Association of XPG rs2094258 polymorphism with gastric cancer prognosis
Xiao-Qin Wang, Paul D Terry, Yang Li, Yue Zhang, Wen-Jing Kou, Ming-Xu Wang
Abstract BACKGROUND The xeroderma pigmentosum group G (XPG) gene at chromosome 13q33 consists of 15 exons, which may be related to the occurrence and development of gastric cancer (GC).AIM To examine the association of several common single nucleotide polymorphisms(SNPs) of the XPG gene with GC risk and survival.METHODS Five SNPs of XPG (rs2094258, rs751402, rs873601, rs2296147, and rs1047768) were genotyped by PCR restriction fragment length polymorphism in 956 histologically confirmed GC cases and 1012 controls in North China. GC patients were followed for survival status and, if deceased, cause of death. Logistic regression and Cox regression were used for analysing associations of XPG SNPs with risk of GC and prognosis, respectively. For rs2094258, heterozygous model(CT vs CC), homozygous model (TT vs CC), recessive model (TT vs CT + CC), and dominant model (TT + CT vs CC) were analyzed.RESULTS None of the examined loci were statistically associated with GC risk, although rs2296147 was marginally associated with GC risk (P = 0.050). GC patients with the rs2094258 CT + CC genotype showed worse survival than those with the TT genotype (log-rank test, P = 0.028), and patients with the CC genotype had a tendency of unfavourable prognosis compared with those with the TT + CT genotype (log-rank test, P = 0.039). The increase in C alleles of rs2094258 [hazard ratio (HR) = 1.19, 95% confidence interval (CI): 1.02-1.45, P = 0.037] were associated with the long-term survival of GC cases. Other risk factors for survival included tumor differentiation (HR = 4.51, 95%CI: 1.99-8.23, P < 0.001),lymphovascular invasion (HR = 1.97, 95%CI: 1.44-3.01, P < 0.001), and use of chemotherapy (HR = 0.81, 95%CI: 0.63-0.98, P = 0.041).CONCLUSION The XPG rs2094258 polymorphism may be associated with overall survival in GC patients.
Key words: Xeroderma pigmentosum group G; Single nucleotide polymorphisms;rs2094258; Gastric cancer; Prognosis
INTRODUCTION
Gastric cancer (GC) is one of the global leading causes of cancer-related death[1]. The World Health Organization (WHO) estimates that 1.03 million people are diagnosed and 783000 die of the disease each year[2]. The highest age standard incidence per 100000 population is observed in Eastern Asia[3], especially in China, at 22.7 per 100000[4]. Multiple factors contribute to the development of this disease, including environmental and genetic factors, such asHelicobacter pylori(H. pylori) infection,drinking, obesity, a high-salt diet, and various genetic factors[5]. Although GC morbidity and mortality have declined in recent years, the social and economic burden of the disease remains high[1,6,7].
DNA repair genes play a key role in maintaining the genomic DNA stability and integrity. Functional genetic variants of DNA repair genes may change the host DNA repair ability and thus affect tumor prognosis[8]. DNA repair genes participate in many crucial pathways including nucleotide excision repair (NER), which is involved in the repair of some types of DNA damage. Recent evidence suggests that NER factors function in processes that facilitate mRNA synthesis or shape threedimensional chromatin structure[9]. Xeroderma pigmentosum group G (XPG or ERCC5) plays a key role in NER repair, as it can recognize DNA damage and initiate the NER process[10]. XPG has 3’-junction cutting ability on bubble substrates, resulting in 3’-incision in the human double-incision) repair system, and non-catalytically XPG is needed for subsequent 5’-incision by XPF-ERCC1[11].
Some studies have demonstrated that the single nucleotide polymorphisms (SNPs)ofXPGmay affect the development of cancer, such as lung cancer[12,13], colorectal cancer[14,15], breast cancer[16], neuroblastoma[17], Hodgkin’s lymphoma[18], and oral squamous cell carcinoma[19]. However, only a few studies have explored the associations betweenXPGgene SNPs and GC, and those studies show inconsistent results. Some studies reported no associations[20-22], while others demonstrated varying degrees of association[23,24]. Furthermore, few studies have explored the potential prognostic importance ofXPGSNPs in GC patients[25]. Therefore, we examined GC risk and survival in relation to common functionalXPGSNPs in a case-control study in North China.
