我国北方岩溶泉域刚毛藻的系统发育及形态学研究
胡变芳 吉莉 陈乐 冯佳 史胜利

Phylogenetic and morphological profile of Cladophora fracta (Cladophorophyceae, Chlorophyta) from karst spings, in North China [J].
Guihaia, 2019, 39(1): 53-61.
Phylogenetic and morphological profile of Cladophora fracta (Cladophorophyceae, Chlorophyta) from karst springs, in North China
HU Bianfang1 , JI Li2* , CHEN Le3 , FENG Jia 4 , SHI Shengli4
( 1. Department of Biology, Jinzhong University, Jinzhong 030600, Shanxi, China; 2. College of Environment and Safety, Taiyuan University of Science and Technology, Taiyuan 030024, China; 3. School of Pharmaceutical Science, Shanxi Medical University, Taiyuan 030001, China; 4. School of Life Science, Shanxi University, Taiyuan 030006, China )
Abstract: Cladophora fracta, a filamentous green macroalgal epiphyte on rhodoliths, is described from five karst springs in North China. Although Cladophora species frequently appear in karst system, their genetic diversities, biogeographical affinities and physiological properties have not been well investigated in these environments. The specific objectives of this study were as follows: (1) Describe the habitat of the cladophora-like algae form the five karst springs; (2) Identify the thallus to species level based on a combination of morphological characteristics and molecular sequence; (3) Explore the morphological influence of habitat. To elucidate the biogeographical patterns in Cladophora, both morphological and molecular evidence were compared of Cladophora specimens across five study sites. Analyses of partial small subunit (SSU) and large subunit (LSU) genes revealed that the studied 50 Cladophora specimens were genetically identical species and a total of thirteen ribotypes were detected. The molecular sequencing results indicated that the examined species was highly homologous with C. vagabunda, though they shared few morphological features. The genus did not form a monophyletic clade but in three different clades both in SSU and LSU trees. The microscopic structure was more consistent with that of C. fracta. The Cladophora from the five karst springs did not show significant variation in cell dimensions. However, the species exhibited larger cell diameters than those reported from lakes. In addition, the rhizoid-like branches are only observed in two locations (XA and ST). Considering the morphological characteristics, we therefore hold our species as C. fracta.
Key words: Cladophora, karst spring, phylogeny, green algae, ribosomal DNA, Cladophoraceae
CLC number: Q949.2
Document code: A
Article ID: 1000-3142(2019)01-0053-09
摘 要: 脆弱刚毛藻(Cladophora fracta)是一种大型丝状绿藻,生境分布广泛。然而,对于岩溶泉域分布的刚毛藻研究较少,它们的遗传多样性、生物地理亲缘性和生理特性都有待于深入研究。该研究对我国北方地区五个典型岩溶泉域的50个脆弱刚毛藻样本进行了形态学和分子系统学描述。主要研究目标:(1)对我国北方地区五个典型岩溶泉的刚毛藻生境进行描述;(2)根据形态学特征和分子序列对藻体进行鉴定;(3)探究生境对藻体生理特性的影响。结果表明:基于SSU和LSU序列的结果,发现所分析的50株刚毛藻个体为同一种,同时还发现了13个不同的核糖体基因型。基于SSU和LSU的系统发育树,刚毛藻属均未能形成单系分支,分布在三个不同的分支上。13個样本基因型在SSU和LSU树中的位置相似,与Cladophora vagabunda有很高的序列同源性,但是形态特征却差异很大。从显微结构结果来看,五个岩溶泉域采集到的刚毛藻在细胞直径上无显著差异,藻体的形态特征与脆弱刚毛藻相一致。但是,岩溶泉域采集的藻体细胞直径比文献报道中在湖泊和河流中采集的脆弱刚毛藻直径要大。另外,仅在两个地点(XA和ST)采集的标本中发现有假根状分枝。因此,基于形态学和分子序列的结果,将这五个泉域的刚毛藻鉴定为脆弱刚毛藻(Cladophora fracta)。
关键词: 刚毛藻属, 岩溶泉, 系统发育, 绿藻, rDNA, 刚毛藻科
Cladophora is a large and common green macroalgal genus belonging to the Cladophoraceae in the Cladophorales, whose members consist of branched or unbranched uniseriate filaments, and absence of akinete. Species of the green algal genus Cladophora (Cladophorales, Chlorophyta) are widely distributed from marine to freshwater habitats worldwide (Leliaert et al., 2009) and extend into cold temperature and polar waters (Boedeker et al., 2012). Most species grow attached to rocky substrate by rhizoidal cells, but they also can form extensive free-floating masses in eutrophic waters.
