Physiological Responses of Contrasting Rice Genotypes to Salt Stress at Reproductive Stage
Maria Elisa B. Gerona, Marjorie P. Deocampo, James A. Egdane, Abdelbagi M. Ismail, Maribel L. Dionisio-Sese
Physiological Responses of Contrasting Rice Genotypes to Salt Stress at Reproductive Stage
Maria Elisa B. Gerona1, 2, Marjorie P. Deocampo3, James A. Egdane3, Abdelbagi M. Ismail3, Maribel L. Dionisio-Sese2
(Division of Natural Sciences and Mathematics, University of the Philippines Visayas Tacloban College, Leyte 6500, the Philippines; Institute of Biological Sciences, University of the Philippines Los Baños College, Laguna 4031, the Philippines; International Rice Research Institute, DAPO Box 7777, Metro Manila, the Philippines)
Salinity is a major abiotic stress affecting plant growth and productivity. Considerable genetic variation is present in rice in response to salt stress, with higher sensitivity during early seedling and reproductive stage. In this study, physiological changes in leaves and developing panicles of rice genotypes (IR686, Sadri, Rc222, CSR28, IR670 and Pokkali) contrasting in salt tolerance at the reproductive stage were evaluated in greenhouse experiment under salt stress. The results showed that IR670 and the tolerant-check Pokkali maintained lower Na+/K+ratio, less reduction in chlorophyll concentration, lower malondialdehyde (MDA) production, higher concentrations of reduced ascorbate (reduced AsA), higher proline accumulation and lower percentage reduction in pollen viability than the salt-sensitive genotypes under salt stress. The higher concentration of reduced AsA suggests an efficient ROS-scavenging system. Physiological measurements and pollen viability analysis revealed that Sadri (moderately tolerant at the seedling stage) is sensitive to salt stress at the flowering stage. The findings will be useful in breeding salt tolerant varieties at both seedling and reproductive stages by selecting appropriate genotypes and phenotypes.
Na+/K+ratio; proline; reproductive stage; rice; salinity stress; reduced ascorbate; pollen viability
Soil salinity is one of the most serious constraints for rice production worldwide. This problem is further worsening because of climate change causing sea level rise and more frequent coastal storms incidences leading to salt intrusion in agricultural lands. Besides, poor water management practices, like poor drainage in irrigated areas, cause secondary salinization (Wassmann et al, 2004; Ismail et al, 2007; Qureshi and Al-Falahi, 2015). Salt-affected areas are estimated at over 800 million hectares worldwide, which is equivalent of more than 6% of the world’s total land area. Based on the United States Department of Agriculture Salinity Laboratory, saline soils are defined as having an electrical conductivity (EC) of 4 dS/m (about 40 mmol/L NaCl) or more as a result of excess of sodium ions, with predominant anions of chloride and sulfate.
The detrimental effect of salinity on plant growth and productivity is associated with low water potential of the root medium since increase in soil salt concentration decreases osmotic potential and ability of plants to take up water. Earlier studies on cereal crops conducted at the seedling stage suggested salt exclusion from leaves as the most important tolerance mechanism (Genc et al, 2007; James et al, 2011; Platten et al, 2013; Adem et al, 2014; Ismail and Horie, 2017). Selective ion uptake by compartmentation and roots of harmful ions in older tissues such as older leaves and leaf sheaths reduce Na+accumulation and prevent its building up to toxic concentrations in photosynthetically active tissues. Ravikiran et al (2018) showed that chlorophyll concentration is lower under saline condition in a set of 192 diverse rice genotypes at the seedling stage. Moradi and Ismail (2007) suggested that the reactive oxygen species (ROS) scavenging systems including reduced ascorbate (AsA) might play a key role in mitigating salt stress effects in rice at the early seedling stage. Höller et al (2015) reported an increase in AsA levels in leaves of rice plants under salt stress at the vegetative stage, which might be due to the physiological role of AsA in reproductive development. Accumulation of excessive ROS disrupts cellular metabolism through oxidative damage of lipids. Lipid peroxidation, monitored by malondialdehyde (MDA) concentration, is an effective indicator of cellular oxidative damage and its level has been reported to be negatively correlated with salinity tolerance, associated with upregulation of ROS-scavenging system (Dionisio-Sese and Tobita, 1998; Moradi and Ismail, 2007; Kanawapee et al, 2013). In addition to the upregulation of ROS-scavenging systems, synthesisand accumulation of compatible solutes such as proline, which stabilizes the structure of proteins, is another strategy for tolerance of salinity (Szabados and Savouré, 2010). Kibria et al (2017) reported significant increase of proline accumulation in the salt-tolerant genotype BINA dhan 10 during the vegetative stage, which might be associated with its salt tolerance.
Of the cereal crops, rice (L.) is the most sensitive to salinity (Munns and Tester, 2008). Most rice genotypes exhibit variable responses to salinity stress, which is also dependent on the developmental stage at which genotypes are exposed (Kanawapee et al, 2013; Wankhade and Sanz, 2013; Ismail and Horie, 2017). Several studies reported that rice is tolerant during germination, and then becomes sensitive at the early seedling stage, whereas gains tolerance at the vegetative stage and then becomes sensitive at the reproductive stage (Ismail et al, 2007; Hakim et al, 2010; Palao et al, 2013). Moreover, salinity tolerance at the seedling stage in rice is poorly associated with tolerance at the reproductive stage (Moradi et al, 2003; Singh and Flowers, 2010). Salinity tolerance at the reproductive stage is important in areas where high salt stress is expected later in the season, because at this stage pollination and formation of grains occur that directly contribute to economic yield. Therefore, in view of the recent changes in climatic conditions and variability, there are urgent needs to exploit natural variation in rice for salt tolerance at the reproductive stage, for development of more resilient, salt-tolerant genotypes.
As indicated above, numerous studies conducted on salinity tolerance at the seedling stage established that the key traits for salt tolerance include higher seedling vigor and tissue tolerance, salt compartmentation and exclusion of ions in older tissues (Hakim et al, 2010; Chunthaburee et al, 2016), and on responses and mechanisms associated with salinity tolerance during the seedling stage, but little is known how about the physiological mechanisms associated with salt tolerance during reproduction. In this study, physiological responses under salinity stress were evaluated during the reproductive stage in a set of contrasting rice genotypes. Information on these mechanisms will improve the efficiency of evaluation and enhance selection during breeding of salt-tolerant genotypes.
MATERIALS AND METHODS
Rice materials and growth conditions
Six rice genotypes (IR686, Sadri, CSR28, IR670, Rc222 and Pokkali) contrasting in tolerance of salt stress were selected. IR670 and IR686 are breeding lines with 1% and 90% reduction in grain yield, respectively, under salt stress (EC = 6 dS/m) during the reproductive stage (Moradi et al, 2003), CSR28 is highly tolerant of alkalinity/sodicity at the seedling stage (Krishnamurthy et al, 2015), and Sadri is an Iranian cultivar with moderate tolerance to salinity (Mohammadi-Nejad et al, 2008). Rc222, a salt sensitive, high-yielding variety from the Philippines, and Pokkali, a salt tolerant landrace from India, were used as the checks.