MATERIALS AND METHODS
Study subjects
From September 2010 to June 2013, pathologically confirmed incident GC cases were selected from the First Affiliated Hospital of Xi’an Jiaotong University. During the same time period, controls were recruited from the Physical Examination Centre of the First Affiliated Hospital of Xi’an Jiaotong University. The cases and controls were matched by sex, age (within 5 years), and residential district. Socio-demographic and clinical data were collected during recruitment, such as alcohol consumption and smoking status. TNM staging of GC tumors was done according to the WHO standard.H. pyloriinfection status was tested by ELISA. The present study protocol was approved by the Institutional Review Board of Health Science Center of Xi’an Jiaotong University. Informed consent was obtained from all study participants.
Follow-up
Cases were followed for survival status and chemotherapy data every 3 mo within the first year and then annually afterwards. Causes and dates of death were recorded,and survival time was calculated from the date of recruitment. Survival time was calculated from time of recruitment to date of death, date of last contact (for those lost to follow-up), or to the last contact with living subjects at the end of the study.
Genotyping
Peripheral blood samples from all cases and controls were collected by the investigators. The TIANamp Blood DNA Kit (Tiangen, Beijing, China) was used for DNA extraction. XPG rs2094258, rs751402, rs873601, rs2296147, and rs1047768 polymorphism genotyping was performed by PCR restriction fragment length polymorphism (PCR-RFLP). The conditions of PCR amplification were: (1)Denaturation at 95 °C for 5 min; (2) 30 cycles of denaturation at 94 °C for 60 s,annealing at 60 °C for 60 s, and extension at 72 °C for 60 s; and (3) Extension at 72 °C for 10 min. PCR products were confirmed by agarose gel electrophoresis. Ten percent of the samples were randomly selected for repeated genotyping and the results were 100 percent consistent.
Statistical analysis
The socio-demographic data and clinical data between case and control subjects were compared by the chi-square test. A goodness-of-fit chi-squared test was also used to analyze whether the SNP (XPG rs2094258, rs751402, rs2296147, rs1047768 and rs873601) distributions conform to the Hardy-Weinberg equilibrium (HWE) in controls. Odds ratios (ORs) and 95% confidence intervals (CIs) for examined SNPs were analyzed by Logistic regression method and adjusted by age, gender, andH.pyloriinfection status. The heterozygous model, homozygous model, recessive model,and dominant model were all analyzed for five SNPs. For rs2094258, heterozygous model, homozygous model, recessive model, and dominant model were CTvsCC, TTvsCC, TTvsCT + CC, TT + CTvsCC, respectively. Kaplan-Meier method and logrank test were used for plotting cases’ survival curves and for comparisons,respectively.
Multivariate Cox regression was used for exploring possible prognostic factors,which included gender, age, drinking, smoking,H. pylori, TNM stage, tumor differentiation, lymphovascular invasion, neural invasion, and chemotherapy. SPSS 24.0 statistical software was used for all statistical analyses (Statistical Package for the Social Sciences, version 24, SSPS Inc, Chicago, IL, United States). All statistical tests were two-sided, withP< 0.05 as the boundary value.
RESULTS
The majority of study participants were male and less than age 60, with no significant differences in these factors between cases and controls (Table 1). All five loci in the control group were consistent with the Hardy-Weinberg equilibrium. The rate ofH.pyloriinfection in GC patients was significantly higher than that in the control group(70.6%vs53.6%,P< 0.001). None of the associations between studied loci (rs2094258,rs751402, rs2296147, rs1047768, and rs873601) and GC risk was statistically significant(Table 2). However, the rs2296147 CC genotype frequency was higher in the GC case group than that in the control group (5.9%vs3.9%,P= 0.050). After adjustment for age, sex, andH. pyloristatus, this genotype was marginally associated with a slightly higher risk of GC (OR = 1.40, 95%CI: 0.97-2.50,P= 0.061) than the TT genotype. The CC genotype was found to be marginally associated with a higher risk of GC (OR =1.36, 95%CI = 0.99-2.49,P= 0.053) in the recessive model (CCvsCT + TT).
