Preparation of a Salmonella LAMP Detection Kit
Yuping WAN Xiaosheng WU Yu ZHANG Meihong DU Zhaoqin WANG Caiwei FENG Zhengmiao ZHAO Fangyang HE

Abstract [Objectives] This study was conducted to establish a method for rapid detection of Salmonella at the grassroots level, and a corresponding product was developed.
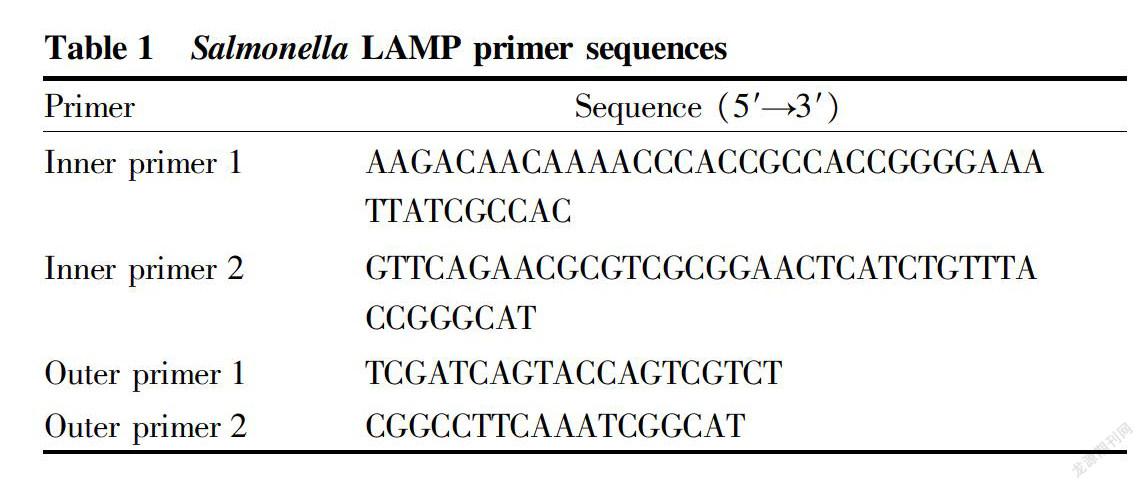
[Methods] By analyzing the invA gene of Salmonella, two pairs of efficient, rapid and specific amplification primers were designed and screened for the six conserved domains of the gene, and the LAMP detection kit for Salmonella based on this method was produced.
[Results] The method is simple, sensitive, rapid and specific. The minimum detection concentration is 102-103 CFU/ml.
[Conclusions] It can be used in the selfinspection of food production enterprises and the onsite sampling inspection and rapid detection of large quantities of samples by grassroots food safety inspection and supervision departments.
Key words LAMP; Salmonella; Rapid detection
Received: July 11, 2019Accepted: September 28, 2019
Supported by Guangxi Key R&D Program (GK AB16380271).
Yuping WAN (1982-), male, P. R. China, senior engineer, master, devoted to research about Food safety testing technology.
*Corresponding author. Email: beijingqinbang@163.com.
Salmonella is a kind of common zoonotic pathogen that infects many animals, including mammals, birds, reptiles, fish, amphibians, and insects[1]. It is mainly infected through food and drinking water. Human and animal infections may be asymptomatic, or may be manifested as clinically morbid deaths. The infection of Salmonella may aggravate morbidity or mortality. According to statistics, among the bacterial food poisoning in countries around the world, food poisoning caused by Salmonella often rank first, accounting for 52% of all bacterial food poisoning[2], and Salmonella is the first in the inland areas of China. Salmonellacaused food poisonings are commonly because of eating animal foods contaminated with Salmonella by mistake, and less common in plant foods[3].
At present, the detection methods of the bacteria mainly include the traditional isolation and identification method, immunological method and PCR method[4-7]. The three methods have their own advantages and disadvantages, among which the traditional isolation and identification method has higher accuracy, but more steps, complicated process and longer detection period, which are not conducive to the monitoring of production process, the quality control of the final product and the management of government departments. The immunological detection method has a short cycle, but poor sensitivity and specificity. The PCR method has short detection time and high sensitivity, but requires high requirements for instruments. It is of great practical significance to study new rapid, accurate and easytouse Salmonella detection methods.
