Circulating microRNAs and liquid biopsy: murine xenograft models for technical validation of clinical protocols
Jessica Gasparello, Matteo Allegretti, Chiara Papi, Elena Giordani, Patrizio Giacomini, Roberto Gambari,3, Alessia Finotti
1Department of Life Sciences and Biotechnology, Ferrara University, Ferrara 44121, Italy.
2Oncogenomics and Epigenetics, IRCCS Regina Elena National Cancer Institute, Rome 00144, Italy.
3Interuniversity Consortium for Biotechnology (CIB), Trieste 34149, Italy.
Abstract In oncology, liquid biopsy is applied to detect with high efficiency clinically relevant analytes, such as tumor cells, cell-free nucleic acids, and exosomes in peripheral blood and other body fluids of cancer patients. Liquid biopsy is considered one of the most advanced non-invasive diagnostic systems useful, in the next future, for enabling personalized treatments in precision medicine. Medical actions include, but are not limited to, early diagnosis, staging, prognosis, anticipation (lead time) and prediction of therapy responses, as well as follow up. Experimental system for validation of the proposed liquid biopsy approaches is highly needed. In this review article we will discuss the establishment of xenotransplanted mouse model systems for the validation of liquid biopsy protocols aimed to identify changes in the miRNA plasma content. Human colon cancer HT-29 and LoVo cells have been xenotransplanted and miR-221-3p and miR-222-3p have been comparatively analyzed in cultured HT-29 and LoVo cells, xenotransplants and plasma samples.
Keywords: Liquid biopsy, circulating tumor RNA, microRNA, xenograft
LIQUID BIOPSY: A NEW FRONTIER FOR CANCER DIAGNOSTICS
In the field of cancer diagnosis and treatment, liquid biopsy is a new diagnostic tool that investigates circulating tumor cells (CTC) and/or cell-free nucleic acids in the peripheral blood [Figure 1A]. Liquid biopsy is considered one of the most advanced non-invasive diagnostic systems. It provides key molecular information relevant to important clinical decisions and, being “longitudinal” (it can be repeated as many times as needed), it fits the idea of precision medicine possibly more than other “static” techniques based on the analysis of tissue nucleic acids[1-5]. Diagnostic actions made possible by liquid biopsy include, but are not limited to, early diagnosis, staging, prognosis, prediction of therapy response and follow up during therapeutic intervention[6-11].
In addition to the use of CTCs[12-15]and circulating tumor DNA (ctDNA)[16-18], other important targets for liquid biopsy are circulating microRNAs (miRNAs)[19-24], a family of small (19 to 25 nucleotides in length) noncoding RNAs playing important roles in controlling post-transcriptional gene expression. Regulatory miRNAs reduce protein synthesis through selective interactions with complementary sequences of target messenger RNAs (mRNAs)[25-27]. Single or multiple mRNAs can be targeted at their 3'-UTR, CDS, 5'-UTR sequences, and it is calculated that more than 60% of human mRNAs are microRNA targets[26]. Dysregulation of microRNAs has been associated with a variety of human pathologies, including cancer[28-31]. In this case miRNAs behave both as tumor promoters (oncomiRNAs and metastamiRNAs) and tumor suppressor molecules[29], depending on their mRNA targets (oncosuppressor mRNAs or mRNA coding oncoproteins, respectively) with opposing activity on cancer cells. Based on this, it is not surprising that circulating cellfree miRNAs have been actively investigated as liquid biopsy analytes. OncomiRNAs are abundant in several extracellular body fluids, where they are protected and stabilized by exosome-like structures and small intraluminal vesicles produced by a variety of cells (including cancer cells)[32-36]. Hence, elevated levels of several miRNAs (including miR-221, miR-222, miR-141, miR-92a, miR-21, miR-155, miR- 506, miR-4316, miR-4772-3p, and miR-29a) are present in the blood from patients with colorectal carcinomas (CRC) and may contribute to diagnosis and prognosis[21,37-42]. Furthermore, is well established that miRNAs may help in monitoring therapeutic approaches. For instance, Ogata-Kawataet al.[22]reported that serum exosomal miRNA levels (let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a) were higher in CRC patients than controls, were already detectable at early disease stages, and that they were significantly downregulated after surgical resection.