As the number of C alleles of rs2094258 increased, the survival rate of GC cases decreased (log-rank test,P= 0.032) (Table 3). Among all the cases, mortality was 45.4% with rs2094258 TT genotype (no C allele), 53.7% with CT genotype (one C allele), and 59.4% with CC genotype (two C alleles). The other four SNP loci (rs751402,Rs2296147, rs1047768, and rs873601) had no significant correlation with overall survival in patients with GC. For rs2094258, survival curves varied significantly with genotype (log-rank test,P= 0.032) (Figure 1A). GC patients with the rs2094258 CT +CC genotype showed a lower survival than patients with the TT genotype (log-rank test,P= 0.028) (Figure 1B). Cases with the CC genotype had a poorer prognosis than those with the TT + CT genotype (log-rank test,P= 0.039) (Figure 1C).
In univariate survival analysis, age (P= 0.011), TNM stage (P< 0.001), no chemotherapy (P< 0.001), poor differentiation (P< 0.001), neural invasion (P< 0.001),and lymphovascular invasion (P< 0.001) were positively associated with the 5-year survival of GC cases (Table 4). Gender andH. pyloristatus were not significantly associated with the 5-year survival of GC cases. In multivariate analysis, the increase in the number of C alleles of rs2094258 (hazard ratio [HR] = 1.19, 95%CI: 1.02-1.45,P=0.037), chemotherapy (HR = 0.81, 95%CI: 0.63-0.98;P= 0.041), differentiation (HR =4.51, 95%CI: 1.99-8.23,P< 0.001), and lymphovascular invasion (HR = 1.97, 95%CI:1.44-3.01,P< 0.001) were associated with survival in GC cases (Table 5).
DISCUSSION
We genotyped five functional SNPs in theXPGgene involved in the NER pathway and assessed their associations with GC risk and survival in China. Although rs2296147 was marginally associated with risk of GC (P= 0.05), we found no statistically significant associations. However, as the number of C alleles of rs2094258 increased, the survival of GC cases showed a decreasing trend. Poor differentiation,lymphovascular invasion, no chemotherapy, and increase in the number of C alleles of rs2094258 were associated with decreased survival among cases.
Few studies have explored the association ofXPGSNPs with survival in GC patients. Our results are similar to those of Liuet al[25], who analysed the association betweenXPGSNPs and survival among 373 GC patients in China. That study found that in univariate model, the survival rate of those with theXPGrs2094258 AG genotype was higher than that of wild-type GG carriers (HR = 0.59, 95%CI: 0.39-0.90,P= 0.014), and that in multivariate analysis, the AA + AG genotype presented a significant survival advantage over the GG genotype (adjusted HR = 0.65, 95%CI:0.44-0.97)[25]. Liuet al[25]also found that the AA + AG genotype of theXPGrs2094258 polymorphism improved survival in those with the following characteristics: Age more than 60, lymphatic metastasis, TNM stage III-IV, Borrmann III-IV, and diffusetype gastric tumors[25].
Consistent with our study, a recent meta-analysis did not observe an overall association between theXPGrs2094258 SNPs and GC risk[26]. Some studies have found associations between the rs2094258 polymorphism and GC risk[22,26-28]. For example,Yanget al[23]assessed threeXPGSNPs (rs2296147T>C, rs2094258C>T, and rs873601G>A) and found that the rs2094258 C>T polymorphism was associated with an increased GC risk. Meanwhile,H. pylori-infected individuals with the rs2094258 TT genotype had a much higher GC risk (OR = 2.13, 95%CI: 1.22-3.35,Pfor interaction =0.030)[23]. However,H. pyloristatus did not modify associations with this SNP in our data. Varying cofactors among study populations, small sample size, the use of different PCR methods, or the low penetrance of this SNP may account for the inconsistent findings[15].
XPGrs2296147 was not associated with GC in the present study, a finding consistent with those of other studies[24,25]. However, significant associations between rs2296147 polymorphisms and GC risk were observed in Asian populations in numerous studies in one meta-analysis (CTvsTT: OR = 0.93, 95%CI: 0.87-0.99,P=0.036)[29]. Further, the rs2296147 CC genotype was associated with a reduced risk of GC in a Chinese population (OR = 0.52, 95%CI: 0.27-0.97)[23]. These differences may be due to regional differences, small sample size, or heterogeneity of clinical feature[26].