In evaluating the effect of salt stress during the reproductive stage, this experiment was conducted at about 7–10 d before panicle initiation and continuing through harvest based on the method of Moradi and Ismail (2007). The experiment was carried out in a greenhouse under the natural light of a day/night air temperature in the range of about 25 ºCto 35 ºC and light intensity in the range of 600–1000 µmol/(m2∙s), using a randomized complete block design with three replications. Surface-sterilized seeds were soaked overnight and pre-germinated at 30 ºC for 48 h, then sown in 1 L pots filled with fertilized soil. The pots were kept in concrete tanks filled with tap water maintained at 3 cm below the soil surface. Two weeks after seeding, the water level was raised to about 1–2 cm above the soil surface. When seedlings were 28-day-old, water was siphoned and drained from the concrete tanks for 12 h. The tanks were then flooded with either tap water (control) or saline solution (NaCl), with EC of 5 dS/m for 3 d, then raised to 10 dS/m until harvest. EC of the saline solution was monitored regularly and adjusted when necessary using NaCl and tap water. Vegetative and reproductive parts were sampled during flowering stage for various measurements.
Measurements of sodium and potassium concentrations
The vegetative (the first, second and third leaves from the top and their corresponding leaf sheaths) and reproductive parts (main stalk and branches with spikelets) were harvested for the determination of Na+and K+concentrations. Harvesting was performed at flowering (61, 62, 61, 63, 61 and 64 d after seeding for Rc222, IR686, Sadri, CSR28, IR670 and Pokkali, respectively). Dried samples (20 mg) were placed in tubes containing 10 mL of 0.1 mol/L N-acetic acid and heated in a water bath at 85 ºC for 2 h. Extracted tissues were cooled at room temperature, left overnight and filtered using Whatman filter paper No. 1. Sodium and potassium concentrations were determined using an atomic absorption spectrometer (PerkinElmer, Massachusetts, USA).
Measurement of chlorophyll concentration
Freeze-dried leaves (the first, second and third leaves from the top) of each genotype were used for the determination of chlorophyll concentration. About 10 mg leaf materials were cut and placed in tubes for chlorophyll extraction using a ratio of 1.0 mg dry leaf material to 1 mL of 95% ethanol and left overnight in a dark cabinet. A total of 200 µL aliquot of extracted chlorophyll was loaded into microplate wells for spectral readings at 649 and 664 nm using a UV spectrophotometer (BMG Labtech, Ortenberg, Germany) with 95% ethanol as blank. Chlorophyll concentration was calculated following the formula of Lichtenthaler and Buschmann (2001).
Determination of lipid peroxidation
Lipid peroxidation was estimated by measuring the amount of MDA formation based on the method of Hodges et al (1999) with slight modifications. The first leaves and developing panicles were collected and kept at -80 ºC until measurements. Samples were ground to fine powder with liquid nitrogen, and a total of 0.2 g subsample was homogenized with 2 mL of 50 mmol/L potassium phosphate buffer (ice-cold, pH 7.0) and centrifuged at 22 000 ×at 4 ºCfor 30 min. Solutions (0.5 mL) with thiobarbituric acid (TBA) or with 50% trichloroacetic acid (TCA) and 0.01% butylated hydroxytoluene were added to a tube containing 0.5 mL plant extract. The solution with TBA has the same chemical constituents as the solution with TCA, except for plus 0.5% TBA. The reaction mixture was heated at 95 ºC for 25 min and cooled quickly on an ice bath, then centrifuged at 10 000×. The absorbance of the supernatant was determined at 440, 532 and 600 nm using a UV spectrophotometer (BMG Labtech, Ortenberg, Germany).
Determination of AsA concentration
AsA concentrations in leaves and panicles were analyzed based on the method of Shigeoka et al (1979) using subsamples from the same tissues for MDA measurements. Approximately 0.2 g fine tissue powder was homogenized with 6% TCA (ice-cold), and then centrifuged at 24 000 ×at 4ºCfor 20 min. The supernatant obtained was used for total AsA and oxidized AsA assays. Reduced AsA was calculated as the difference between total AsA and oxidized AsA. The concentration of AsA (µmol/g) was determined using a standard curve.
Measurement of proline concentration
Proline concentration was quantified following the modified method of Bates et al (1973). About 0.2 g fine tissue powder was homogenized in 1 mL of 3% sulfosalicylic acid and centrifuged at 5 000 ×for 10 min. A total of 200 µL aliquot was added to a reaction mixture containing glacial acetic acid and acidic ninhydrin. The mixture was incubated at 96 ºC for 60 min, and the reaction was then terminated quickly by placing the sample in an ice bath. Toluene was added to the cooled mixture and absorbance of the chromophore was read at 520 nm using a UV spectrophotometer (BMG Labtech, Ortenberg, Germany).
Assessment of pollen viability
To determine pollen viability, the potassium iodide method was used (Sarhadi et al, 2012). Panicles were collected randomly during heading stage (54, 56, 54, 55, 55 and 56 d after seeding for Rc222, IR686, Sadri, CSR28, IR670 and Pokkali, respectively). Samples were placed in vials with 70% ethanol and stored at 4 ºC. The spikelets were dissected to expose the anthers, which were then crushed thoroughly to release the pollen and stained with 1% I2-KI solution. Pollen was then mounted on slides and viewed under a microscope (Axioplan 2, Zeiss, Germany). Pollen grains stained black were considered viable, and those stained yellow or light colored were counted as sterile. Pollen viability was calculated by dividing the number of fertile pollen grains by the total number and presented as percentage.
Measurements at maturity stage
Aboveground biomass (shoot dry weight) and yield components including panicle length (cm), number of panicles per plant, and percentages of filled and unfilled grains were measured at the maturity stage (91, 95, 90, 96, 90 and 97 d after seeding for Rc222, IR686, Sadri, CSR28, IR670 and Pokkali, respectively). Five plants from each genotype in each replicate were harvested. Panicle length (cm) was measured from the neck node to the tip of the panicle. Panicles were then threshed to determine the number of filled and unfilled (partially filled and sterile spikelets) grains and oven-dried to reduce moisture content for measurement of grain yield per plant (14% moisture content). Shoots of individual plants were oven-dried at 70 ºC to constant weight to determine shoot dry weight (g).
Statistical analysis
Statistical analysis was performed for each parameter based on a randomized complete block design model with three replications using Statistical Tool for Agricultural Research. The Fisher’s least significance difference (LSD) was performed at the0.05 significance level to determine specific pairwise differences between means. Associations among parameters were examined using the Pearson correlation analysis.
RESULTS
Effects of salinity on yield components
Significant differences in shoot dry weight, grain yield and yield components were observed between normal and salt stress (Table 1). On average, salinity reduced shoot dry weight, pollen viability and grain yield by about 45%, 37% and 52%, respectively. Average shoot dry weight at maturity of the tolerant genotypes (CSR28, IR670 and Pokkali) decreased by 18%, whereas that of the sensitive genotypes (Rc222, IR686 and Sadri) decreased by 75%. Variation among genotypes was very high, ranging from 14% to 79%. Average pollen viability was severely affected by salinity, with an average reduction in salt-sensitive genotypes of 68%, but with much less reduction in the tolerant genotypes of only 7%. On average, salinity stress decreased grain yield by 52%, with a range of 14% to 83% among genotypes (Table 1). The highest reduction percentage in grain yield under saline condition was observed in Sadri, and the lowest was in IR670.