TECHNOLOGIES FOR MICRORNA ANALYSIS
In order to quantify miRNAs in the plasma and other body fluids isolated from cancer patients, several types of technologies for RNA analysis have been proposed[43-51]. Quantitative real-time PCR (RT-qPCR)[52], NGS RNA sequencing[53], miRNA microarray analysis[54], and digital PCR[55]are the most used [Table 1] and can be employed not only for tissue or cells but also for highly diluted samples, such as body fluids. One of the major limits of RT-qPCR and ddPCR is the limited number of miRNAs that can be quantified for single run. This problem was partially solved by introduction of TaqMan low density arrays, that allows to quantify the content of a significant number of miRNAs (about 700 miRNAs) using PCR-based methods[56]. In addition to these methodologies, other technologies have been described for direct miRNA detection from serum samples. For example, Chapinet al.[57]proposed rolling circle amplification (RCA) based on the use of a universal adapter ligated to the targets captured on encoded gel microparticles. The system allows the multiplexed profiling of miRNA at sub-femtomolar concentration. Interestingly, Williamset al.[58]proposed a miRNA detection technique able to amplify miRNAs directly in body fluids, avoiding upstream sample preparation. The technique, based on isothermal target amplification, has a sensitivity positioned in the femtomolar range. Other conventional technologies normally proposed for miRNA detection in cells or tissues, such as northern blotting[59], are not suitable for miRNA detection in body fluids, due to the low sensitivity of the technology, requiring therefore large amounts of RNA. Other unconventional miRNA detection techniques have been proposed in recent years such as bead-based flow cytometry[60]but at the moment they are employed only for miRNA detection in tissues or cellular samples.
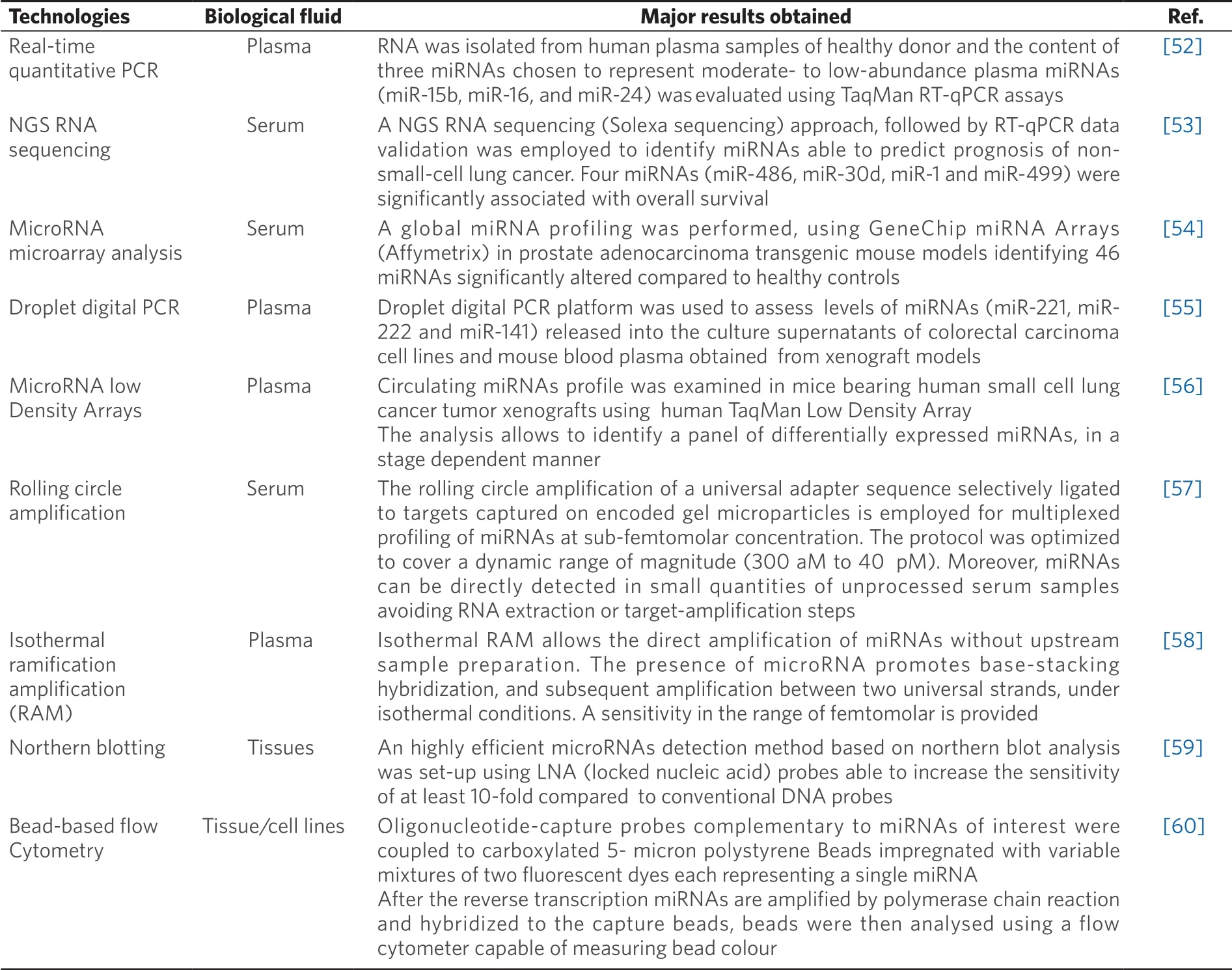
Table 1. Technologies applied to miRNA detection
EXPERIMENTAL MODEL SYSTEMS FOR LIQUID BIOPSY
Given the fast pace of technological evolution and the clinical complexity of human cancers, there is an increasing need for experimentalin vivosystems and associated validation technologies. A robust analysis of bio-fluids must acknowledge the fact that liquid biopsy is a complex strategy requiring the achievements of several key points, including pre-analytical steps, post-analytical optimization, and careful selection of optimal analytes for specific biological queries.In vivomodel systems might be very useful to address and isolate these many individual variables (that are both technical and biological), and validate complex multistep approaches. It is surprising, in this respect, that