入侵昆虫基因组研究进展
黄聪 李有志 杨念婉


摘要 随着全球贸易的加速发展,入侵物种对农林业、生态环境及人类健康的威胁日益严重。基因组学研究为阐明外来有害生物入侵的分子机制与生态适应性过程以及研发新型防控技术提供了新手段、新平台与大数据。本文综述了入侵昆虫基因组学研究的发展现状,系统总结了基因/基因家族、转座子/重复序列等基因组信息在决定昆虫入侵性中的重要作用,着力探讨了基因组学研究在助力害虫RNAi、昆虫不育技术 (SIT)、化学生态防治和物理防治等防控新技术/新产品开发方面的潜力,并展望了基因组学研究应用于入侵昆虫综合防控的前景。
关键词 入侵昆虫; 基因组; 入侵性
中图分类号: Q 963 文献标识码: A DOI: 10.16688/j.zwbh.2019050
Abstract With the accelerated development of global trade, invasive species seriously threaten agriculture, forestry, ecological environment and human health. Genomic study provides novel tools, platforms and big data for elucidating the molecular mechanisms and ecological adaptation of the invasive alien species (IAS), and developing new prevention and control technologies. We reviewed the status of invasive insect genomic study, systematically summarized the important roles of genes, gene families, transposons and repetitive sequences in determining insect invasiveness. We also analyzed the potential of genomic study in facilitating the development of novel pest control technologies or products such as RNAi, sterile insect technique (SIT), chemical ecology and physical methods, and envisioned the prospect of the application of genomic study for the integrated management of invasive pests.
Key words invasive insect; genome; invasiveness