Loopmediated isothermal amplification (LAMP) is a new type of constanttemperature nucleic acid amplification technology developed by Notomi et al.[8] on the basis of PCR technology in 2000. Its amplification can be performed at a constant temperature without the need for thermal cycling, and DNA can be detected at very low detection limits compared with other methods. Because of its high specificity, high sensitivity, low cost and easy operation, this method has been widely used in the experimental diagnosis of infectious diseases[9-11]. This study applied the LAMP method to food safety testing without the use of large or highpriced instruments. The method is suitable for rapid screening and testing of food safety, can be applied in primary laboratories as it can meet the needs of pathogenic bacteria detection in food, and can ultimately prevent Salmonellacontaminated food from entering the market to ensure food safety.
Materials and Methods
Materials and reagents
Test strains
S. typhimurium ATCC 14028; S. anatum CICC21498; Salmonella CMCC(B)50001; S. enteritis CMCC(B)50041; S. cholerae suis CMCC(B)101859; S. enteritis CICC21490; Staphylococcus epidermidis CMCC(B)26069; Staphylococcus aureus CICC10477; Staphylococcus aureus ATCC6538; Escherichia coli ATCC 25922; Vibrio parahaemolyticus ATCC 17802; Shigella flexneri ATCC2457T; Listeria monocytogenes CMCC(B)54002; Yersinia enterocolitica CMCC(B)52225; Enterobacter sakazakii; Serratia marcescens; Proteus penneri; P. mirabilis; S. haemolyticus; Micrococcus; Enterococcus faecium.
Instruments and equipment
Thermostat water bath (USA WEALTEC CORR company); fluorescence quantitative PCR instrument (Thermo Fisher Scientific, America); biochemical incubator; autoclave; 4 ℃ centrifuge; ultraclean workbench.
Test reagent
Normal saline; secondary enrichment liquids corresponding to pathogens; brain infusion; sterile swab; API reagent strip; 7.5% sodium chloride; broth medium; buffered peptone water medium; Salmonella chromogenic medium; blood plate; Bst DNA polymerase; nutrient agar; trypticase soyyeast extract agar medium; Salmonella Latex test kit (Guangdong Huankai Microbial Sci.&Tech. Co., Ltd.); combined enrichment liquid (Beijing Liangrun Biological Technology Co., Ltd.).
Methods
Primer design
GallegosRobles et al.[12] designed PCR primers based on the Salmonella invA gene, and the established PCR method can amplify Salmonellaspecific fragments, and failed in the specific amplification for nonSalmonella bacteria. Therefore, this study used the invA gene that determines the intestinal invasiveness of Salmonella[12], and analyzed the nucleotide sequence of the gene using biological online software. The primer design software Primer Explorer V4.0 was used to screen and synthesize LAMP primers: inner primer 1, outer primer 1, inner primer 2, outer primer 2, as shown in Table 1.
Preparation of template DNA
According to the national standard GB 4789.42016 "National Food Safety Standard Food Microbiological Examination: Salmonella", the samples to be tested should be treated and enriched.
Preparation of enrichment liquid template DNA: A certain amount of the prepared sample enrichment liquid (1 ml) was centrifuged at 10 000 r/min for 2 min in a 1.5 ml centrifuge tube, and the supernatant was discarded. Then, 100 μl of DNA extracting solution was added into the centrifuge tube, which was then heated in a boiling water bath for 10 min. Finally, the liquid was centrifuged at 10 000 r/min for 2 min, obtaining the supernatant as the template DNA.
Preparation of suspected colony template DNA: Single colonies were picked and added to 100 μl of DNA extracting solution. The obtained liquid was oscillated and mixed, and boiled in water for 10 min. Centrifugation was then performed at 10 000 r/min for 2 min, obtaining the supernatant as the template DNA.
The DNA extracting solution contained 0.5 mol/L TrisHCl, 0.05 mol/L disodium ethylenediamine tetraacetate and 0.5% Triton X100.