only few reports are available focusing on the use of animal models.In vivomodel systems for the detection of circulating tumor cells and DNA have been proposed[61-63]. These include injection of cancer cells into their orthotopic site of origin (e.g., a “recap” of natural tumor onset), injection of tumor cells directly into the bloodstream of the animal (to recap distant seeding), genetically engineered mouse and mice xenografted with patient-derived tumors[64-69](to mimic “true” tumors). As to the analysis of circulating miRNAs in these experimental settings, several reports are available[52,54-56,70,71][Table 2].
Mice xenografted with human tumor cell lines or patient-derived tumor
The pattern of circulating miRNAs has been extensively studied in mice xenografted with tumor cells and patient-derived tumors. Different independent studies firmly demonstrated that miRNAs released in the circulation by the tumor xenograft are distinct from the “background” mouse miRNAs pattern. This is a key point, since pre-existing miRNAs present in the mouse body fluids are identical in sequence to most human miRNAs and hence they might be a powerful confounding parameter, possibly altering conclusions and implications of any circulating miRNA signature. In this respect, the use of laboratory mouse strains has the advantage that its “background” mouse miRNA pattern is stable and easily quantifiable. Mitchellet al.[52]demonstrated that several miRNAs originating from xenografted human prostate cancer cells are present in the mouse circulation (one of the most interesting being miR-141), and are readily measured in plasma, allowing a clear distinction between tumor xenografted and control tumor-free mice. Waterset al.[70]observed a complex miRNAs dysregulation in the circulation of athymic nude mice subcutaneously injected with MDA-MB-231 cells. Some miRNAs (such as miR-10b) were undetectable in the circulation, others (miR-195 and miR-497) were significantly decreased, miR-221 content did not change, and a positive correlation was observed between miR-497 and miR-195. This study highlighted distinct roles of miRNA subsets in the circulation and in disease dissemination and progression, all of which may be candidates as molecular targets for diagnosis as well as design of systemic therapy. Gasparelloet al.[55]studied liquid biopsy in mice bearing CRC xenografts, demonstrating gateways regulating the levels of circulating tumor-derived miRNAs (ctmiRNAs), e.g., cell-specific roadblocks that determine whether a given cell xenotransplants releases or retains a specific miRNA. These roadblocks are often not present in cultured cells, and build “barriers” to detection in a liquid biopsy format.