Significant effects on panicle length, number panicle per plant and percentage of filled grains were observed between salinity, genotypes and their interaction (Table 1). Panicles were shorter by about 28% under salt stress, with enormous variation among genotypes ranging from 13% to 40%. Large variation was also evident in genotype responses to salinity, ranging from a reduction of 12% in panicle length in CSR28 to 40% in Sadri. Similarly, higher percentage of filled grains was observed in tolerant genotypes with a decrease of 17%, compared to sensitive oneswith 85% reduction. Genotypic variation in percentage of filled grains ranges from a smaller decrease of about 7% (IR670) to a drastic reduction of up to 87% (Sadri). Salinity stress decreased percentage of filled grains in all genotypes, with more effects on sensitive ones.
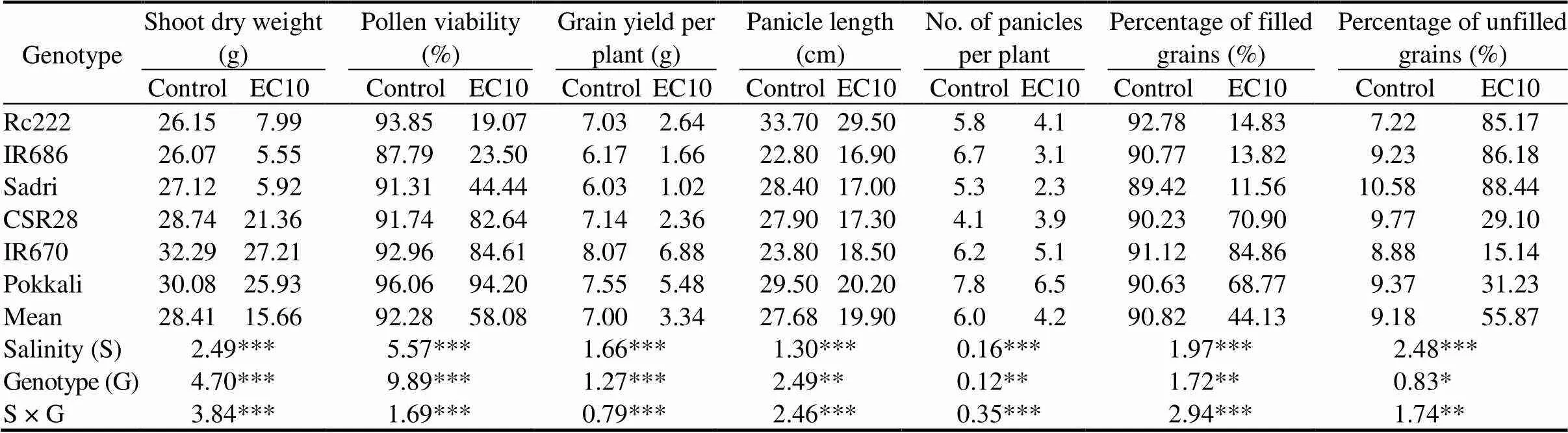
Table 1. Shoot dry weight, pollen viability, grain yield and yield components of six rice genotypes under control and salt stressconditions.
EC10, Salt stress condition with electrical conductivity of 10 dS/m.
Data are means of three replications; *, ** and ***,Significance at the 0.05, 0.01 and 0.001 levels by theFisher’s least significant difference, respectively.
Correlation coefficients among yield attributes and pollen viability at reproductive stage were analyzed. Pollen viability correlated positively with percentage of filled grains (= 0.79***) and grain yield (= 0.80***), but negatively with unfilled grains (= -0.67***) (data not shown).
Variation in Na+ concentration and Na+/K+ ratio in the vegetative and reproductive tissues
Significant changes in Na+concentration (Table 2) and Na+/K+ratio (Table 3) in both vegetative and reproductive parts were observed in salinity, genotype and their interaction. Sodium concentrations in vegetative and reproductive parts of the sensitive genotypes were significantly higher than those of the tolerant genotypes. Sodium accumulated to a much greater extent in panicle main stalks than in branches with spikelets, with higher accumulation in salt- sensitive genotypes. Na+/K+ratio was higher in panicle main stalk than in the branches with spikelets under salt stress (Table 3). In contrast, the gradient of potassium concentration was opposite to that of the sodium concentration. Due to the higher accumulation of sodium and reduction in potassium content, salt stress resulted in higher Na+/K+ratios in older leaves of the sensitive genotypes than in the tolerant genotypes. The salt-sensitive genotypes Rc222, IR686 and Sadri showed an increasing ratio with leaf age in both leaf blades and leaf sheaths, which was not observed in the salt-tolerant genotypes.
Total chlorophyll concentration at the flowering stage
Salinity decreased chlorophyll concentration in leaves. On average, total chlorophyll concentrations in the first leaf, the second leaf and third leaf from the top of the salt-sensitive genotypes decreased by 21%, 47% and 50%, respectively, under salt stress, and the tolerant ones showed relatively smaller corresponding reductions of 21%, 28% and 40% (Fig. 1). Genotypic differences as well as interactions with salinity were also significant. Substantial genotypic variation in total chlorophyll concentration of the first leaves was observed, ranging from a slight decrease of 10% in IR670 to greater reduction of 36% in Rc222.
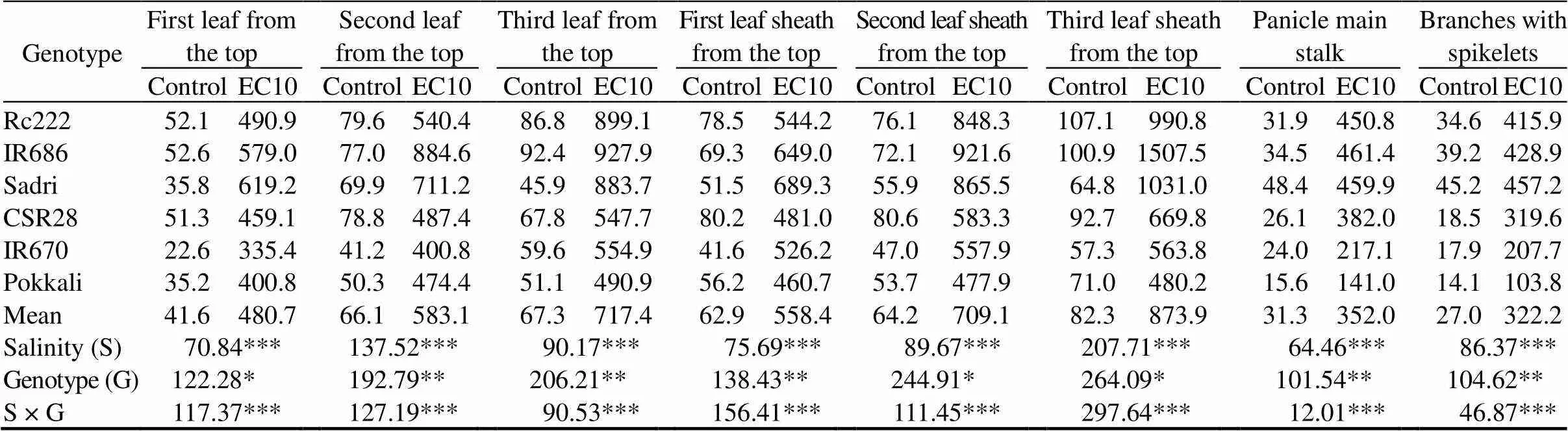
Table 2. Na+ concentrations of six rice genotypes at the flowering stage. mmol/kg
EC10, Salt stress condition with electrical conductivity of 10 dS/m.