Establishment of LAMP reaction system
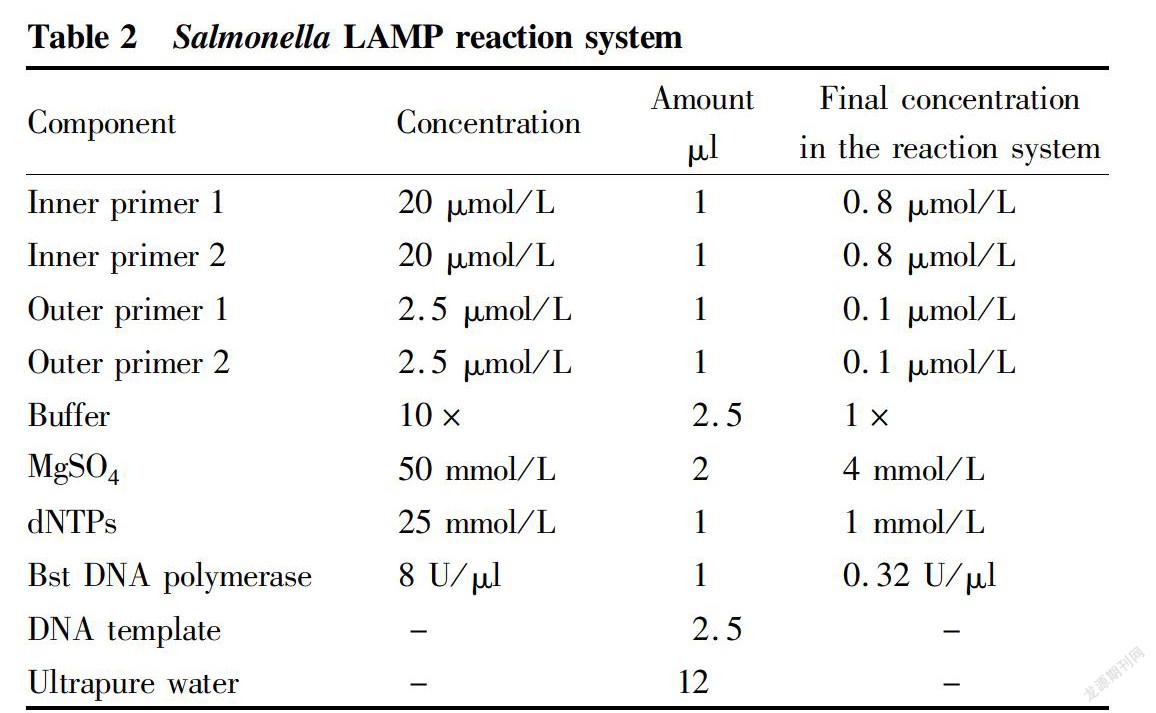
The Salmonella LAMP reaction system is shown in Table 2.
The prepared reaction system was covered with a suitable amount of sealing liquid (sterilized liquid paraffin) to prevent contamination, and a negative control and positive control were also done. The negative control was the DNA extracting solution, and the positive control was a DNA extract of Salmonella standard strain. Both controls were prepared into 25 μl of LAMP reaction system. Amplification was performed at 65 ℃ for 60 min.
After the amplification was completed, 1-2 μl of 1 000×SYBR Green I was added, followed by mixing. Under natural light, the color development was observed by naked eyes. Under the premise that the negative control is orange and the positive control is green, the sample to be tested is judged to be positive in green, that is, the test sample contains Salmonella, and the sample to be tested is judged to be negative in orange, that is, the sample does not contain Salmonella. If the negative positive control does not match the above, the test result is invalid, and retesting should be carried out.