Genetically engineered mouse model systems
Genetically engineered mouse models (GEMMs) manipulate target oncogene or tumor suppressor expression in mice in order to promote tumor development. Transgenic and knockout GEMMs have provided important models for identifying tumor-associated and metastasis-associated genes that can lead to tumor formation and disease progression. In addition, GEMMs have been applied to the development of liquid biopsy methods based on the analysis of circulating microRNAs. Selthet al.[54]performed a global miRNA profiling and identified a set of miRNAs exhibiting significantly altered serum levels in transgenic mice models of prostate cancer (i.e., Transgenic Adenocarcinoma of Mouse Prostate mice). Global miRNA profiling identified 46 miRNAs at significantly altered levels in the serum of mice with advanced prostate cancer compared to healthy mice used as controls. Interestingly, four miRNAs altered in mice (mmu-miR-141, mmu-miR-298, mmu-miR-346 and mmu-miR-375) were also found to be expressed at higher levels in the serum of patients with metastatic prostate cancer compared with control subjects. Moreover, three of these (hsa-miR-141, hsa-miR-298 and hsa-miR-375) were upregulated in prostate tumors compared with normal prostate tissue, suggesting that they are directly released from the tumor into the blood as disease progresses. This study was the first to demonstrate that specific serum miRNAs (miR-141, miR-298 and miR-375) are common between human prostate cancer and a mouse model of the disease, highlighting the potential of such models for the discovery of novel biomarkers.
Zhanget al.[71]investigated FOXP3-inducible breast cancer cells, Foxp3 heterozygous Scurfy mutant (Foxp3 sf/+) female mice, and patients with breast cancer for characterization of the formation and regulation of the miR-200 family in breast cancer cells and circulation. While levels of miR-200c and miR-141 were lower in Foxp3 sf/+ tumor cells than in normal breast epithelial cells, plasma levels of miR-200c and miR-141 in the Foxp3 sf/+ mice increased during tumor progression and metastasis. Interestingly, the levels of miR-200c and miR-141 were higher in plasma from patients with metastatic breast cancer than in plasma from those with localized breast cancer, with benign breast tumors, with a family history of breast cancer, or from healthy controls. The conclusion of the work reported by Zhanget al.[71]supports the concept that miR-200c and miR-141 are regulated by a FOXP3-KAT2B axis in breast cancer cells, and circulating levels of miR-200c and miR-141 are potential biomarkers for early detection of breast cancer metastasis. Moreover, they highlight the idea that roadblocks evolve during the natural history of tumors.