The NER pathway belongs to the DNA repair pathways, and its functions include removing exogenous or endogenous DNA damage or adduct between chains and recruiting proliferating-cell nuclear antigen to the damage site for the subsequent gap-filling DNA synthesis[30]. DNA repair usually includes two stages of excision and repair synthesis[31]. TheXPGgene locating at chromosome 13q33 consists of 15 exons[10], which is considered to cut the DNA at the 3’ terminus, initiate transcriptioncoupled DNA repair, and participate in RNA polymerase II transcription[32]. In the process of DNA repair, XPG binds to XPB as part of the transcription factor IIH(TFIIH) complex and strongly interacts with the TFIIH complex, a multi-subunit complex located at the intersection of transcription and DNA repair[33], which is involved in the DNA demethylation induced by overexpression of Gadd45a[10].Meanwhile, XPG-related nucleases are used hierarchically for the excision of doublestranded rDNA break resection[34]. Recent report suggests that XPG incises the R-loop structure and participates in the RAD52-dependent resolution of DNA-RNA hybrids[35]. The rs1047768, rs751402, and rs2296147 are located in the exon 2, proximal promoter, and 5' untranslated region of the gene, respectively[36,37]. Based on the dbSNP database, the rs2094258 located at theXPGgene intron region participates in the initiation of the transcription-coupled DNA repair.
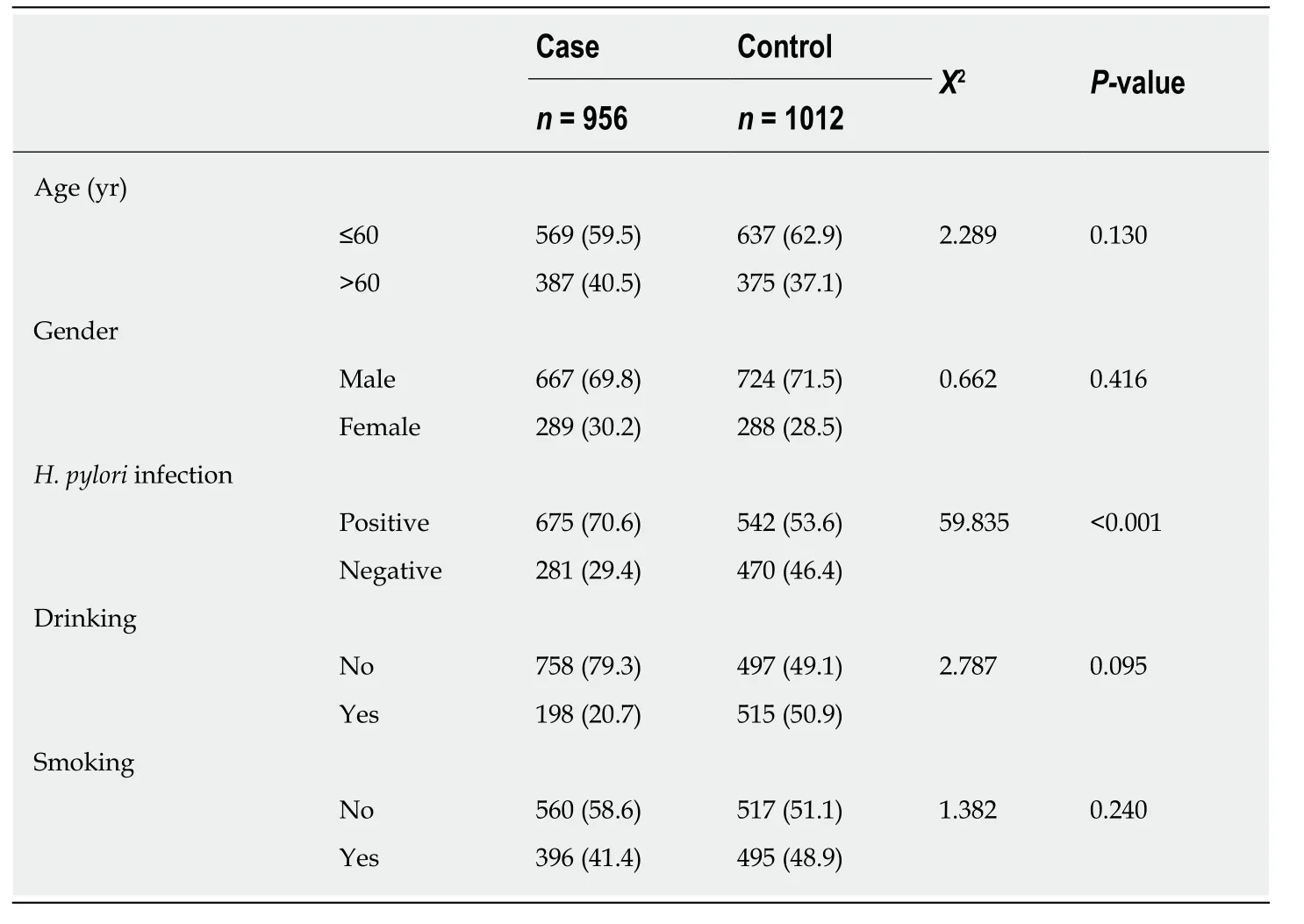
Table 1 Demographic characteristics of the participants