Kit specificity test
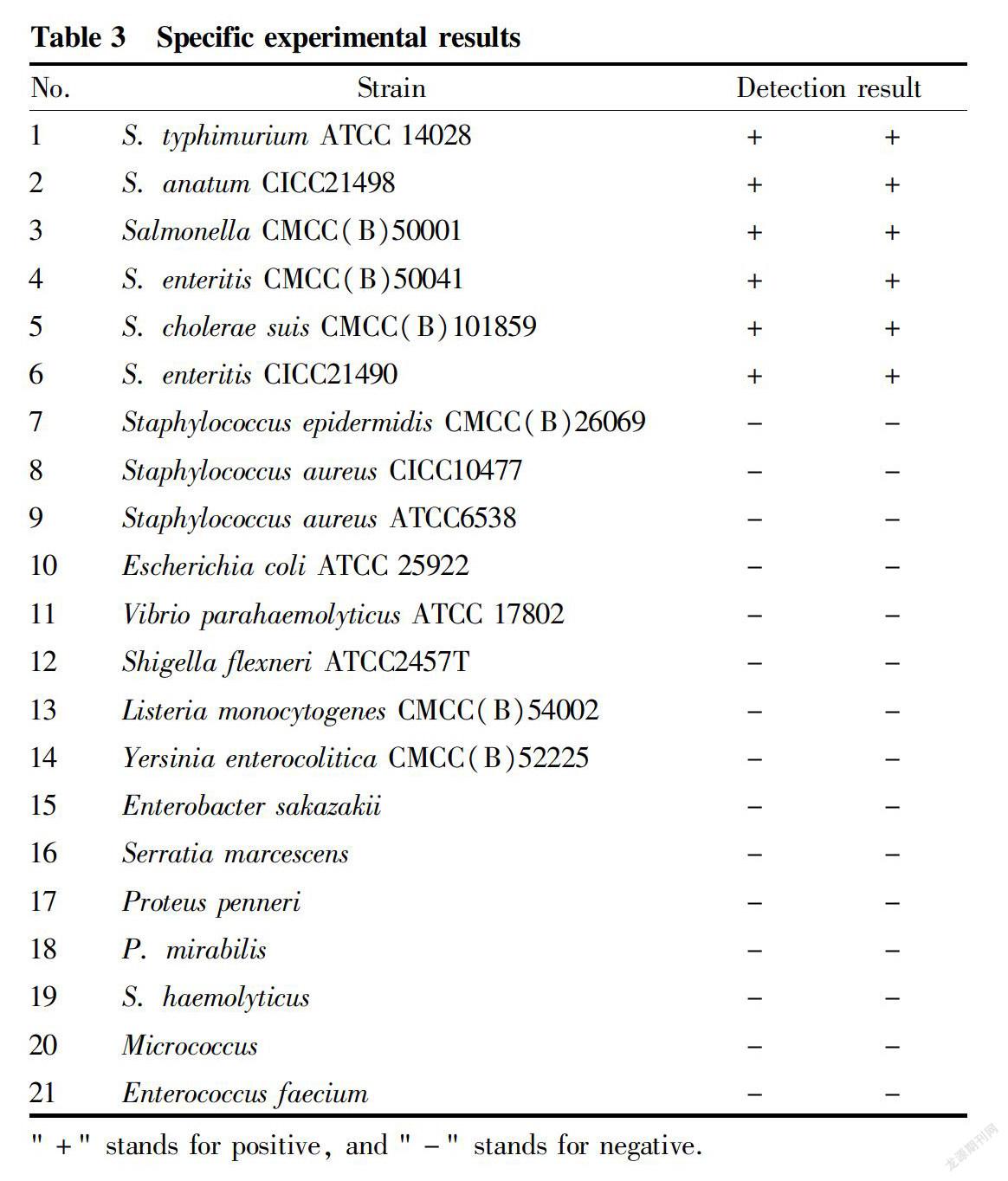
The common 21 foodborne pathogenic strains and highlyhomologous strains were tested. Salmonella DNA was set as a positive control, and DNA extracting solution was set as a negative control. Each sample was tested in duplicate.
Kit sensitivity test
After counting, the bacterial liquid was diluted 10 times, and then detected by the established LAMP detection method.
Compliance test between kit and national standard test method
Different concentrations of Salmonella were used to treat pork, dumplings, dried meat floss, pasteurized milk and milk powder as samples. The treated samples were tested by the methods specified in the national standard GB 4789.4-2016 "National Food Safety Standard Food Microbiological Examination: Salmonella". The test results of the two methods were compared. Each sample was tested in triplicate, and a negative control and a positive control were also set.
Results and Analysis
Kit specificity test
In the test results of 21 common pathogenic bacteria, Salmonella showed positive results, and the rest were negative, indicating that the Salmonella LAMP detection method established in this study has good specificity (Table 3).