The taxonomy of Cladophora species has been problematic because of the wide degree of morphological variation in response of different environmental conditons (Hayakawa et al., 2012). The Cladophorales is polyphyletic with representatives in three main clades (Siphonocladus clade, Cladophora clade and Aegagropila clade) (Hanyuda et al., 2002; Leliaert et al., 2003; Yoshii et al., 2004), and the fourth lineage (Okellyaceae including only one species) has been proposed as a sister to the three main clades (Leliaert et al., 2009; Boedeker et al., 2012). More recently, molecular phylogenetic studies based on SSU and LSU rDNA sequences have provided the insight into the relationships within the Cladophorales (Hanyuda et al., 2002; Leliaert et al., 2003, 2007; Boedeker et al., 2012; Ichihara et al., 2013). However, the systematic relationship of genera remain poorly understood. Mole- cular phylogeny revealed that the traditional family and genus level classifications did not reflect the phylogenetic relationships (Leliaert et al., 2007, 2009). The Cladophora species distribute in all three lineages rather than form a monophyletic group.
Genus Cladophora contains a heterogeneous group of species that are very hard to tell apart and classify, mainly because of the high morphological plasticity and cryptic diversity (Gestinari et al., 2010). Cladophora is one of the most species-rich genera of green macroalgae and the species of the genus are morphologically highly variable (van den Hoek & Chihara, 2000). It is also difficult to define stable taxonomic characteristics, since these characters are influenced by habitat, age and environmental conditions. However, studies on the habitat and ecology of genus Cladophora remain rare, especially about those in karst springs.