外来入侵物种(invasive alien species, IAS) 是指分布在原产地以外、建立了能够自我维持的种群并对当地的经济、生态和社会安全造成威胁的物种[1]。在过去的200年间,全球外来入侵物种数量持续增长;尤其近50年来,随着全球贸易的加速发展,外来物种入侵的增长越发加剧,全球16 926个新增外来物种中有超过三分之一 (37%)发生在1970年-2014年间[2]。此外,对全球首次记录的外来物种风险评估发现,其中约16%的物种具有一定的入侵潜力[3],一旦被有意或无意地引入原产地以外的生境,不仅会危及当地经济和生态,也会威胁到人类健康、食物资源以及国家安全[4]。昆虫是世界上种类最多的动物群体,占所有生物种类50%以上,因此,外来入侵昆虫对人类造成的危害十分严重,据统计,每年由于外来入侵昆虫导致的全球经济和人类健康损失分别超过700亿美元和69亿美元[5]。作为世界上最大的两个经济体,中国和美国是遭受外来入侵生物威胁最严重的两个国家,年度经济损失分别达到189亿和400亿美元[4,6]。
明确入侵机制有利于高效预警、阻截和治理外来入侵昆虫。基因组学技术的迅速发展为深入挖掘入侵昆虫的入侵途径和适应性机制带来了契机,科学家们正试图利用组学数据来研究入侵生物学[7-9]。基因组包含生物体的全部遗传信息,一切与入侵分子机制相关的遗传密码均贮藏在基因组中,通过对入侵生物进行基因组测序和重测序,深入挖掘分析组学数据有利于解密入侵物种入侵的分子基础、遗传机制和进化过程。本文对目前已完成基因组测序的入侵昆虫进行了梳理,分析了入侵昆虫基因组测序进展,总结了利用基因组测序技术揭示昆虫入侵分子机制的研究思路,以及探讨了基因组研究在害虫防控新技术/新产品开发中的推动作用。
1 入侵昆虫基因组测序现状
1.1 入侵昆虫基因组测序及动态
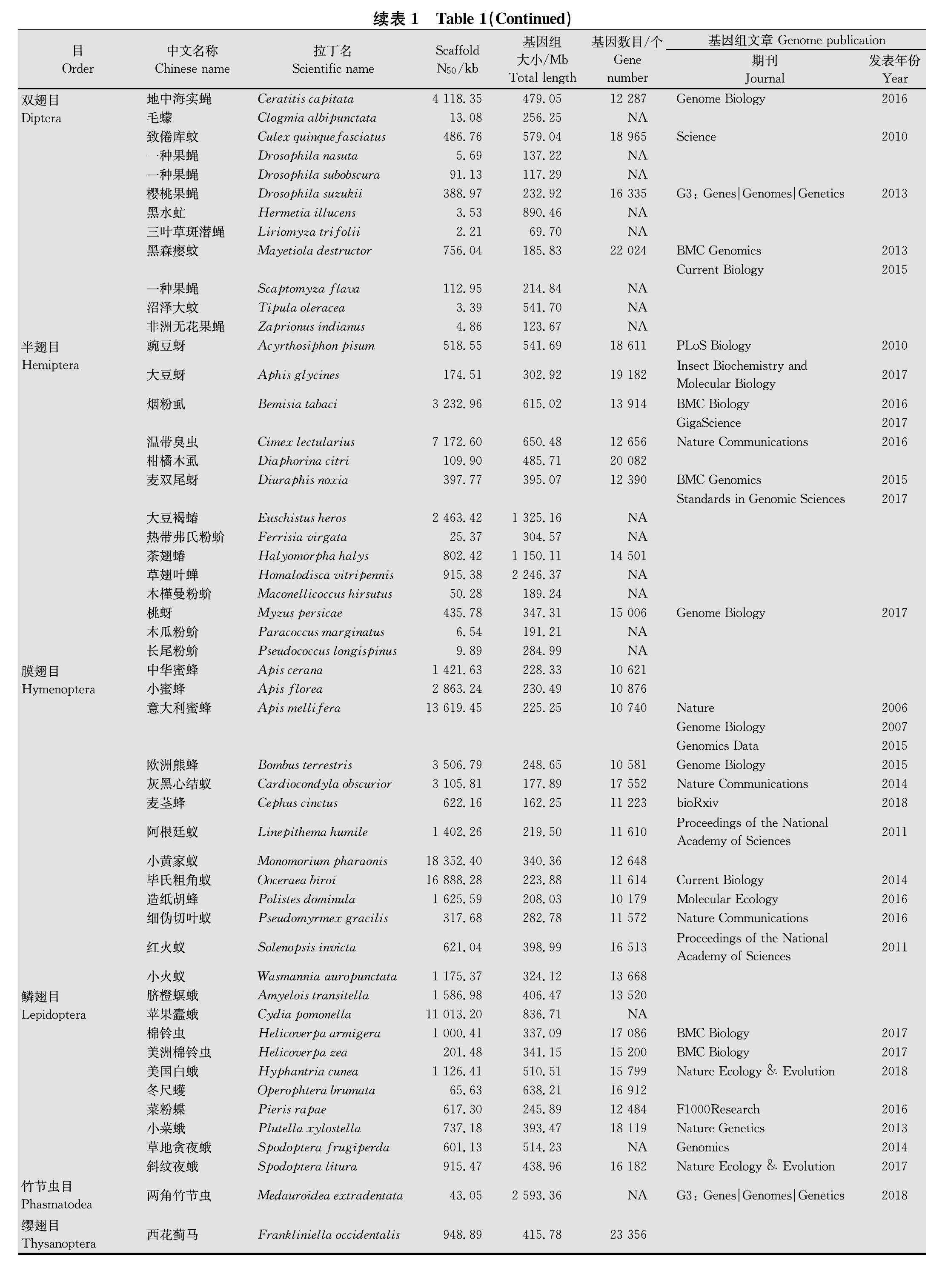
组学技术的发展极大地促进了昆虫学的研究,随着测序技术的不断革新,近年来越来越多的昆虫基因组被测序,据统计,2016年12月底之前已完成基因组拼接的昆虫215种[10],而2017年12月增至近260种[11],截至2018年12月,NCBI中已有331个完整拼接的昆虫基因组,其中入侵昆虫67种 (20.2%) (图1,表1)。入侵昆虫鉴定参考中国外来入侵物种数据库(http:∥www.chinaias.cn/wjPart/index.aspx)、GBIF (http:∥data.gbif.org)、CABI(www.cabi.org/isc)和IOBIS (http:∥www.iobis.org/)等数据库。
分析67种入侵昆虫基因组提交至NCBI的时间动态可以看出,2011年以前入侵昆虫基因组测序进展缓慢,每年新增入侵昆虫基因组数目基本为1个,这与整个昆虫基因組测序动态基本一致[10],2011年以后随着测序技术的革新,完成基因组测序的入侵昆虫数量有了大幅增长,到2015年达到顶峰,新提交14个入侵昆虫基因组数据,随后,入侵昆虫基因组测序进展速度放慢,每年新增7~10个 (图1a)。