Kit sensitivity test
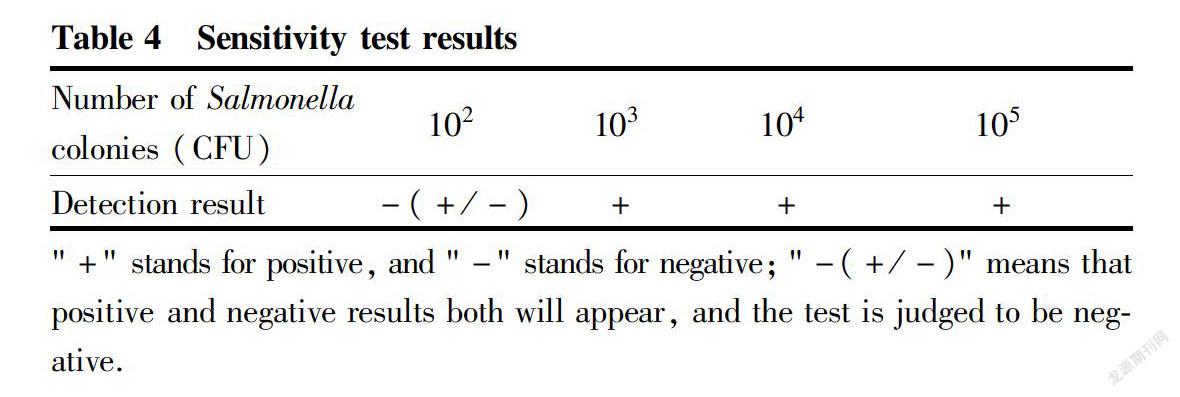
The sensitivity test results of the Salmonella LAMP assay for 102-105 CFU colonies are shown in Table 4. A positive response was observed when the Salmonella concentration was 102 CFU/ml. However, due to template extraction, dilution error, etc., the order of magnitude of sensitivity may be 103 CFU/ml. Therefore, the minimum detectable concentration of Salmonella in this method was 102-103 CFU/ml.
Compliance test between kit and national standard test method
For pork, dumplings, dried meat floss, pasteurized milk and milk powder, which are more serious for Salmonella infection, they were added with different concentrations of Salmonella. The results of the three groups of repeated tests are shown in Table 5. The results of the LAMP method for different samples were consistent with the national standard method, and the coincidence rate was 100%.
Yuping WAN et al. Preparation of a Salmonella LAMP Detection Kit
Conclusions and Discussion
The LAMP detection method can complete the gene amplification in 60 min at 65 ℃, and the whole process takes about 150 min from the preparation of the template to the result. The entire experiment takes about 15 h plus the enrichment of sample, while the traditional method generally takes more than 4 d. The genes for detection of Salmonella are mainly classified into three types: genusspecific primer genes, serogroupspecific primer genes, and serotypespecific primer genes[13]. According to the genotyping of primers for Salmonella detection, there are mainly nuc genes and inv genes. The inv gene family of Salmonella is the most genusspecific, and encodes intestinal invasion proteins that determine the intestinal invasiveness of Salmonella. It includes a series of genes such as invA, invB, invC, invD and invE, which are widely distributed in Salmonella[14-20], and the protein encoded by the invA gene is the most pathogenic[21]. The LAMP method is effective in reducing cumbersome personnel operations and eliminating the need for expensive test equipment. The Salmonella LAMP test kit developed in this study can be used for qualitative detection of Salmonella in food or quantitative detection together with a reader. The minimum detectable concentration of this kit is 102-103 CFU/ml, and the sensitivity and specificity are better.
In this study, we designed specific primers according to the invA gene of Salmonella can accurately bind to the target gene, and established the Salmonella nucleic acid detection method which can realize visual rapid detection with high efficiency, high sensitivity and high specificity by adding SYBR Green I for staining. The established method is applicable to the popularization and promotion in laboratories and grassroots units at all levels.
References
[1] ZHU SM, WU JJ, XU C, et al. Rapid detection of Salmonella spp. by loopmediated isothermal amplification method[J]. Modern Food Science & Technology, 2008, 24(7): 725-730.
[2] Centers for Disease Control and Prevention. Surveillance for foodborne disease outbreaks United States, 2006. Morbidity and Mortality Weekly Report, 2009, 58(22): 609-615.
[3] RIEMANN H, HIMATHONGKHAM S, WILLOUGHBY D, et al. A survey for Salmonella by drag swabbing manure piles in California egg ranches[J]. Avian Dis. 1998. 42:67-71.
[4] PANG XY, MAN CX, ZHAO YM, et al. Rapid detection of Salmonella in raw meat samples by loopmediated isothermal amplification[J].Food and Nutrition in China, 2015, 21(08):12-15.