In this study, we collected Cladophora-like filamentous green algae from five karst springs in North China. We performed molecular phylogenetic analyses based on nuclear-encoded small subunit (SSU), partial large subunit (LSU) sequences and the combined SSU and LSU sequences, as well as morphological observation to reveal the diversity of these filamentous green algae from karst springs. The specific objectives of this study were as follows: (1) Describe the habitat of the Cladophora-like algae form the five karst springs; (2) Identify the thallus to species level based on a combination of morphological characteristics and molecular sequence; (3) Explore the morphological influence of habitat.
1 Materials and Methods
1.1 Plant materials
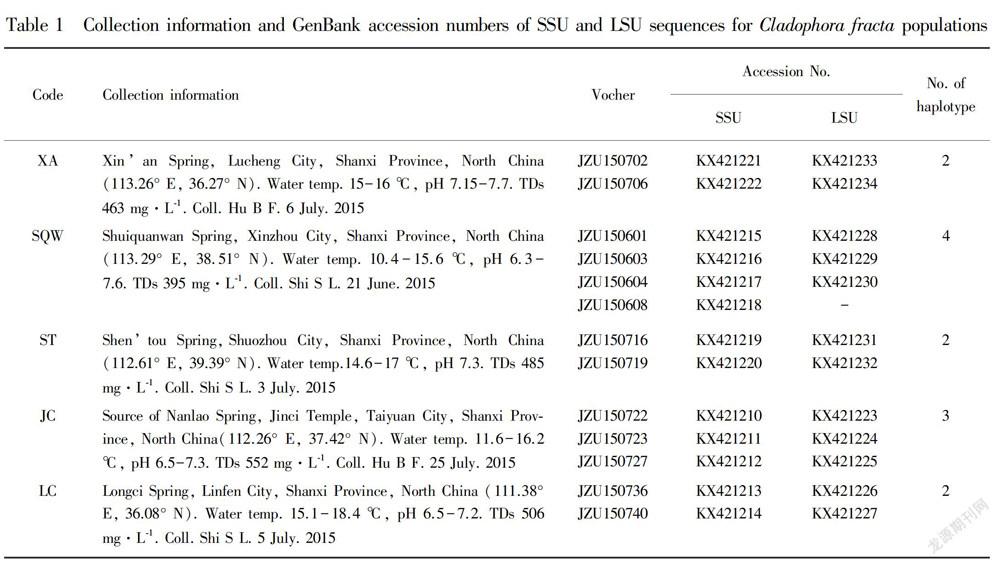
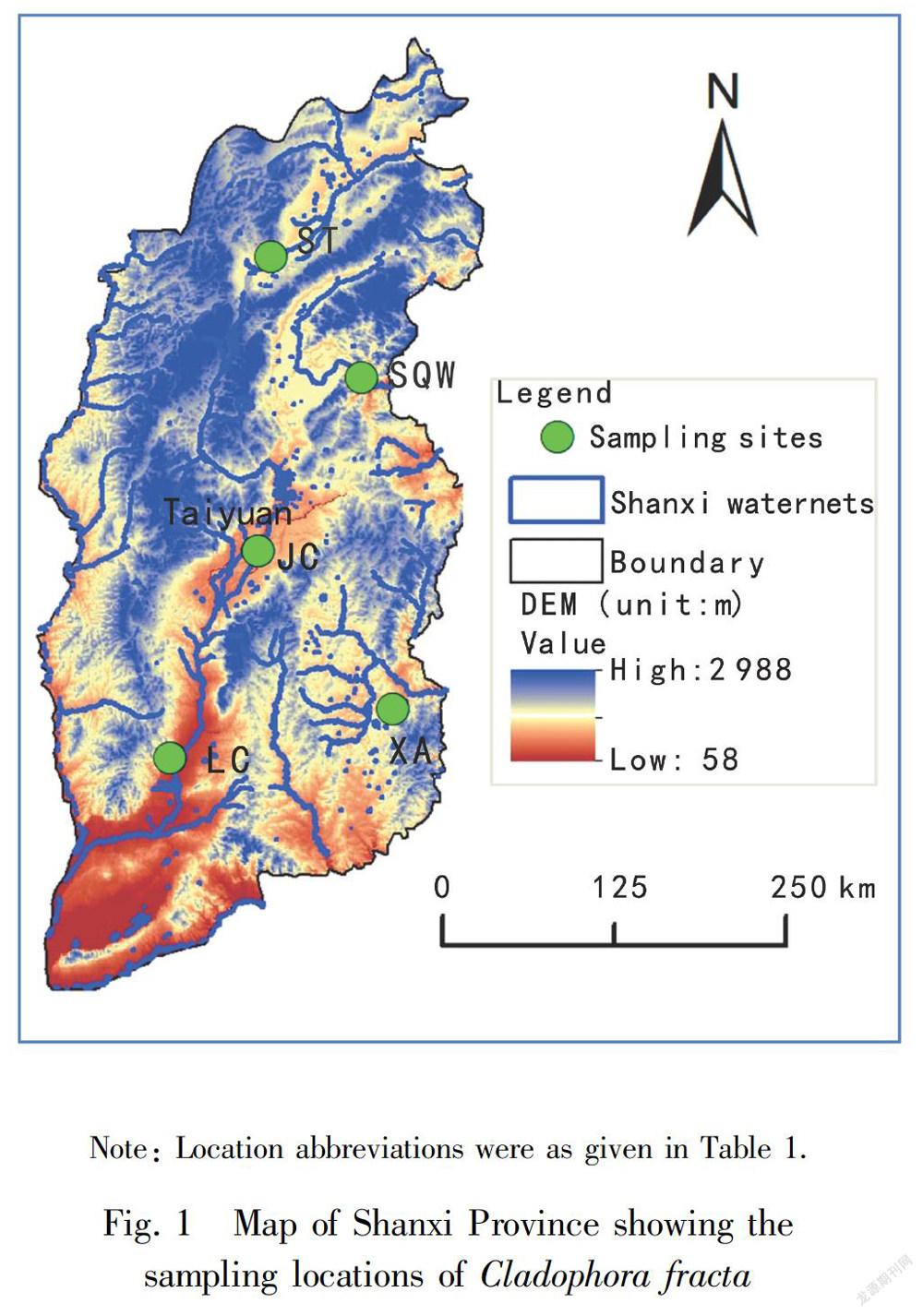
A field survey was conducted at five typical karst springs in Shanxi Province, China in June and July 2015 to identify the habitat of Cladophora. Fig. 1 showed the sampling locations of Cladophora in Shanxi Province. Water temperature and pH were measured on site with a pH/EC/TDS/Temperature Tester (HI98129, HHANNA Instruments Inc., Italia). Specimens of Cladophora were collected from the five streams and Table 1 gave detail information on the sampling sites. Voucher species were deposited at Herbarium of Jinzhong University (JZU) and the voucher information was also given in Table 1. Specimens used for the morphological studies were preserved in freshwater containing 4% formalin or kept alive in freshwater, and those used for the molecular studies were frozen at -20 ℃. External features of thalli were observed under an Olympus BX51 bright field microscope (Olympus Co., Tokyo, Japan).
