2期
- Journal of Ocean University of China的其它文章
- Abyssal Circulation in the Philippine Sea
- Numerical Study of Storm Surge Inundation in the Southwestern Hangzhou Bay Region During Typhoon Chan-Hom in 2015
- Interannual Variability and Scenarios Projection of Sea Ice in Bohai Sea Part I: Variation Characteristics and Interannual Hindcast
- Probability Distribution of the Hull Motion and Mooring Line Tension of Two Floating Systems
- Suppression of Vortex-Induced Vibration by Fairings on Marine Risers
- The Characteristics of Storm Wave Behavior and Its Effect on Cage Culture Using the ADCIRC+SWAN Model in Houshui Bay, China