入侵昆虫在长期与寄主植物互作过程中,为扩大寄主范围及适应多样化的寄主,基因组中与寄主定位、化合物识别、食物消化和解毒代谢相关的基因家族发生了扩张,甚至进化出种系特异的基因家族。红火蚁Solenopsis invicta和阿根廷蚁是两种杂食性入侵昆虫,与近缘非入侵蚂蚁比较发现入侵蚂蚁具有更强的气味识别和侦测能力[32],这得益于它们基因组中气味受体的大量拷贝,如红火蚁基因组中拥有超过400个气味受体,阿根廷蚁基因组中气味受体也多达367个。由于寄主多样性,多食性的入侵昆虫通常具有发达的味觉感受系统以识别不同寄主植物的化合物成分,如多食性的棉铃虫、斜纹夜蛾和美国白蛾Hyphantria cunea基因组中味觉受体(GR)发生大量扩张,分别为213、237和147個GRs,远高于单食性和寡食性昆虫的45~76个[33-36]。值得注意的是,与抗药性相关的解毒代谢类基因家族也被认为与一些入侵昆虫的寄主适应性和多食性有关,如棉铃虫、烟粉虱和斜纹夜蛾基因组中与解毒代谢相关的基因家族 (如P450s、UGTs、GSTs和羧酸酯酶等)的扩张[15,26,30]。多食性的入侵昆虫取食寄主植物后,为了消化各类寄主植物,与消化相关的基因家族也发生了特异性扩张,如棉铃虫基因组中与食物消化相关的丝氨酸蛋白酶(主要是胰蛋白酶和胰凝乳蛋白酶)基因家族发生了显著扩张;光肩星天牛和两角竹节虫基因组中与编码降解植物细胞壁的相关酶的基因家族发生扩张[31, 37];美国白蛾基因组中参与碳水化合物代谢通路相关的基因家族发生了扩张[38]。
2.2 转座子/重复序列与昆虫入侵性
转座子是真核生物基因组进化的主要驱动力[39],许多物种在不利环境下的适应性进化都离不开转座子的作用[40-41],在进化压力胁迫下转座子活性可引起物种基因组大小改变[42-43]、物种分化[44]、基因复制[45] 和新基因产生[46] 等,以适应各种进化压力。研究表明由转座子导致的入侵物种基因组大小的改变、基因组结构的变化等可引起入侵种的快速适应性进化[47-49]。即入侵物种进入新环境后往往由少数遗传多样性很低的建群种迅速扩张种群,即“奠基者效应”,但遗传多样性低的建群种如何迅速适应新环境并扩散分布,一直以来困扰着入侵生物学者。对转座子的研究表明,新环境压力诱导的转座子活性会改变基因的作用,促进结构变异,进而产生迅速适应新环境的特性[50]。
昆虫基因组中也存在大量的转座子[51],可以引起基因组大小进化[52],或调节昆虫寿命[53]。近期研究成果表明转座子在入侵昆虫入侵性中也起着重要作用,能够促进入侵昆虫的适应性进化。通过基因组测序发现入侵蚂蚁灰黑心结蚁基因组在已测序蚁科昆虫中属于最小的,其基因组中含有7.18%的转座子,但集中在高密度的转座子岛中,转座子岛表现出更高的进化速率和序列多样性,该岛中的基因较非转座子岛区域的基因拷贝数变异发生更普遍,具有更高的进化速率,如与幼虫发育、抗药性、繁殖和化学感受相关的基因变异有助于对新环境的适应,促进入侵的奠基者种群迅速扩张[14]。在蚊科中,广泛分布的入侵种白纹伊蚊拥有目前已知蚊科昆虫中最大的基因组,主要原因是其基因组含有68%的重复序列,其转座子含量是本地限制性分布种冈比亚按蚊Anopheles gambiae 的10倍,这种高含量的转座子被认为与其入侵性有关[29]。随后的研究也证实了这一猜想,研究人员为了揭示白纹伊蚊同时适应温带和热带气候环境的生态可塑性机制,对140个来自越南本地种和欧洲入侵种的白纹伊蚊雌成虫进行高通量测序,分析发现生活在温带气候的欧洲种群具有更高的转座子插入频率,是入侵后的适应性进化的结果,对这些高频率转座子插入区域的基因分析发现与滞育、磷脂和保幼激素代谢通路相关的基因可能与入侵种对温度的适应性进化相关[54]。