1.2 DNA methods
Specimens were frozen in liquid nitrogen until use and the total DNA was extracted with Aqua-SPIN Plant gDNA Isolation Mini Kit (Watson Biotechnologies, Inc) following the manufacturer’s instructions. Molecular phylogenetic analyses were carried out based on nuclear-encoded small subunit (SSU) and partial large subunit (LSU), and both genes were combined in partitioned alignment. The primer pairs used for amplifying SSU were based upon Teng (2011) (18S rDNAF: 5′-AAT GGC TCG GTA AAT CAG TT-3′ and 18S rDNAR: 5′-AGT TGA TGA CTC GCG CTT AC-3′). LSU primers were based upon Leliaert et al. (2007) (C1: 5′-ACC CGC TGA ATT TAA GCA TATC-3′ and D2: 5′-TCC GTG TTT CAA GAC GG-3′). The standard polymerase chain reaction (PCR) analyses were carried out on a thermocycler (MyCycler Thermal, BIO-RAD, USA). For SSU sequences, PCR amplification was carried out as follows: initial denaturation at 95 ℃ for 5 min, 35 cycles at 95 ℃ for 30 s, 56.9 ℃ for 30 s and 72 ℃ for 1 min 30 s, and a final extension at 72 ℃ for 7 min. For LSU, PCR amplification was carried out as follows: initial denaturation at 95 ℃ for 5 min, 35 cycles at 95 ℃ for 30s, 63.8 ℃ for 30 s and 72 ℃ for 30 s, and a final extension at 72 ℃ for 7 min. PCR product was purified with Gel Extraction Mini Kit (Watson Biotechnologies, Inc) according to the manufacturer’s recommendation for direct sequencing. The PCR products were commercially sequenced by Shanghai Personal Biotechnology Co., Ltd. Additonal taxa from which obtained from Genbank were shown on Fig. 2.