Data are means of three replications; *, ** and ***,Significance at the 0.05, 0.01 and 0.001 levels by the Fisher’s least significant difference, respectively.
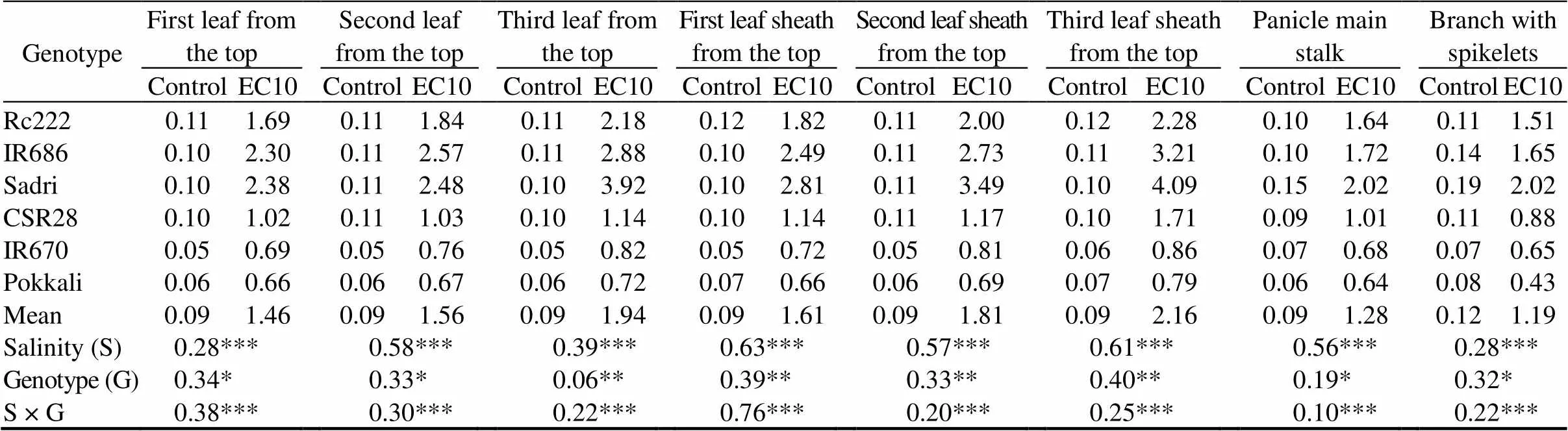
Table 3.Na+/K+ratio of six rice genotypes at the flowering stage. mmol/kg
EC10, Salt stress condition with electrical conductivity of 10 dS/m.
Data are means of three replications; *, ** and ***,Significance at the 0.05, 0.01 and 0.001 levels by the Fisher’s least significant difference, respectively.
Effects of salinity on MDA and AsA concentrations at the flowering stage
Salinity affected MDA and AsA concentrations to varying extent based on the genotypes (Fig. 2). On average, MDA concentration in the first leaf (Fig. 2-A) and developing panicles (Fig. 2-D) of the salt-sensitive genotypes increased by 32% and 150%, respectively. The salt-tolerant genotypes showed respective increases of only 95% and 50%. This indicates that the extent of lipid peroxidation in first leaves and developing panicles of sensitive genotypes was significantly higher under salt stress than those of tolerant genotypes. Significant differences were observed due to genotypes or their interactions with salinity. Reduced AsA concentrations in first leaves (Fig. 2-B) and developing panicles (Fig. 2-E) of salt-sensitive genotypes decreased by about 92% and 95%, respectively, with lower corresponding reductions in tolerant genotypes of 54% and 64%. A strong negative correlation was observed between MDA and total AsA (= -0.62**) and between MDA and reduced AsA (= -0.61**) measured in the first leaves under salt stress. However, correlations of MDA in developing panicles was slightly weaker and negative with the total AsA (= -0.54*) and reduced AsA (= -0.54*) under salt stress.
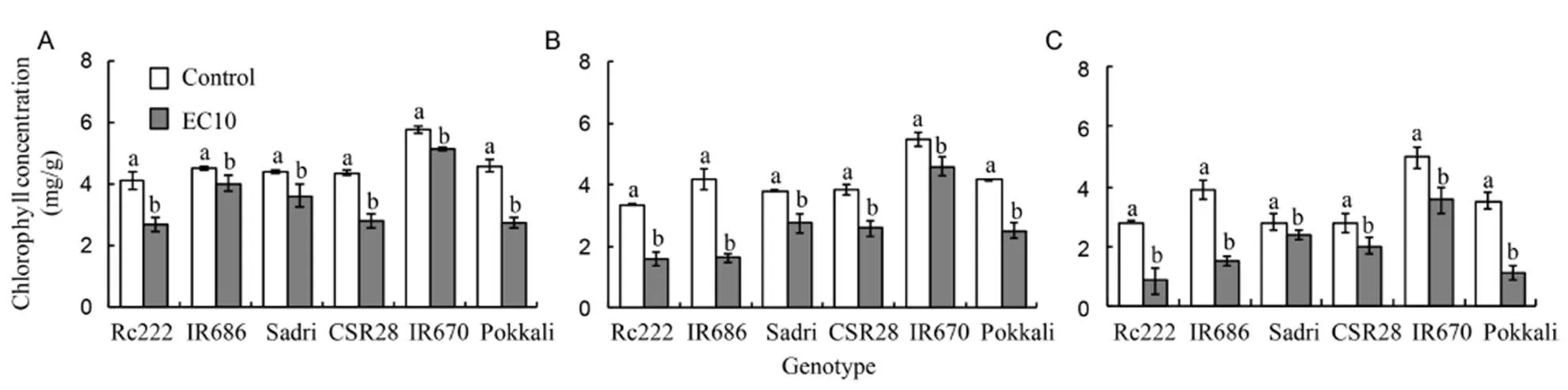
Fig. 1. Chlorophyll concentrations of the first (A), second (B) and third leaves (C) from the top in six rice genotypes at the flowering stage.