3 基因组研究助力害虫防控新技术/新产品开发 通过分析基因组数据可以获得入侵昆虫的全部遗传信息,挖掘与其重要生物学特性相关的基因,进而揭示其入侵性的分子机制。目前,化学农药仍然是害虫防治最主要的手段,如前所述,入侵昆虫往往具有很强的适应性进化能力,迅速产生抗药性,因此,开发新型绿色安全的害虫防治新途径尤为重要。
3.1 基因组研究助力RNAi防治技术
利用RNAi技术控制害虫被证明是一项行之有效的方法,具有广阔的应用前景[55-57]。研究表明将双链RNAs (dsRNAs) 加入到入侵昆虫玉米根萤叶甲Diabrotica virgifera virgifera人工饲料中可显著引起幼虫发育畸形和死亡[56],近期研究结果也表明,转基因马铃薯(干扰马铃薯甲虫Leptinotarsa decemlineata蜕皮相关的蜕皮激素受体基因)对马铃薯甲虫具有明显的抗性,可显著减少马铃薯甲虫造成的危害[58]。目前,玉米根萤叶甲和马铃薯甲虫基因组均已测序和拼接完成,通过基因组分析,我们可以鉴定出更多可用于RNAi的靶基因,开发针对入侵昆虫的绿色安全防治技术。值得注意的是,马铃薯甲虫基因组文章中已发现更多的可用于RNAi的靶基因[59],黑森瘿蚊Mayetiola destructor基因组分析也鉴定出一些具有潜在利用价值的RNAi靶基因[60],相信在不久的将来,RNAi技术防治入侵害虫必定会取得巨大的成功。
3.2 基因组研究助力SIT防治技术
不育昆虫释放技术(sterile insect technique, SIT)是一种环境友好型和可持续的害虫防控技术。该技术包括两个难题,一是如何快速、准确地获得单一雄虫,二是如何导致雄虫不育。昆虫转基因技术和基因编辑技术(如CRISPR/Cas9)为SIT技术的改进提供了契机。利用转基因技术可对害虫基因组进行遗传修饰,获得单一雄虫[61],如通过与受四环素调控的tetoff基因表达系统构建地中海实蝇雌性特异致死系统可实现雌性特异的早期发育致死[62],基因组研究为该技术提供了更多的候选致死基因,如通过基因组分析,研究人员鉴定出地中海实蝇基因组中的RHG促细胞凋亡基因 (reaper、hid和grim)等致死基因,可用于指导该技术的应用[25]。此外,实蝇类害虫SIT技术中也常用遗传突变的白蛹来区分雌雄虫[63-64],然而利用遗传学方法获得突变的白蛹品系效率低、耗时长,其遗传分子机制尚不清楚,基因组学研究为解决这一科学问题提供了技术支持,如研究人员通过基因组测序组装获得高质量的染色体级别的瓜实蝇Bactrocera cucurbitae基因组,分析表明导致白蛹的突变位点位于3号染色体的42 Mb位置附近,结果对于指导其他物种获取类似的突变品系具有重大意义[65]。
[6] PAINI D R, SHEPPARD A W, COOK D C, et al. Global threat to agriculture from invasive species [J]. Proceedings of the National Academy of Sciences, 2016, 113(27): 7575-7579.