1.3 Sequence analyses
The Clustal-X 2.0 software was used to align the sequences (Thompson et al., 1997), and the program jModeltest (Posada, 2008; Guindon & Gascuel, 2003) was used to determine parameters for the maximum likelihood and Bayesian analyses. For the SSU, the model was as follows: GTR + I + G distance model, portion of invariable sites=0.592 0; gamma distribution=0.486 0; base frequencies A=0.246 7, C=0.221 0, G=0.286 9, T= 0.245 5; and rate matrix A-C=0.783 2, A-G=2.434 9 , A-T=2.096 1, C-G=0.613 3, C-T=6.101 0 . For the LSU gene, the model was as follows: GTR + G distance model, gamma distribution=0.378 0; base frequencies A= 0.208 6, C=0.284 6, G=0.305 6, T= 0.201 2; and rate matrix A-C=0.842 7, A-G=2.347 0 , A-T=1.569 8 , C-G=0.478 6, C-T=4.404 0 . For the combination of the genes, the model was as follows: for the SSU partition portion, the model was as follows: GTR+I+G distance model, portion of invariable sites=0.592 0; gamma distribution=0.486 0; base frequencies A=0.246 7, C=0.221 0 , G=0.286 9, T= 0.245 5 ; and rate matrix A-C=0.783 2 , A-G=2.434 9 , A-T=2.096 1 , C-G=0.613 3, C-T=6.101 0 . For the LSU partition, the model was as follows: GTR + G distance model, gamma distribution=0.394 0; base frequencies A=0.211 5, C=0.285 0, G=0.299 5, T=0.203 9 ; and rate matrix A-C=0.900 2, A-G=2.917 3 , A-T=1.856 0 , C-G=0.569 4, C-T=5.437 1 . The Maximum Likelihood (ML) analyses were conducted using PhyML 3.0 (Guindon & Gascuel, 2003). The robustness of trees obtained from ML analyses was estimated using bootstrap resampling with 1 000 replicates (Felsenstein, 1985). The Bayesian analyses of the combined data were conducted using MrBayes version 3.1.2 (Huelsenbeck et al., 1996; Ronquist & Huelsenbeck, 2003). A MCMCMC (Metropolis-coupled Markov chain Monte Carlo) algorithm running four Markov chains simultaneously was used to estimate the posterior probability of the phylogenetic trees. The Markov chains were started from a random tree and run for 10 000 000 generations sampling every 1 000 generations for a total of 10 000 samples each run. The first 2 500 samples from each run were discarded as burn-in. The consensus tree was reconstructed after burn-in of 25% generations.
2 Results and Analysis
2.1 Habitat
Karst spring, part of a karst system, is ususlly the end of a cave system at the place where a river cave reaches the Earth’s surface. Shanxi Province is the most abundant and typical region of karst system in North China, where the exposed karst region was 2.6×104 km2 , occupying nearly 17.5% of the total area of the whole province (Han et al., 1993). There were 18 large karst springs with the average discharge being more than 1.0 m3 ·s-1 , including the five investigated karst springs. Water temperature of the five streams ranged from 10.4 to 18.6 ℃ in July (except that for SQW was in June). As expected the water temperature showed some seasonal and geographical fluctuations. The pH of the five collecting sites was more consistent over the sampling period, ranging from 6.3 to 7.7. Total dissolved solids (TDS) showed not much fluctuation over the sampling sites, varying from a minimum of 395 mg·L-1 at SQW where the water temperature was the lowest to a maximum of 552 mg·L-1 at JC where the water temperature was the highest.
2.2 Morphological observations
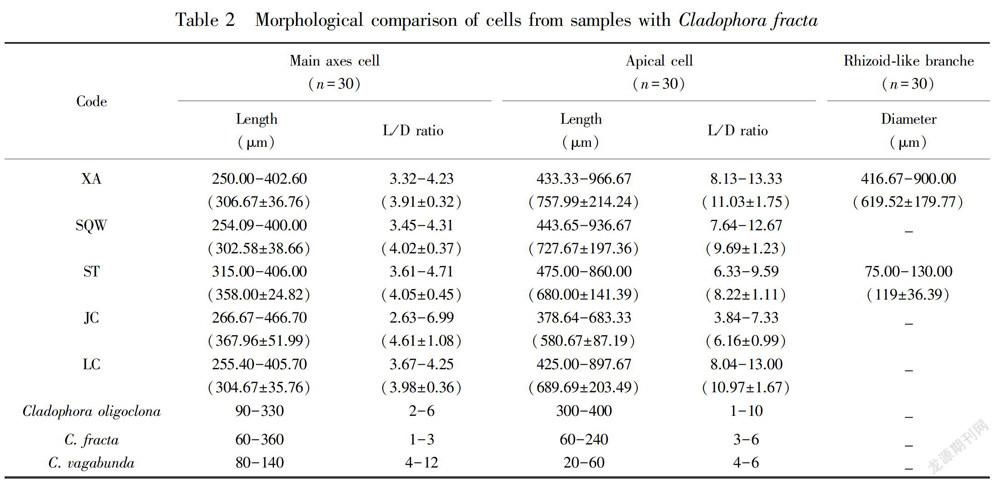
The morphological analyses were based on three specimens of each location.Plants form bright green, clustered, floating or mostly attached to the rock substrates, and measured at about 10 cm in height. Thalli are composed of irregularly branched, uniseriate filaments. Cells are almost pear shaped with irregular swelling. The main axes of the cells were about 250.00-466.70 μm in length, and the length/width ratio was highly variable, in the range of 2.63-6.99 in regions with few cell divisions. The apical cells were about 378.64-966.67 μm in length, and the length/width ratio was also highly variable, in the range of 3.84-13.33. The rhizoid-like branches were only observed in XA and ST. Although there were morphological variances in the samples from five sites, when we carefully checked the diagnostic characters and compared to the previous reports (Hu & Wei, 2006), the other features were common in all individuals detected in this study. Therefore, the samples were identified as Cladophora fracta.