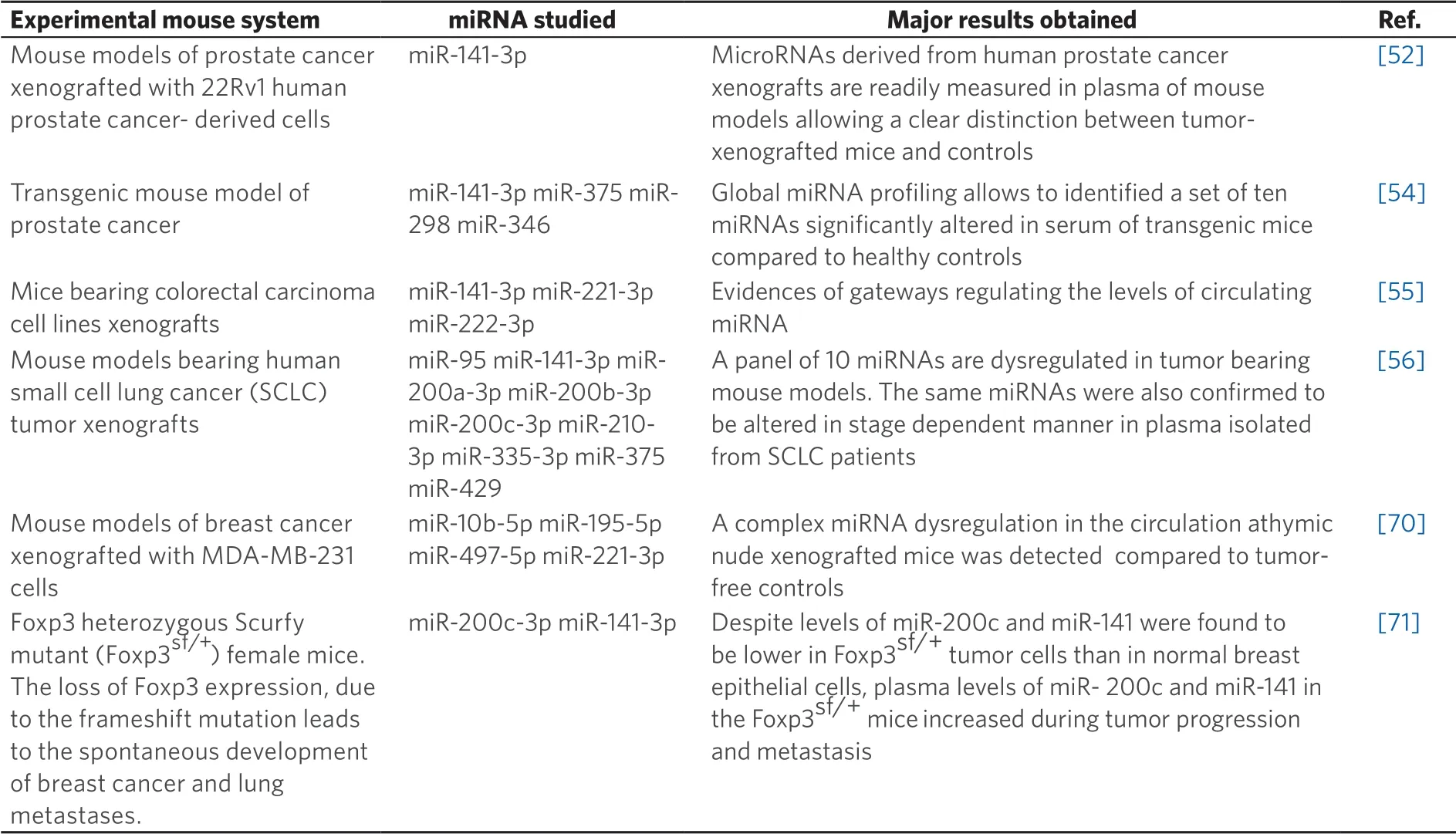
Table 2. Examples of experimental systems to study miRNA content in liquid biopsy
LIQUID BIOPSY IN MICE BEARING COLORECTAL CARCINOMA XENOGRAFTS OBTAINED AFTER IMPLANTATION OF HT-29 AND LOVO CARCINOMA CELLS
Analysis of miRNA content has been recently performed in mice xenografted with colon cancer cell lines[55]. Among the different xenografted models the one based on the implantation of the HT-29 and LoVo CRC cells was found the most efficient for miRNA detection. The HT-29 cells were derived from a KRAS-WT, differentiated colorectal adenocarcinoma[72,73], while LoVo cells (originally described as Dukes’ type C, grade IV) harbor a heterozygous KRAS c.38G>A mutation (G13D)[74].