[7] RIUS M, BOURNE S, HORNSBY H G, et al. Applications of nextgeneration sequencing to the study of biological invasions [J]. Current Zoology, 2015, 61(3): 488-504.
[8] KIRK H, DORN S, MAZZI D. Molecular genetics and genomics generate new insights into invertebrate pest invasions [J]. Evolutionary Applications, 2013, 6(5): 842-856.
[9] TAY W T, GORDON K. Going globalgenomic insights into insect invasions [J]. Current Opinion in Insect Science, 2018, 31: 1-8.
[10] 尹傳林, 李美珍, 贺康, 等. 昆虫基因组及数据库研究进展[J]. 环境昆虫学报, 2017,39(1): 1-18.
[11] RICHARDS S, CHILDERS A, CHILDERS C. Editorial overview: Insect genomics: Arthropod genomic resources for the 21st century: It only counts if its in the database![J]. Current Opinion in Insect Science, 2018, 25: ivvii.
[12] MATTHEWS B J, DUDCHENKO O, KINGAN S B, et al. Improved reference genome of Aedes aegypti informs arbovirus vector control [J]. Nature, 2018, 563(7732): 501-507.
[13] HUEY R B, GILCHRIST G W, HENDRY A P. Using invasive species to study evolution[M]∥SAX D F, STACHOWICZ J J, GAINES S D. Species invasions: Insights to ecology, evolution and biogeography. Sunderland: Sinauer Associates. 2005: 139-164.
[14] SCHRADER L, KIM J W, ENCE D, et al. Transposable element islands facilitate adaptation to novel environments in an invasive species [J]. Nature Communications, 2014, 5: 5495.
[15] PEARCE S L, CLARKE D F, EAST P D, et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species [J]. BMC Biology, 2017, 15(1): 63.
[16] WANG Huidong, SHI Yu, WANG Lu, et al. CYP6AE gene cluster knockout in Helicoverpa armigera reveals role in detoxification of phytochemicals and insecticides [J/OL]. Nature Communications, 2018, DOI:10.1038/s41467018072266.
[17] JIN Lin, WANG Jing, GUAN Fang, et al. Dominant point mutation in a tetraspanin gene associated with fieldevolved resistance of cotton bollworm to transgenic Bt cotton [J]. Proceedings of the National Academy of Sciences, 2018, 115(46): 11760-11765.
[18] HUFBAUER R A, TORCHIN M E. Integrating ecological and evolutionary theory of biological invasions [M]∥NENTWIG W. Biological invasions. Berlin: Springer. 2008: 79-96.
[19] BLOSSEY B, NOTZOLD R. Evolution of increased competitive ability in invasive nonindigenous plantsa hypothesis[J]. Journal of Ecology, 1995, 83(5): 887-889.
[20] BENFEY P N, MITCHELLOLDS T. From genotype to phenotype: systems biology meets natural variation[J]. Science, 2008, 320(5875): 495-497.
[21] SUZUKI H C, OZAKI K, MAKINO T, et al. Evolution of gustatory receptor gene family provides insights into adaptation to diverse host plants in nymphalid butterflies [J]. Genome Biology and Evolution, 2018, 10(9): 2240.