2.3 Datasets and alignments
The SSU and partial LSU regions of the isolated material were deposited in to the GenBank and the accession numbers are given in Table 1. Fifty Cladophora specimens examined, a total of 13 ribotypes were detected (Table 1).The alignments of the two ribosomal genes, SSU and partial LSU, were 1 417 bp and 495 bp in length, respectively. The SSU fragment was approximately three times as long as the partial LSU region but it contained about the same number of variable and parsimony-informative characters (SSU: 322/144; LSU: 330/155). It seems that the pairwise sequence divergence in the SSU was significantly lower than in the LSU (Leliaert et al., 2007). The alignment of SSU sequences and partial LSU sequences was 1 912 bp in length, including 282 parsimony-informative sites among 651 variable sites. The pairwise distances between studies haplotypes were less than 2% both in SSU andpartial LSU sequences. No significant saturation was tested in either SSU or LSU regions, according to the Iss statistic (Xia & Xie, 2001).
2.4 Phylogenetic analyses
Phylogenetic analyses using BI for the combined SSU-partial LSU data (Fig. 2) showed better performance in terms of resolution than the separate SSU and partial LSU analyses (trees not shown). The overall trees had similar topology for the analyzed taxa, but differed in the placement of a few taxa. The Cladophora haplotypes occupied a separate well-supported (0.99/0.91) position in most analyses sister with Cladophora vagabunda. The placement of C. sterrocladi, Siphonocladus tropicus and Valonia aegagropila differed among different analyses. Without forming a monophyletic clade, the Siphonocladus tropicus and Valonia aegagropila clade positioned on the main Cladophora branch in the SSU trees (0.88) or sister with Pithophora clade in the LSU trees (0.82/0.78).
3 Discussion
Combining sequences from different data sets has long been questioned in phylogenetic analyses (Huelsenbeck et al., 1996; Leliaert et al., 2007), while the combination of multiple-gene data sets show better resolved and supported trees, compared with single-gene partitions (Leliaert et al., 2007; Boedeker et al., 2012). In the present study, the combined SSU+ LSU data were found to be superior to the individual partition trees, on account of the SSU fragment contained less variable and parsimony-informative characters and as its length was approximately 3 times as long as the partial LSU region. The taxonomy of genus Cladophora is problematic (Leliaert et al., 2007, 2009). Phylogenetically, all the ribotypes did not correspond with any previously described Cladophora fracta specimens, because we didn’t get any available sequence data of Cladophora fracta from genbank. However, our molecular phylogenetic analysis showed a well supported Cladophora clade.