The limitations of this study are as follows. First, we could not explore and determine the exact mechanism by whichXPGSNPs influence GC survival. Second,this study only examined five functional SNPs and did not include all the SNPs in theXPGgene that might play a key role in GC development. Third, selection bias and information bias are inherent threats to case-control studies, and cannot be ruled out in our study. Genetic factors have been shown to play key roles in the development,progression, and prognosis of GC. If confirmed by other studies, the results of our study suggest thatXPGrs2094258 polymorphisms may serve as genetic biomarkers for GC prognosis, and may provide clues to biological underpinnings of GC progression.

Table 2 Single nucleotide polymorphism and genotype distribution between gastric cancer cases and controls
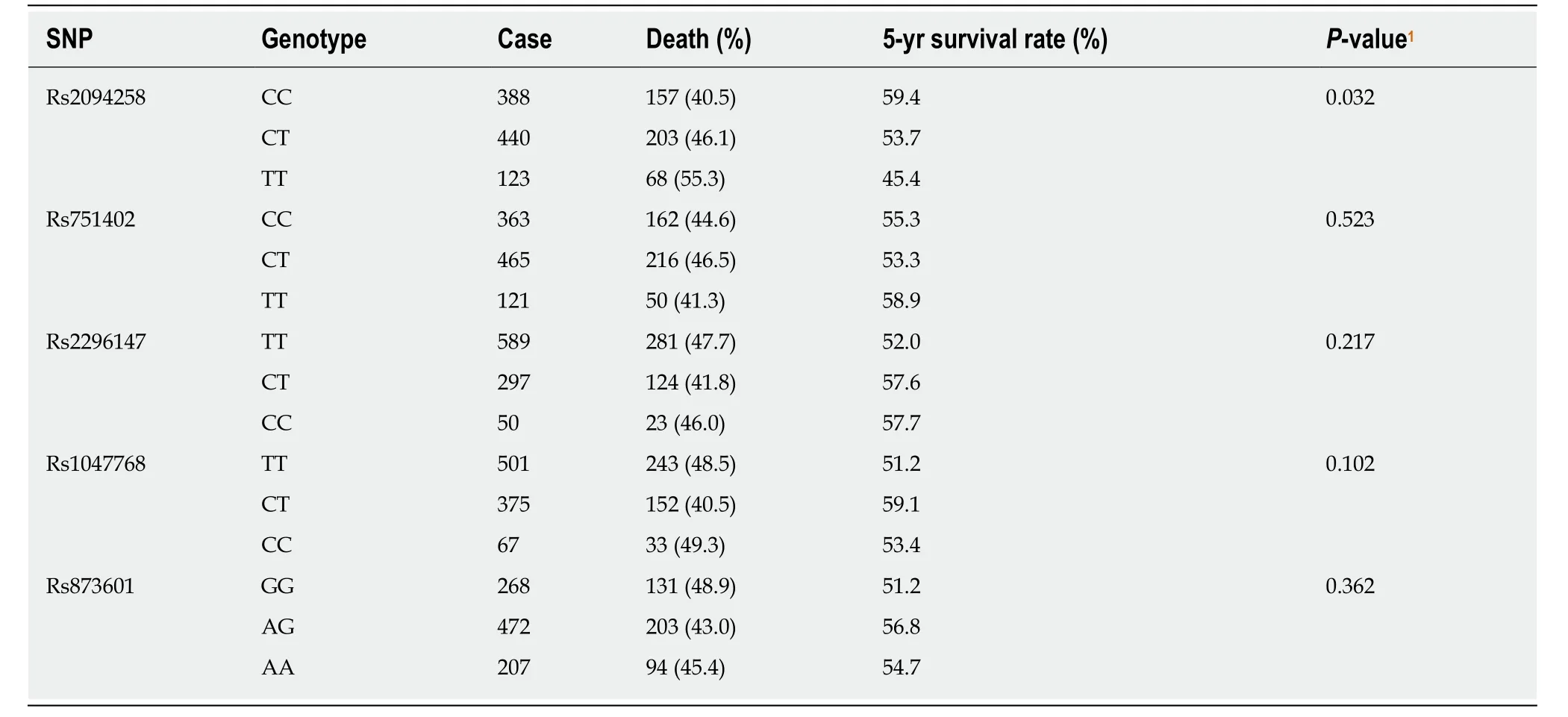
Table 3 Genotypes of single nucleotide polymorphism and 5-year survival status of gastric cancer cases

Table 4 Univariate analysis of associations between influencing factors and 5-year survival of gastric cancer cases

Table 5 Multivariate analysis of gastric cancer survival
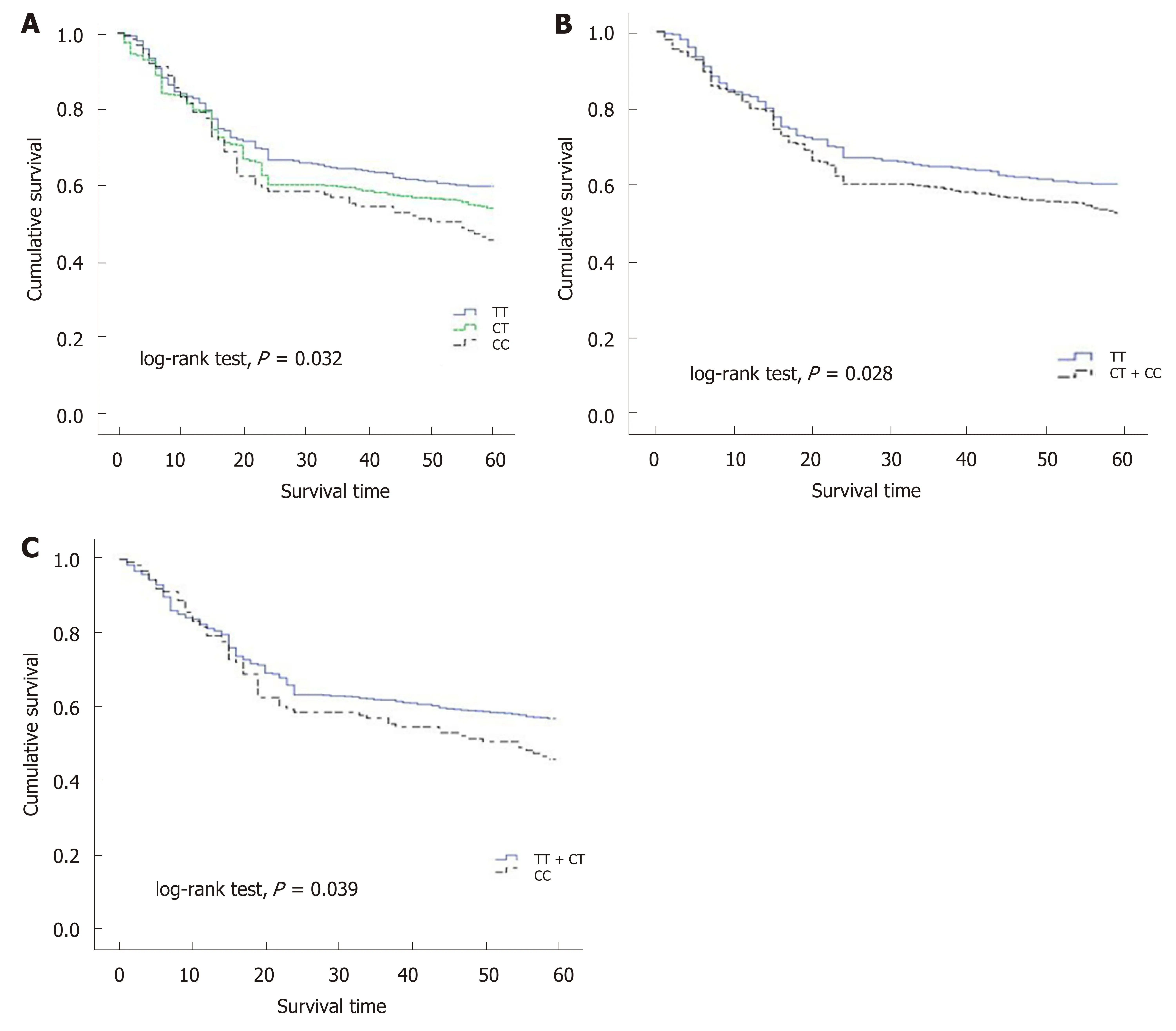
Figure 1 Survival plots of gastric cancer cases stratified by rs2094258 genotype. A: Compared with the survival of cases with no C allele (genotype TT, blue line), cases with one C allele (genotype CT, green line) demonstrated a decreased survival rate and cases with two C alleles (genotype CC, dotted line) exhibited the lowest survival rate; B: Compared with the survival of cases with no C allele (genotype TT, blue line), cases with recessive allele (genotype CT + CC, dotted line)demonstrated a decreased survival rate; C: In the dominant model, case with CC allele showed a lower survival rate compared with cases with dominant allele.