[5] PARK SH, AYDIN M, KHATIWARA A, et al. Current and emerging technologies for rapid detection and characterization of Salmonella in poultry and poultry products [J]. Food microbiology, 2014, 38: 250-262.
[6] RASOOLY A, HEROLD KE. Food microbial pathogen detection and analysis using DNA microarray technologies [J]. Food borne pathogens and disease, 2008, 5(4): 531-550.
[7] GALLEGOSROBLES MA, et al. PCR detection and microbiological isolation of Salmonella spp. from fresh beef and cantaloupes. [J]. Journal of Food Science, 2009, 74 (1).
[8] NOTOMI T, et al. Loopmediated isothermal amplification of DNA[J]. Nucleic Acids Research, 2000, 28(12): 63.
[9] ARYAN E, MAKVANDI M,FARAJZADEH A, et al. Clinical value of IS6110based loopmediated isothermal amplification for detection of Mycobacterium tuberculosis complex in respiratory specimens [L]. J Infect, 2013, 66(6): 487-493
[10] MINAMI M, OHTA M, OHKURA T, et al. Use of a combination of brushing technique and the loop mediated isot hermal amplification method as a novel. rapid, and safe system for detection of helibacter pylori [J] Clin Microbiol, 2006, 44(11): 4032-4037.
[11] YANO A, ISHIMARU R, HUJIKATA R. Rapid and sensitive detection of heatlabile I and heatstable I enterotoxin genes of enterotoxin genic Escherichia coli by loop mediated isothermal amplification [J]. J Microbiol Methods, 2007.68(2): 414-420.
[12] ZHANG HW, YE LM, PENG YS, et al. Development of LAMP detection of Salmonella in food[J]. Food Research and Development, 2009, 30(5): 115-118.
[13] HUANG JH, SUN YH, CHEN R, et al. Development of loopmediate isothermal amplification assay for Salmonella detection in food[J]. Journal of Tianjin University, 2012, 45(5): 468-472.
[14] ZHONG WJ, ZHAO QM, ZHANG CH, et al. Establishment of rapid PCR detection method for Salmonella in food[J]. Chinese Journal of Zoonoses, 2007, 23(12): 1216-1221.
[15] ZHONG WJ, ZHAO QM, ZHANG CH, et al. Development of polymerase chain methods for Salmonella detection in foods[J]. Chinese Journal of Zoonoses, 2007, 23(12): 1216-1221(in Chinese).
[16] IHIRA MASARU, OHTA AKANE, SUGATA KEN, et al. Loopmediated isothermal amplification for discriminating between human herpesvirus 6 A and B [J]. Journal of Virological Methods, 2008, 154(1/2): 223-225.
[17] CURTIS KA, RUDOLPH DL, OWEN SM. Rapid detection of HIV1 by reverse transcription, loopmediated isothermal amplification (RTLAMP)[J]. Journal of Virological Methods, 2008, 151(2): 264-270.
[18] GAO HW, LEI ZW, JIA JT, et al. Application of loopmediated isothermal amplification for detection of Yersinia enterocolitica in pork meat [J]. Journal of Microbiological Methods, 2008, 77(2): 198-201.
[19] MORI Y, NAGAMINE K, TOMITA N, et al. Detection of loopmediate isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation[J]. Biochem Biophys Res Commun, 2001, 289(1): 150-154.
[20] BLTE M, JAKOB P. The use of a PCRgenerated invA probe for the detection of Salmonella spp. in artificially and naturally contaminated foods [J]. International Journal of Food Microbiology, 1995, 26(3): 335-344.
[21] KIKO HARAKUDO, et al. Loopmediated isothermal amplification for the rapid detection of Salmonella[J]. FEMS Microbiology Letters, 2005, 253: 155-161.
- 农业生物技术(英文版)的其它文章
- Observation on Cardiac Opening of the Inferior Vena Cava in Goat Fetuses
- Evaluation on Application and Spraying Effect of AirAssisted Sprayer in Apple Orchard with Dwarfing Rootstocks
- Problems in the Development of Traditional Chinese Medicinal Materials Planting Industry in Shiyan City and Countermeasures
- Study on Practical Mature Age of Individual Pinus thunbergii×P. densiflora
- Effects of Different Densityreducing Methods on Canopy Microenvironment, Tree Growth and Fruit Quality in Closed Apple Orchard
- Development of Whole Potato Flour Fish Noodles