The inability to match our plants with a described taxon is confirmed by morphological data. Our phylogenetic analyses show that all the ribotypes formed a single clade sister with Cladophora vagabunda. However, C. vagabunda has distinct morphological features. For instances, the filament cells are cylindrical, with 80-140 μm in diameter and 4-12 μm in length; the branch lets taper to 40 μm in diameter, slightly constricted at the junction with main axes; the apices are straight above, curved or sickle-shaped below; and the apical cells are 20-60 μm in diameter, and 5-11 μm long (Russell & Balazs, 2000), but it shares similar length/diameter ratios in the main axes cells with our samples. It was reported that C. vagabunda shares morphological similarity with C. glomerata, while differing from each other on the cell diameter and cell wall thickness which are easily affected by the salinity regimes. C. vagabunda and C. glomerata also showed an incredibly close relationship in the phylogenetic trees (Hayakawa et al., 2012), and both have been the most often mentioned species in eutrophic freshwaters (Whitton, 1970). C. fracta is separated from C. glomerata by the typically more slender filaments and the more pronounced tendency of the akinetes to swell into the pear-shaped structures. Considering the morphological characteristics, we therefore hold our species as Cladophora fracta.
Table 2 summarizes a morphological comparison between the analyzed taxa and similar Cladophora species. The cell sizes were similar among/within the analyzed ribotypes; however, the ST samples distinguished from the XA individuals by a thinner rhizoid-like branch (75-130 μm), while XA samples exhibited the largest length/diameter ratio in the apical cells. The rhizoid-like branches were not observed in the SQW, JC and LC specimens. Although Cladophora species frequently appear in karst springs, their genetic diversity, phenological patterns and physiological properties have not been well investigated in these environments. In this study, collections from the five karst springs showed a unique morphological characteristic. Our samples collected from the five karst springs did not exhibit significant variation in cell dimensions, but the cell diameters of the individuals from the five locations were larger than those reported for the cells from lakes (Whitton, 1970), indicating that the habitat conditions may have an effect. A similar tendency was observed in cell length and cell wall thickness. Perhaps these differences may have been caused by regional differences. Sinha (1968) had concluded that the number of nucleoli and chromocentre like bodies can be correlated with the degree of polyploidy which makes species larger in size. Moreover, morphological plasticity is related to diverse habitats and different salinity levels (Nienhuis, 1975; Hayakawa et al., 2012; Ichihara et al., 2013). Some frameworks, however, still lack enough sequence data to support. It is clear that additional sampling of Cladophora species from different salinity levels as well as karst springs will be needed to further clarify the diversity and plasticity of the species.
Acknowledgements We thank Dr.Don Zhao (Professor Elton Z. and Lois G. Huff Chair of Environmental Engineering, Auburn University, Auburn, USA) for his critical reviews of the manuscript and editorial assistance with the English. Also we want to acknowledge the anonymous reviewers that enhanced the quality of this manuscript.
References:
BO RDEKER C, O’KELLY CJ, STAR W, et al., 2012. Molecular phylogeny and taxonomy of the Aegagropila clade (Cladophorales, Ulvophyceae), including the description of Aegagropila gen, nov. and Pithophora gen. nov [J]. J Phycol, 48(3): 808-825.
FELSENSTEIN J, 1985. Confidence limits on phylogenies: An approach using the bootstrap [J]. Evolution, 39(4): 783- 791.
GESTINARI LMDS, PEREIRA SMB, YONESHIGUE-VALENTIN Y, 2010. Distribution of Cladophora species (Cladophorales, Chlorophyta) along the Brazilian Coast [J]. Phytotaxa, 14(3): 22-42.
GUINDON S, GASCUEL O, 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood [J]. Systematic Biol, 52(5): 696-704.
HANYUDA T, WAKANA I, ARAI S, et al., 2002. Phylogenetic relationships within Cladophorales (Ulvophyceae, Chlorophyta) inferred from 18S rRNA gene sequences, with special reference to Aegagropila linnaei [J]. J Phycol, 38(3): 564-571.
HAYAKAWA Y, OGAWA T, YOSHIKAWA S, et al., 2012. Genetic and ecophysiological diversity of Cladophora (Cladophorales, Ulvophyceae) in various salinity regimes [J]. Phycol Res, 60(2): 86-97.