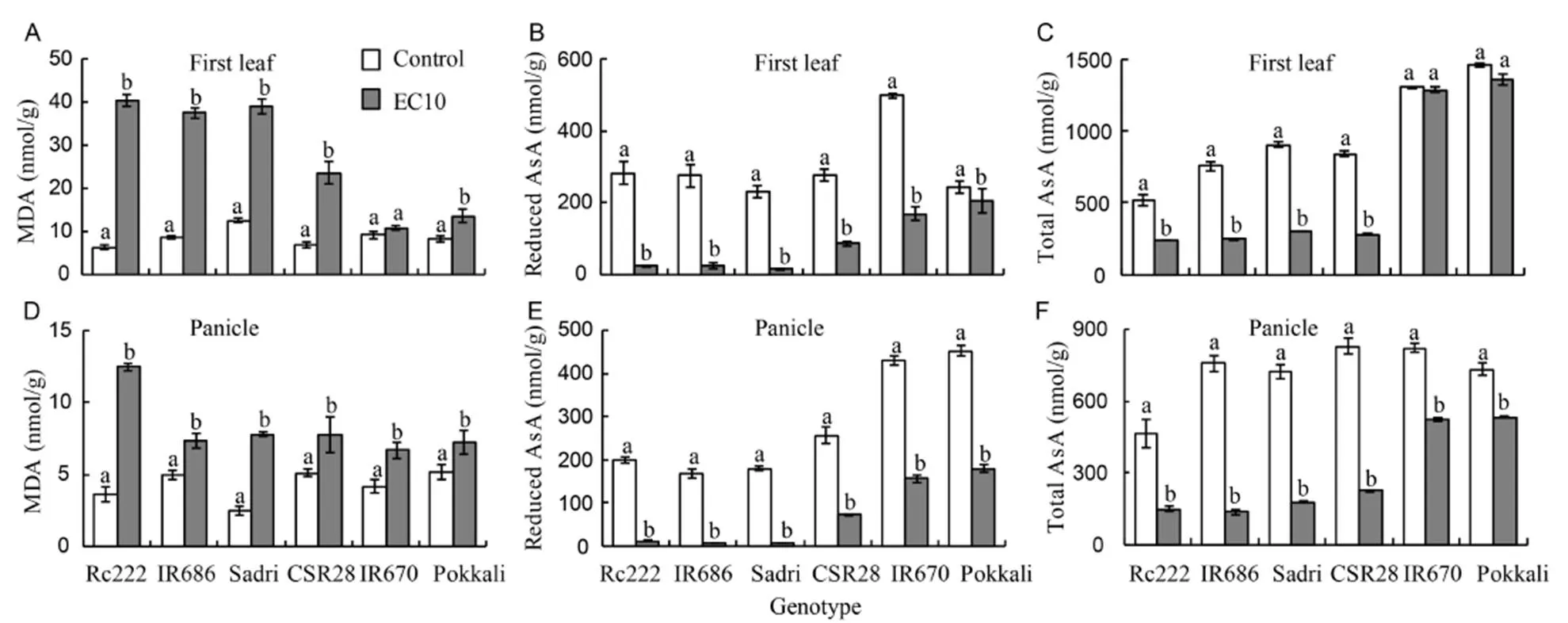
Fig. 2. Malondiadehyde (MDA) and ascorbate (AsA) concentrations in six rice genotypes at the flowering stage.
A, MDA in the first leaf. B, Reduced AsA in the first leaf. C, Total AsA in the first leaf. D, MDA in the developing panicle. E, Reduced AsA in the developing panicle. F, Total AsA in the developing panicle.
EC10, Salt stress condition with electrical conductivity of 10 dS/m.
Data are Mean ± SE (= 3). Within groups, the same lowercase letter(s) indicate no significant difference at< 0.05 by the Fisher’s least significant difference.
Proline accumulation at flowering stage
Proline concentrations were measured in the first leaves and developing panicles at the flowering stage to determine their association with salinity tolerance. Salt stress substantially increased proline concentrations in the first leaves and developing panicles, by about 36 and 3 times, respectively, across all genotypes. Its concentrations in the first leaves and developing panicles of salt-sensitive genotypes increased on average, by about 16 and 2 times, respectively, but the increase was much higher in tolerant genotypes, by about 35 and 2 times, respectively. Two of the tolerant genotypes IR670 and Pokkali showed much higher increase in proline concentration in their first leaves than in the panicles, however, this effect was not apparent in the tolerant genotype CSR28 (Fig. 3).
DISCUSSION
Effects of salinity on rice yield and yield components
Salt stress significantly reduced growth, yield and yield components (Tables 1 and 2), but the varying extents depended on the genotype. Salt stress negatively affected several aspects of yield components, especially the percentage of filled grains and pollen viability, leading to significant reduction in grain yield. Several studies on cereals and other commercially important crops reported genotypic differences in response to salinity stress (James et al, 2002; Oyiga et al, 2016, 2017). The increase in the percentage of unfilled grains in Rc222 and IR686 may be due to the reduction in pollen viability and limited carbohydrate translocation to the panicles (Abdullah et al, 2001), or in part due, to the higher accumulation of sodium in floral parts. Moreover, reduction in the percentage of filled grains may also be attributed to competition for carbohydrate supply between vegetative growth and developing panicles and also among spikelets within panicle (Zeng and Shannon, 2000), because the source capacity decreases under salt stress. Rao et al (2008) reported similar decrease in grain yield in 25 rice genotypes due to salt stress, which could be attributed to a combination of factors, including reduced photosynthetic carbon assimilation (Moradi and Ismail, 2007) and reduced assimilate supply to developing grains (Yang et al, 2000), resulting in reduced percentage of grain filling and high sterility (Abdullah et al, 2001). Salinity also affects rice phenology to varying extent based on genotypes. In this study, rice genotypes were observed to have accelerated maturity. The heading dates were earlier (54–56 d from seeding). This can be attributed to shorter period of exposure to salinity, and hence, lesser accumulation of salt. Early- maturing genotypes may avoid damage if they pass through the most critical stages when salt accumulation is sufficiently high and seriously affects photosynthesis and reduces the fertility and yield. However, the observed delay of reproductive development (days from heading to maturity) which is 36–40 d maybe due to a reduction in growth caused by osmotic stress and reduced photosynthesis and carbohydrate supply under salt stress. This delay of growth duration is within the range of salinity-induced delay in reproductive development (Moradi et al, 2003). The greater damage caused by salinity stress during the later period of reproductive stage may have contributed on the observed delay of reproductive stage. Moreover, a salinity-induced delay has been reported (Khatun et al, 1995; Ansari et al, 2001). This genetic variability in the extent of the delay in reproductive development under salt stress could be useful for breeding.