[22] BASS C, FIELD L M. Gene amplification and insecticide resistance [J].Pest Management Science,2011,67(8):886-890.
[23] BOULAIN H, LEGEAI F, GUY E, et al. Fast evolution and lineagespecific gene family expansions of aphid salivary effectors driven by interactions with hostplants [J]. Genome Biology and Evolution, 2018, 10(6): 1554-1572.
[24] LIU Deguang, TRUMBLE J T. Comparative fitness of invasive and native populations of the potato psyllid (Bactericera cockerelli)[J]. Entomologia Experimentalis Et Applicata, 2007, 123(1): 35-42.
[25] PAPANICOLAOU A, SCHETELIG M F, ARENSBURGER P, et al. The whole genome sequence of the Mediterranean fruit fly, Ceratitis capitata (Wiedemann), reveals insights into the biology and adaptive evolution of a highly invasive pest species [J]. Genome Biology, 2016, 17(1): 192.
[26] CHEN Wenbo, HASEGAWA D K, KAUR N, et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance[J/OL].BMC Biology,2016,14(1):110.DOI:10.1186/s129150160321y.
[27] XIE Wen, YANG Xin, CHEN Chunhai, et al. The invasive MED/Q Bemisia tabaci genome: a tale of gene loss and gene gain [J]. BMC Genomics, 2018, 19(1): 68.DOI:10.1186/s1286401841489.
[28] SMITH C D, ZIMIN A, HOLT C, et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile) [J].Proceedings of the National Academy of Sciences, 2011, 108(14): 5673-5678.
[29] CHEN Xiaoguang, JIANG Xuanting, GU Jinbao, et al. Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution[J]. Proceedings of the National Academy of Sciences of the United States of America, 2015, 112(44): E5907E5915.
[30] CHENG Tingcai, WU Jiaqi, WU Yuqian, et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest [J]. Nature Ecology & Evolution, 2017, 1(11): 1747-1756.
[31] MCKENNA D D, SCULLY E D, PAUCHET Y, et al. Genome of the Asian longhorned beetle (Anoplophora glabripennis), a globally significant invasive species, reveals key functional and evolutionary innovations at the beetleplant interface [J]. Genome Biology, 2016, 17(1): 227.DOI:10.1186/s1305901610888.
[32] HOLWAY D A.Competitive mechanisms underlying the displacement of native ants by the invasive Argentine ant [J]. Ecology, 1999, 80(1): 238-251.
[33] YOU Minsheng, YUE Zhen, HE Weiyi, et al. A heterozygous moth genome provides insights into herbivory and detoxification [J]. Nature Genetics, 2013, 45(2): 220-225.
[34] BRISCOE A D, MACIASMUNOZ A, KOZAK K M, et al. Female behaviour drives expression and evolution of gustatory receptors in butterflies [J/OL].PLoS Genetics,2013,9(7): e1003620.
[35] GUO Huizhen, CHENG Tingcai, CHEN Zhiwei, et al. Expression map of a complete set of gustatory receptor genes in chemosensory organs of Bombyx mori [J]. Insect Biochemistry and Molecular Biology, 2017, 82: 74-82.
[36] KOENIG C, HIRSH A, BUCKS S, et al. A reference gene set for chemosensory receptor genes of Manduca sexta[J]. Insect Biochemistry and Molecular Biology, 2015, 66: 51-63.
[37] BRAND P, LIN Wei, JOHNSON B R. The draft genome of the invasive walking stick, Medauroidea extradentata, reveals extensive lineagespecific gene family expansions of cell wall degrading enzymes in Phasmatodea[J]. G3: Genes|Genomes|Genetics, 2018, 8(5): 1403-1408.
[38] WU Ningning, ZHANG Sufang, LI Xiaowei, et al. Fall webworm genomes yield insights into rapid adaptation of invasive species [J]. Nature Ecology & Evolution, 2018, 3: 105-115.