TUMOR XENOGRAFTS AND PLASMA PREPARATION
Figure 1B shows the study workflow based on the tumor xenografts as models for liquid biopsy to assess plasma levels of circulating miRNAs. In this study workflow, miRNAs are compared considering (1)in vitrocultured tumor cells; (2) tumor xenografts; and (3) blood plasma samples. The HT-29 and LoVo cell lines were selected as proxies of clinically evident cancers and sources of soluble analytes. MicroRNAs were extracted from both cell lines, matched tumor tissue and blood plasma samples and were then subjected to ddPCR and RT-qPCR analysis. Tumor xenotransplants were established by inoculating HT-29 and LoVo cells in the flank of 4-month old Nu/CD1 mice. Tumors were taken at sacrifice along with blood. Frozen tissues were used as the source of miRNAs. For the analysis of ctmiRNA, blood plasma was treated to disrupt exosomes and denature miRNA-binding proteins with QIAzol Lysis Reagent. After the addition of 400 amoles of celmiR-39-3p (an equalizer), total RNA was purified and reverse transcribed. Finally, droplet digital PCR (RTddPCR) assays for microRNA content analysis were performed to quantify the levels of miR-221-3p[37,75]and miR-222-3p[76]. Droplets were analyzed using the QX200 Droplet Reader, and data analysis was performed with QuantaSoft version 1.7.4 (Bio-Rad, Hercules, CA, USA).
TUMOR XENOGRAFTS AND PLASMA PREPARATION: MAJOR RESULTS
The main point of this study is focused on determining whether the pattern of plasma miRNA content recapitulates HT-29 and LoVo cells and xenografted tumors. A representative example of miR-222-3p content is shown in Figure 2A and all the quantitative data for miR-221-3p and miR-222-3p are presented in Figure 2B. The miRNA levels were independently assessed by RT-qPCR and ddPCR results, obtaining similar results, as reported elsewhere[55]. Of course, in the quantitative analysis shown in Figure 2B and concerning the plasma miRNA quantitation, we have taken into account the fact that cross-species miRNA homology might influence ourin vivoresults. Accordingly, we quantitated baseline, endogenous miR-221-3p and miR-222-3p levels in tumor-free, healthy nude mice. As expected (the sequences of mouse miR-221-3p and miR-222-3p are identical to those found in human cells) both RT-ddPCR and RT-qPCR demonstrated that circulating miRNAs were detectable even in the absence of tumor growth. However, the differences between tumor-bearing and tumor-free mice were clearly appreciable for both miRNAs. Figure 2B shows that the miR-222-3p content is higher than miR-221-3p content in HT-29 and LoVo cells, and in tumor and plasma samples isolated from HT-29 and LoVo xenografted mice, despite the miR-222/miR-221 ratio is much higher in plasma in comparison with cell and tumors. This is consistent with the “gateway” effect mentioned above and discussed in deep in Gasparelloet al[55]. This issue is particularly of interest, since detailed knowledge of the molecular mechanisms underlying the release of circulating analytes is still lacking. Alternatively, thein vivoresponse of xenotransplanted mice to tumor cell injection might contribute to the reported unbalanced content of miR-222/miR-221. The proposed system is expected to help in verifying the underlying cellular mechanisms.
CONCLUSION
Circulating miRNAs have been recently used as biological markers for early diagnosis, prognosis, prediction of response to therapy and clinical outcome, particularly in a liquid biopsy setting[1-11,77-79]. Liquid biopsy is a powerful tool applicable to all or most human cancers, including colorectal, lung, melanoma, and breast neoplasms[80,81]. From a more general viewpoint, tumor-xenotransplanted mice and otherin vivomodels may have an important role because they resolve biological variables from technical variables (such as handling and storage of biological fluids, pre-analytical processing, as well as DNA and RNA isolation protocols) that might affect efficient marker detection by liquid biopsy[82,83]. Liquid biopsy of cancer is mainly based on the analysis of circulating tumor cells and/or cell-free nucleic acids in the peripheral blood of cancer patients, as well as in other body fluids suitable for diagnostic assessment. Among these, cerebrospinal fluid for tumors of the central nervous system, saliva for tumors affecting the head and neck, pleural effusion in the case of respiratory tract cancers and urine for urinary tract cancers. We propose thatin vivoxenotransplant models monitoring miRNAs may find application in all the body fluids, contributing to assess the relevance of clinical liquid biopsy. The importance ofin vivomodel systems adds to the established role of liquid biopsy in complementing key limitations of surgical tissue biopsy. These include, but are not limited to: (1) invasiveness and inherent patient compliance; (2) a static representation of the tumor pathology strictly limited to the tumor tissue sampling; (3) ethical and practical issues preventing repeated tissue biopsy, particularly at unaccessible (or difficult to access) body sites; (4) tumor heterogeneity, especially during progression and metastatic dissemination (making multiple sampling necessary); (5) easier and real-time patient monitoring by non-invasive liquid biopsy. Therefore, although liquid biopsy approach still suffers from important drawbacks (fragmentation of cfDNA, instability of RNA, low yield of isolated samples to be analyzed and variable presence of normal DNA and RNA), this approach is generally deemed of great potential interest for future applications, patent development, and clinical trials, and mouse xenotransplants may be an important “shortcut” to application and technical streamlining.