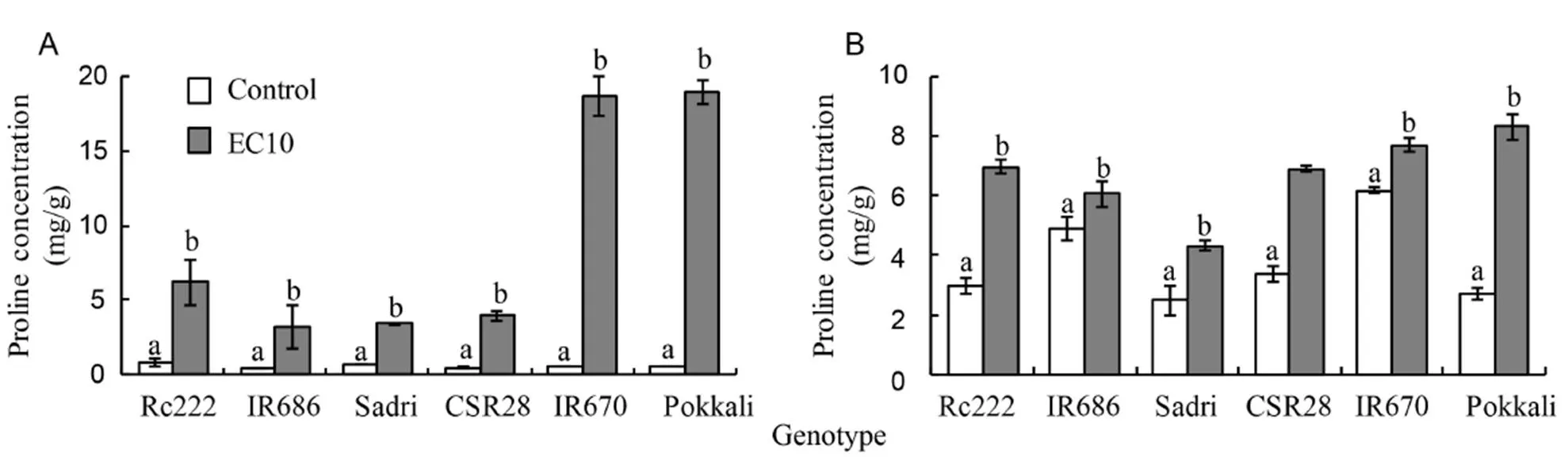
Fig. 3. Proline concentration in the first leaves (A) and panicles (B) of six rice genotypes at the flowering stage.
Physiological changes at reproductive stage
Reduction in photosynthetic carbon assimilation could also be attributed to the observed reduction in total chlorophyll concentration across genotypes under salt stress at the flowering stage (Fig. 1). In sorghum, genotypic differences in the extent of chlorophyll content retention were observed greater reduction as soil salinity increases when exposed to salt stress with EC of 4, 8 and 12 dS/m (Kumari et al, 2016). This reduction in chlorophyll concentration was also reported in several other plant species, including mustard (Mittal et al, 2012), sugarcane (Cha-um et al, 2012), rice (Saeedipour, 2014a), wheat (Zhu et al, 2016), cabbage (Sanoubar et al, 2016) and bean (Taïbi et al, 2016). The reduction in chlorophyll concentration could probably be one of the reasons for the reduction in photosynthesis according to the previous reports (Dionisio-Sese and Tobita, 2000; Moradi and Ismail, 2007; Morales et al, 2012). Accumulation of sodium to toxic levels in salt sensitive genotypes was reported to have several deleterious effects including disruption of membrane stability, causing detachment of plasma membrane from the cell wall (Mitsuya et al, 2002), altered orientation of the grana, swelling of thylakoids and distortion of grana lamellae (Zahra et al, 2014). The ability of the salt-tolerant genotypes to retain their chlorophyll, particularly in upper leaves, is important mechanism to maintain photosynthetic capacity even under salt stress.
The genotypes used in this study showed significant genetic variation in Na+concentration and Na+/K+ratio in both vegetative and reproductive parts under salt stress, with higher concentrations in older tissues than in younger ones. On average, the salt-sensitive genotypes and the moderately tolerant Sadri showed significant increase in Na+/K+ratio primarily due to higher Na+accumulation than the tolerant genotypes. The data agree with the view that there is a strong association between salt exclusion and salt tolerance (Munns et al, 2006; Platten et al, 2013). In durum wheat, genotypic differences in Na+transport depend on variation in the capacity of the leaf sheath to extract and sequester Na+as it entered the leaf (Davenport, 2005). The salt tolerant genotypes have higher Na+exclusion capacity than the sensitive ones, and greater sequestration ability excluding salts from entering active leaves. Responses of these salt-tolerant genotypes to salt stress could be attributed to better Na+efflux from roots to the rhizosphere through the well-recognized SOS1-dependent exclusion system, Na+sequestration in vacuoles is regulated by Na+/K+antiporters, and Na+loading and unloading at the xylem (Hasegawa et al, 2000; Ismail and Horie, 2017; Saddiq et al, 2017). Several studies concluded that the main mechanism preventing Na+accumulation in leaves involves the combined action of transporters mediating Na+unloading from the root and xylem, minimizing the transfer and accumulation of physiologically toxic Na+in shoots, and eventually in photosynthetic tissues, mediated by the high-affinity K+transporter (HKT) proteins (Hauser and Horie, 2010; Suzuki et al, 2016). It can also be partly due to the ability of the salt-tolerant genotypes to either maintain K+content or even increase its K+concentration in the presence of salt (Dionisio-Sese and Tobita, 2000). In addition, K+efflux systems have been of major interest because of their deep relevance for K+retention during salt stress. Numerous studies on barley and wheat varieties highlight the importance of K+retention ability (i.e., lower K+efflux activity) under salt stress. Strong correlation between higher K+content in leaf mesophyll and salt tolerance suggests that K+retention in photosynthetic cells can greatly contribute in maintaining low cytosolic Na+/K+ratios and thus increase in salt tolerance (Shabala and Cuin, 2008; Wu et al, 2014, 2015). The distinct susceptibilities of rice genotypes to vegetative and reproductive tissues’ Na+accumulation resulting in varying Na+/K+ratio may have brought about the differential responses in total chlorophyll concentration, lipid peroxidation and yield components. However, the other possibility that the varying salt movement and growth stage duration under salt stress is a consequence of salt stress effect on rice phenology (Moradi et al, 2003) cannot be entirely ruled out.
Previous studies showed that spikelet development is extremely sensitive to changes in sodium concentration (Asch et al, 1999; Saeedipour, 2014b). Two different pathways of uptake into the panicle exist for sodium, one independent of transpiration and the other driven by panicle transpiration (Asch et al, 1999). In wheat and barley, the control of salt transport to the reproductive apex is exerted through loading of the phloem (Munns and Rawson, 1999). Munns et al (1986) had shown that the phloem in salt-stressed barley excludes Na+and Cl-while keeping K+concentrations higher. In this study, Na+/K+ratios in the reproductive stage of rice genotypes were lower than those in vegetative stage under salt stress. These results are in line with the report of Abdullah et al (2001) that sodium accumulation in rice genotypes decreases progressively while moving up the plant towards the reproductive apex, with K+concentration gradient opposite to Na+concentration. The ability of the salt-tolerant genotypes to exhibit lower Na+/K+ratios in the reproductive parts is an important trait contributing to salinity tolerance, especially during pollen development, pollination and seed formation under salt stress in comparison with that of the salt-sensitive ones.