[39] BELYAYEV A. Bursts of transposable elements as an evolutionary driving force[J]. Journal of Evolutionary Biology, 2014, 27(12): 2573-2584.
[40] SCHRADER L, SCHMITZ J. The impact of transposable elements in adaptive evolution [J/OL].Molecular Ecology,2018.DOI:10.1111/mec.14794.
[41] LI Ziwen, HOU Xinghui, CHEN Jiafu, et al. Transposable elements contribute to the adaptation of Arabidopsis thaliana[J].Genome Biology and Evolution,2018,10(8):2140-2150.
[42] KREINER J M, WRIGHT S I. A less selfish view of genome size evolution in maize [J/OL].PLoS Genetics,2018,14(5):e1007249.
[43] PELLICER J, HIDALGO O, DODSWORTH S, et al. Genome size diversity and its impact on the evolution of land plants [J]. Genes, 2018, 9(2).DOI:10.3390/genes9020088.
[44] SERRATOCAPUCHINA A,MATUTE D R.The role of transposable elements in speciation [J/OL]. Genes, 2018, 9(5): 254.DOI:10.3390/genes9050254.
[45] CERBIN S, JIANG N. Duplication of host genes by transposable elements [J/OL]. Current Opinion in Genetics & Development, 2018, 49: 63-69.DOI:10.1016/j.gde.2018.03.005.
[46] JOLYLOPEZ Z, HOEN D R, BLANCHETTE M, et al. Phylogenetic and genomic analyses resolve the origin of important plant genes derived from transposable elements [J]. Molecular Biology and Evolution, 2016, 33(8): 1937-1956.
[47] LAVERGNE S, MUENKE N J, MOLOFSKY J. Genome size reduction can trigger rapid phenotypic evolution in invasive plants [J]. Annals of Botany, 2010, 105(1): 109-116.
[48] PYSEK P, SKALOVA H, CUDA J, et al. Small genome separates native and invasive populations in an ecologically important cosmopolitan grass [J]. Ecology, 2018, 99(1): 79-90.
[49] SPECCHIA V, JANZEN S, MARINI G, et al. The potential link between mobile DNA and the invasiveness of the species[J].Journal of RNAi & Gene Silencing,2017,13(1):557-561.
[50] STAPLEY J, SANTURE A W, DENNIS S R. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species [J]. Molecular Ecology, 2015, 24(9): 2241-2252.
[51] PECCOUD J, LOISEAU V, CORDAUX R, et al. Massive horizontal transfer of transposable elements in insects [J].Proceedings of the National Academy of Sciences, 2017, 114(18): 4721-4726.
[52] TALLA V, SUH A, KALSOOM F, et al. Rapid increase in genome size as a consequence of transposable element hyperactivity in woodwhite (Leptidea) butterflies [J]. Genome Biology and Evolution, 2017, 9(10): 2491-2505.
[53] ELSNER D, MEUSEMANN K, KORB J. Longevity and transposon defense, the case of termite reproductives [J]. Proceedings of the National Academy of Sciences, 2018, 115(21): 5504-5509.
[54] GOUBERT C, HENRI H, MINARD G, et al. Highthroughput sequencing of transposable element insertions suggests adaptive evolution of the invasive Asian tiger mosquito towards temperate environments [J].Molecular Ecology,2017,26(15):3968-3981.
[55] BURAND J P, HUNTER W B. RNAi: future in insect management [J]. Journal of Invertebrate Pathology, 2013, 112: S68S74.
[56] BAUM J A, BOGAERT T, CLINTON W, et al. Control of coleopteran insect pests through RNA interference [J]. Nature Biotechnology, 2007, 25(11): 1322-1326.
[57] KATOCH R, SETHI A, THAKUR N, et al. RNAi for insect control: current perspective and future challenges [J]. Applied Biochemistry and Biotechnology, 2013, 171(4): 847-873.