Among possible application of mouse models we suggest: (1) studies on the relationship between the tumor size and the plasma miRNAs content (e.g., miR-222/miR-221 ratios); (2) analysis of the “gateway” hypothesis involved in the selection of released microRNAs (e.g., miR-221 and miR-222); (3) studies concerning the possible local and systemic responses of normal cells and tissues to xenotransplant procedure (tumor seeding); (4) analysis of the effects on miRNA plasma content on the susceptibility to experimental treatment of xenografted mice with physical and/or chemotherapeutic agents; (5) verification of the selectivity of the effects on plasma miRNA content of miRNA targeting and relative delivery approaches; (6) usage as key tools for the comparison of different analytical strategies including, among others, different PCR/RT-qPCR and NGS platforms, instruments and protocols, as well as PCR-free methods[84-86]. Among possible limits of the mouse xenograft model systems here presented are the differences between man and mouse with respect to ctDNA and microRNA dinamics in respect to their vasculature. Therefore we should carefully consider the sharply different ratios between the dimension of implanted tumors, the mouse body weight and the blood volume on one hand and those related the same parameters (i.e., tumor weight, body weight and blood total volume) in CRC patients. In this respect the analysis of the miRNome in liquid biopsy obtained when tumors of different dimensions are employed in mouse xenograft model systems might clarify whether the ratio between tumor size and mouse body weight or blood volume might affect the results. This might also be of interest for developing algorithms in human clinical settings.
DECLARATIONS
Authors’ contributions
Revised and approved the final manuscript: Gasparello J, Allegretti M, Papi C, Giordani E, Giacomini P, Gambari R, Finotti A
Wrote the manuscript: Allegretti M, Giacomini P, Gambari R, Finotti A
Performed the literature search: Allegretti M, Giacomini P, Gambari R, Finotti A
Critically analyzed the existing literature: Allegretti M, Giacomini P, Gambari R, Finotti A
Designed the figures and created the tables: Gasparello J, Papi C, Giordani E, Gambari R, Finotti A
Availability of data and materials
Not applicable.
Financial support and sponsorship
This work was supported by the European Union (EU) Horizon 2020 Research and Innovation Programme: project ULTRAsensitive PLAsmonic devices for early CAncer Diagnosis (ULTRAPLACAD) (633937); Associazione Italiana per la Ricerca sul Cancro (AIRC) (13575) to Gambari R, (14204, 19052) to Giacomini P. Allegretti M is the recipient of a three-year AIRC fellowship (id. 19503). This study was also supported by the Interuniversity Consortium for the Biotechnology, Italy.
Conflicts of interest
The author declared that there are no conflicts of interest.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Copyright
© The Author(s) 2019.
 Journal of Cancer Metastasis and Treatment2019年6期
Journal of Cancer Metastasis and Treatment2019年6期
- Journal of Cancer Metastasis and Treatment的其它文章
- The role of hypoxia-induced factor 1α in breast cancer
- Paxillin serine 178 phosphorylation in control of cell migration and metastasis formation through regulation of EGFR expression in breast cancer
- A new view of the mammary epithelial hierarchy and its implications for breast cancer initiation and metastasis
- Significance of trace eIement quantities in the prostatic secretion of patients with benign prostatic hyperpIasia and prostate cancer
- Cellular plasticity and metastasis in breast cancer: a pre- and post-malignant problem
- Differential expression and function of the endogenous lactate receptor, GPR81, in ERαpositive/HER2-positive epithelial vs. post-EMT triple-negative mesenchymal breast cancer cells