MDA production in leaves and developing panicles increased under salt stress, with significant variability among genotypes as reflected by the significant interaction. Salt-sensitive genotypes showed higher MDA concentration in leaves and developing panicles compared to the tolerant genotypes. The observed lower MDA concentration in leaves and developing panicles of salt-tolerant genotypes is probably because of the ability to maintain higher concentrations of antioxidants to scavenge ROS generated during stress (Fig. 2-A and -D). This improved protection is reflected in the higher concentration of reduced AsA (Fig. 2-B and -E). The accumulation of excessive ROS is expected under salt stress due to the depletion of oxidized NADP+, which acts as the final acceptor of electrons in PSI, consequently increasing the leakage of electrons to O2forming ROS (Abogadallah, 2010). When ROS generation exceed the capacity of the plants to scavenge them, lipid peroxidation in biological membranes increases, causing serious damageto organelles like mitochondria, chloroplasts and plasma membranes. MDA production paralleled changes in Na+/K+ratios by exhibiting higher MDA concentration in leaves of salt-sensitive genotypes (Fig. 2-A). Rice genotypes with higher concentrations of antioxidants have been reported to have greater tolerance of salt stress (Dionisio-Sese and Tobita, 1998; Moradi and Ismail, 2007). AsA is an important metabolite with a vital role as an antioxidant. It can directly scavenge ROS through enzymes of the ascorbate-glutathione cycle involving ascorbate peroxidase, or non- enzymatically by reducing H2O2directly to water (Evans et al, 2016). Apart from its role as an antioxidant, AsA also acts as an electron donor for violaxanthin-epoxidase, an enzyme involved in the xanthophyll cycle to dissipate excess excitation energy (Miyaji et al, 2015). Moreover, AsA influences important enzymatic reactions as it is a co-factor of many metal-containing enzymes (Parida and Das, 2005; Bielen et al, 2013; Gupta and Huang, 2014). The current data clearly support the important role of AsA in reducing membrane damage during salinity stress.
Another well-known adaptive mechanism of most plants to cope with salinity stress is the accumulation of compatible osmolytes such as proline. The accumulation of proline in the first leaves and developing panicles of the six genotypes was assessed at flowering to investigate its association with tolerance. Salt stress significantly increased the proline concentrations in first leaves and developing panicles of all genotypes, with pronounced increase in the salt-tolerant genotypes, IR670 and Pokkali, but not in CSR28 (Fig. 3). It has been widely-reported that proline has a protective role against NaCl-induced oxidative damage in several plant species (Biancucci et al, 2015; Bhusan et al, 2016). Proline is known to be important in protecting subcellular structures and mediating osmotic adjustment under stress conditions (Rao et al, 2013). Furthermore, proline has multifunctional roles including protection against oxidative damage (Hoque et al, 2008), acting as a signaling molecule for plant recovery from stress (Szabados and Savouré, 2010), and in stabilizing proteins (Bozorgmehr and Monhemi, 2015). Moreover, studies on proline metabolism suggest that its biosynthesis is activated during dehydration, whereas rehydration triggers the opposite response (Szabados and Savouré, 2010). Our findings are consistent with a previous report, which indicated that seedlings of tolerant genotypes produce more proline under salt stress (Ghosh et al, 2011). In addition, recent studies using exogenous proline showed that it significantly improves tolerance to salt stress in salt-sensitive rice by maintaining lower Na+/K+ratio and enhancing indigenous proline concentration and antioxidant defense systems (Hasanuzzaman et al, 2012; Bhusan et al, 2016). More importantly, proline has been implicated to have a vital role in pollen development, since it is the most abundant amino acid in the tapetum layer supplying nutrients during microsporogenesis (Mariani et al, 1990; Paupière et al, 2014; Biancucci et al, 2015). A recent study using a proline-deficient mutant ofrevealed that male gametophytes carrying the mutation are severely compromised, indicating that proline is required for pollen development and transmission (Mattioli et al, 2012). Given these roles of proline in pollen development, the observed accumulation of proline in developing panicles may be due to its role in stabilizing proteins (Bozorgmehr and Monhemi, 2015; Schmidt et al, 2016). However, the role of proline in mediating salt stress could be genotype-specific, as shown by the low proline concentration in the first leaves of CSR28, which is tolerant to alkalinity. Moradi and Ismail (2007) also observed an increase in proline concentration in sensitive genotypes in response to salt stress at the vegetative stage. More studies are probably needed to establish the role of proline upregulation in salt stress across genotypes and stages of development.
Kanawapee et al (2013) observed significant increase in chlorophyll and MDA concentrations, but a maintained concentration of proline. However, Kibria et al (2017) reported significant decrease in chlorophyll content, but an increase in proline content with increasing salt concentration. It has been well established that salt stress at the reproductive stage has substantial effect on grain yield. In this study, all rice genotypes showed reduction in pollen viability under salt stress, but to a greater extent in salt-sensitive genotypes. This might be due to the ability of the tolerant genotypes to exclude sodium more effectively at the root level or partitioning it in leaf sheaths and older leaves as reflected by the lower Na+/K+ratio in the reproductive parts (Table 3).
CONCLUSIONs
Salinity induced substantial physiological changes in the vegetative and reproductive tissues of rice during reproductive stage, including higher Na+/K+ratio in both vegetative and reproductive parts, coupled with higher concentration of MDA, less reduced AsA and lesser ability to accumulate proline in first leaves and developing panicles. The greater tolerance of the landrace Pokkali and the breeding line IR670 seemed to be associated with greater capability to exclude Na+, as reflected in lower Na+/K+ratio, higher concentrations of reduced AsA and lower concentrations of MDA under salt stress. This study also showed that greater proline accumulation in the developing panicles of IR670 and Pokkali may contribute to tolerance specifically for pollen development under salt stress. Interestingly, Sadri, which was previously reported as moderately tolerant genotypes at the seedling stage, showed similar responses to that of the salt-sensitive ones at the reproductive stage. The study also highlighted that some of the traits essential for tolerance at the seedling stage are also important during reproduction, however, there are additional traits, e.g. associated with pollen grain viability and seed-setting rate, are important during reproductive stage. These variations in the major traits associated with tolerance during the reproductive stage in rice could be exploited for breeding tolerant genotypes.
Acknowledgements
We thank the International Rice Research Institute, DOST-ASTHRDP and University of the Philippines Visayas for providing support to the first author. And we also thank R. Platten and R. Eugenio for technical assistance during the study.
Abdullah Z, Khan M A, Flowers T J. 2001. Causes of sterility in seed set of rice under salinity stress., 187(1): 25–32.
Abogadallah G M. 2010. Insights into the significance of antioxidative defense under salt stress., 5(4): 369–374.
Adem G D, Roy S J, Zhou M, Bowman J P, Shabala S. 2014. Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley., 14: 113.
Ansari R, Shereen A, Flowers T J. 2001. Identifying rice lines for improved salt tolerance from a mapping population:: Peng S, Hardy B. Rice Research for Food Security and Poverty Alleviation. Los Baños, the Philippines: International Rice Research Institute.