[58] HUSSAIN T, AKSOY E, CALISKAN M E, et al. Transgenic potato lines expressing hairpin RNAi construct of moltingassociated EcR gene exhibit enhanced resistance against Colorado potato beetle (Leptinotarsa decemlineata, Say) [J]. Transgenic Research, 2019, 28(1): 151-164.
[59] SCHOVILLE S D, CHEN Y H, ANDERSSON M N, et al. A model species for agricultural pest genomics: the genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae)[J/OL]. Scientific Reports,2018,8(1):1931.DOI:10.1038/s41598018201541.
[60] KHAJURIA C, WILLIAMS C E, BOUHSSINI M E, et al. Deep sequencing and genomewide analysis reveals the expansion of MicroRNA genes in the gall midge Mayetiola destructor[J].BMC Genomics,2013,14:187.DOI:10.1186/1471216414187.
[61] 武强, 吕志创, 张桂芬, 等. 遗传控制技术在实蝇类害虫中的研究进展[J]. 生物安全学报, 2015, 24(2): 161-170.
[62] FU Guoliang, CONDON K C, EPTON M J, et al. Femalespecific insect lethality engineered using alternative splicing [J]. Nature Biotechnology, 2007, 25(3): 353-357.
[63] ISASAWIN S, AKETARAWONG N, THANAPHUM S. Characterization and evaluation of microsatellite markers in a strain of the oriental fruit fly, Bactrocera dorsalis (Diptera: Tephritidae), with a genetic sexing character used in sterile insect population control [J]. European Journal of Entomology, 2012, 109(3): 331-338.
[64] MCINNIS D O, TAM S, LIM R, et al. Development of a pupal colorbased genetic sexing strain of the melon fly, Bactrocera cucurbitae (Coquillett) (Diptera: Tephritidae)[J]. Annals of the Entomological Society of America,2004,97(5):1026-1033.
[65] SIM S B, GEIB S M. A chromosomescale assembly of the Bactrocera cucurbitae genome provides insight to the genetic basis of white pupae [J]. G3: Genes|Genomes|Genetics, 2017, 7(6): 1927-1940.
[66] KANDUL N P, LIU Junru, SANCHEZ C H, et al. Transforming insect population control with precision guided sterile males with demonstration in flies [J/OL].Nature Communications, 2019, 10(1): 84.DOI:10.1038/s41467018079647.
[67] ZHANG Ruibin, WANG Bing, GROSSI G, et al. Molecular basis of alarm pheromone detection in aphids [J]. Current Biology, 2017, 27(1): 55-61.
[68] JAYANTHI K P, KEMPRAJ V, AURADE R M, et al. Computational reverse chemical ecology: virtual screening and predicting behaviorally active semiochemicals for Bactrocera dorsalis [J/OL]. BMC Genomics, 2014, 15: 209.DOI:10.1186/1471216415209.
[69] LEAL W S, BARBOSA R M, XU Wei, et al. Reverse and conventional chemical ecology approaches for the development of oviposition attractants for Culex mosquitoes[J/OL]. PLoS ONE, 2008, 3(8): e3045.DOI:10.1371/journal.pone.0003045.
[70] 万方浩,严盈,王瑞,等.中国入侵生物学学科的构建与发展[J].生物安全学报,2011,20(1): 1-19.
[71] RICHARDSON D M, PYEK P. Fifty years of invasion ecologythe legacy of Charles Elton[J]. Diversity and Distributions, 2008, 14(2): 161-168.
[72] 羅嘉鹏. 利用组学数据检测昆虫的抗药性和入侵性[D]. 南京: 南京师范大学, 2018.
[73] QIAN Wanqiang, WAN Fanghao. China launches the “IAS1000 Project” [J]. Journal of Integrative Agriculture, 2018, 17(12): 2840-2841.
(责任编辑: 田 喆)