HAN XR, LU RA, LI QS, et al., 1993. Karst system—research on large karst springs in Shanxi [M]. Beijing : Geological Publishing House. [韓行瑞, 鲁荣安, 李庆松, 等, 1993. 岩溶水系统--山西岩溶大泉研究 [M]. 北京: 地质出版社.]
HU HJ, WEI YX, 2006. The freshwater algae of China-syste- matics, taxonomy and ecology [M]. Beijing: Science Press:770. [胡鸿钧, 魏印心, 2006. 中国淡水藻类:系统,分类及生态 [M]. 北京: 科学出版社: 770. ]
HUELSENBECK JP, BULL JJ, CUNNINGHAM CW, 1996. Combining data in phylogenetic analysis [J]. Trends Ecol Evol, 11(4): 152-158.
ICHIHARA K, SHIMADA S, MIYAJI K, 2013. Systematics of Rhizoclonium-like algae (Cladophorales, Chlorophyta) from Japanese brackish waters, based on molecular phylogenetic and morphological analyses [J]. Phycologia, 52(5): 398- 410.
LELIAERT F, BOEDEKER C, PENA V, et al., 2009. Cladophora rhodolithicola sp. nov. (Cladophorales, Chlorophyta), a diminutive species from European maerl beds [J]. Eur J Phycol, 44(2):55-169.
LELIAERT F, ROUSSEAU F, DE REVIERS B, et al., 2003. Phylogeny of the Cladophorophyceae (Chlorophyta) inferred from partial LSU rRNA gene sequences: is the recognition of a separate order Siphonocladales justified? [J]. Eur J Phycol, 38(3): 233-246.
LELIAERT F, DECLERCK O, VERBRUGGEN H, et al., 2007. Molecular phylogeny of the Siphonocladales (Chlorophyta: Chlorophyceae) [J]. Mol Phylogenet Evol, 44(3): 1237-1256.
NIENHUIS PH, 1975. Biosystematics and ecology of Rhizoclonium riparium (Roth) Harvey (Chlorophyceae: Cladophorales) in the estuarine area of the rivers Rhine, Meuse, and Scheldt [M]. Bronder Offset B. Y., Rotterdam: 240.
POSADA D, 2008. jModelTest: Phylogenetic Model Averaging [J]. Mol Biol Evol, 25(7): 1253-1256.
RONQUIST F, HUELSENBECK JP, 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models [J]. Bioinformatics, 19(12): 1572-1574.
RUSSELL DJ, BALAZS GH, 2000. Identification manual for dietary vegetation of the Hawaiian green turtle, Chelonia mydas [M]. NOAA TM-NMFS-SWFSC-294: 49.
SINHA JP, 1968. Cytotaxonomical studies on Cladophora glomerata, four freshwater forms [J]. Int J Cytol, 32(3-4): 507-518.
TENG LH, 2011. Study on morphology and molecular phylogeny of Cladophorales (Chlorophyta) along China sea coast, with Its DNA Barcoding Based on ITS and 18S rDNA Sequences [D]. Beijing: Master Dissertation, Graduate University of Chinese Academy of Sciences: 17.
THOMPSON JD, GIBSON TJ, PLEWNIAK F, et al., 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools [J]. Nucleic Acids Res, 25(25): 4876-4882.
VAN DEN HOEK C, CHIHARA M, 2000. A taxonomic revision of the marine species of Cladophora (Chlorophyta) along the coast of Japan and the Russian Far-East. National Science Museum Monographs [M]. Tokyo: National Science Museum, 19:242.
WHITTON BA, 1970. Biology of Cladophora in freshwaters [J]. Water Res, 4(7): 457-476.
XIA X, XIE Z, 2001. DAMBE: Software package for data analysis in molecular biology and evolution [J]. J Hered, 30(7): 1720-1728.
YOSHII Y, HANYUDA T, WAKANA I, et al., 2004. Carotenoid compositions of the Cladophora balls (Aegagropila linnaei) and some members of the Cladophorales (Ulvophyceae, Chlorophyta): their taxonomic and evolutionary implication [J]. J Phycol, 40(6): 1170- 1177.