ARTICLE HIGHLIGHTS
Research background
Gastric cancer (GC) is one of the leading causes of cancer-related death worldwide, which causes a high social and economic burden. Xeroderma pigmentosum group G (XPG) or ERCC5 may play a key role in DNA repair, thus affecting cancer prognosis. Studies have shown thatXPGSNPs may affect the development of cancer such as lung cancer, colorectal cancer, and breast cancer.
Research motivation
Only a few studies have explored the relationships betweenXPGgene SNPs and GC, and the results are inconsistent.
Research objectives
To examine GC risk and survival in relation to common functionalXPGSNPs through a casecontrol study, which might improve our understanding of GC and provide new therapeutic targets for this malignancy.
Research methods
A total of 956 histologically confirmed GC cases and 1012 controls were matched by sex, age(within 5 years), and residential district. Cases were followed and the survival time was recorded. DNA was extracted from peripheral blood samples of all cases and controls.XPGrs2094258, rs751402, rs2296147, rs1047768, and rs873601 polymorphisms were genotyped using PCR-RFLP. Logistic regression and Cox regression were used for analyzing associations ofXPGSNPs with risk of GC and prognosis, respectively.
Research results
We found that GC patients with the rs2094258 CT + CC genotype showed a worse survival than those with the TT genotype, and patients with the CC genotype had a tendency of unfavorable prognosis compared with those with the TT + CT genotype. The increase in the number of C alleles of rs2094258 was associated with the long-term survival of GC cases. Other risk factors for survival included tumor differentiation, lymphovascular invasion, and use of chemotherapy.However, the exact mechanisms by whichXPGSNPs influence GC survival remain to be solved.
Research conclusions
TheXPGrs2094258 polymorphism may be associated with overall survival in patients with GC.
Research perspectives
If confirmed by other studies, the results of our study suggest thatXPGrs2094258 polymorphisms may serve as genetic biomarkers for GC prognosis, and may provide clues to biological underpinnings of GC progression.
ACKNOWLEDGEMENTS
We thank all the subjects and researchers who participated in this study.
 World Journal of Gastroenterology2019年34期
World Journal of Gastroenterology2019年34期
- World Journal of Gastroenterology的其它文章
- Diagnostic and prognostic potential of tissue and circulating long non-coding RNAs in colorectal tumors
- Autoantibodies: Potential clinical applications in early detection of esophageal squamous cell carcinoma and esophagogastric junction adenocarcinoma
- Role of endoscopic ultrasound in the screening and follow-up of high-risk individuals for familial pancreatic cancer
- Hepatic senescence, the good and the bad
- Optimizing proton pump inhibitors in Helicobacter pylori treatment:Old and new tricks to improve effectiveness
- Regulatory effect of a Chinese herbal medicine formula on nonalcoholic fatty liver disease