Asch F, Dingkuhn M, Wittstock C, Doerffling K. 1999. Sodium and potassium uptake of rice panicles as affected by salinity and season in relation to yield and yield components., 207: 133–145.
Bates L S, Waldren R P, Teare I D. 1973. Rapid determination of free proline for water-stress studies., 39(1): 205–207.
Bhusan D, Das D K, Hossain M, Murata Y, Hoque M A. 2016. Improvement of salt tolerance in rice (L.) by increasing antioxidant defense systems using exogenous application of proline., 10(1): 50–56.
Biancucci M, Mattioli R, Forlani G, Funck D, Costantino P, Trovato M. 2015. Role of proline and GABA in sexual reproduction of angiosperms., 6: 680.
Bielen A, Remans T, Vangronsveld J, Cuypers A. 2013. The influence of metal stress on the availability and redox state of ascorbate, and possible interference with its cellular functions., 14(3): 6382–6413.
Bozorgmehr M R, Monhemi H. 2015. How can a free amino acid stabilize a protein? Insights from molecular dynamics simulation., 44(1): 45–53.
Cha-um S, Chuencharoen S, Mongkolsiriwatana C, Ashraf M, Kirdmanee C. 2012. Screening sugarcane (sp.) genotypes for salt tolerance using multivariate cluster analysis., 110(1): 23–33.
Chunthaburee S, Dongsansuk A, Sanitchon J, Pattanagul W, Theerakulpisut P. 2016. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt tolerance at seedling stage., 23(4): 467–477.
Davenport R, James R A, Zakrisson-Plogander A, Tester M, Munns R. 2005. Control of sodium transport in durum wheat., 137(3): 807–818.
Dionisio-Sese M L, Tobita S. 1998. Antioxidant responses of rice seedlings to salinity stress., 135(1): 1–9.
Dionisio-Sese M L, Tobita S. 2000. Effects of salinity on sodium content and photosynthetic responses of rice seedlings differing in salt tolerance., 157(1): 54–58.
Evans M J, Choi W G, Gilroy S, Morris R J. 2016. A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress., 171(3): 1771–1784.
Genc Y, McDonald G K, Tester M. 2007. Reassessment of tissue Na+concentration as a criterion for salinity tolerance in bread wheat., 30(11): 1486–1498.
Ghosh N, Adak M K, Ghosh P D, Gupta S, Sen Gupta D N, Mandal C. 2011. Differential responses of two rice varieties to salt stress., 5(1): 89–103.
Gupta B, Huang B. 2014. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization., 2014(1): 701596.
Hakim M A, Juraimi A S, Begum M, Hanafi M M, Ismail M R, Selamat A. 2010. Effect of salt stress on germination and early seedling growth of rice (L.)., 9(13): 1911–1918.
Hasanuzzaman M, Hossain M A, da Silva J A T, Fujita M. 2012. Plant response and tolerance to abiotic oxidative stress: Antioxidant defense is a key factor.: Asif M, Kamran A. Crop Stress and Its Management: Perspectives and Strategies. Springer Netherlands: 261–315.
Hasegawa P M, Bressan R A, Zhu J K, Bohnert H J. 2000. Plant cellular and molecular responses to high salinity., 51(1): 463–499.
Hauser F, Horie T. 2010. A conserved primary salt tolerance mechanism mediated by HKT transporters: A mechanism for sodium exclusion and maintenance of high K+/Na+ratio in leaves during salinity stress., 33(4): 552–565.
Hodges D M, DeLong J M, Forney C F, Prange R K. 1999. Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds., 207(4): 604–611.
Höller S, Ueda Y, Wu L, Wang Y, Hajirezaei M R, Ghaffari M R, von Wirèn N, Frei M. 2015. Ascorbate biosynthesis and its involvement in stress tolerance and plant development in rice (L.)., 88(6): 545–560.
Hoque M A, Banu M N A, Nakamura Y, Shimoishi Y, Murata Y. 2008. Proline and glycinebetaine enhance antioxidant defense and methylglyoxal detoxification systems and reduce NaCl-induced damage in cultured tobacco cells., 165(8): 813–824.
Ismail A M, Heuer S, Thomson M J, Wissuwa M. 2007. Genetic and genomic approaches to develop rice germplasm for problem soils., 65(4): 547–570.
Ismail A M, Horie T. 2017. Genomics, physiology, and molecular breeding approaches for improving salt tolerance., 68(1): 405–434.
James R A, Rivelli A R, Munns R, von Caemmerer S. 2002. Factors affecting CO2assimilation, leaf injury and growth in salt-stressed durum wheat., 29(12): 1393–1403.
James R A, Blake C, Byrt C S, Munns R. 2011. Major genes for Na+exclusion,and(wheatand), decrease Na+accumulation in bread wheat leaves under saline and waterlogged conditions., 62(8): 2939–2947.
Kanawapee N, Sanitchon J, Srihaban P, Theerakulpisut P. 2013. Physiological changes during development of rice (L.) varieties differing in salt tolerance under saline field condition., 370: 89–101.
Kibria M G, Hossain M, Murata Y, Hoque M A. 2017. Antioxidant defense mechanisms of salinity tolerance in rice genotype., 24(3): 155–162.
Khatun S, Rizzo C A, Flowers T J. 1995. Genotypic variation in the effect of salinity on fertility in rice., 173(2): 239–250.
Krishnamurthy S L, Sharma S K, Kumar V, Tiwari S, Singh N K. 2015. Analysis of genomic region spanning Saltol using SSR markers in rice genotypes showing differential seedlings stage salt tolerance., 25(3): 331–336.
Kumari P, Arya S, Pahuja S K, Joshi U N, Sharma S K. 2016. Evaluation of forage sorghum genotypes for chlorophyll content under salt stress., 5(3): 1200–1207.
Lichtenthaler H K, Buschmann C. 2001. Chlorophylls and carotenoids: Measurement and characterization by UV-VIS spectroscopy.: Cornforth D P. Current Protocols in Food Analytical Chemistry. New York, USA: John Wiley & Sons: 431–438.
Mariani C, Beuckeleer M D, Truettner J, Leemans J, Goldberg R B. 1990. Induction of male sterility in plants by a chimaeric ribonuclease gene., 347: 737–741.
Mattioli R, Biancucci M, Lonoce C, Costantino P, Trovato M. 2012. Proline is required for male gametophyte development in., 12: 236.
Mitsuya S, Yano K, Kawasaki M, Taniguchi M, Miyak H. 2002. Relationship between the distribution of Na+and the damages caused by salinity in the leaves of rice seedlings grown under a saline condition., 5(4): 269–274.
Mittal S, Kumari N, Sharma V. 2012. Differential response of salt stress on: Photosynthetic performance, pigment, proline, D1 and antioxidant enzymes., 54: 17–26.
Miyaji T, Kuromori T, Takeuchi Y, Yamaji N, Yokosho K, Shimazawa A, Sugimoto E, Omote H, Ma J F, Shinozaki K, Moriyama Y. 2015.is a chloroplast-localized ascorbate transporter in., 6: 5928.
Mohammadi-Nejad G, Arzani A, Rezai A M, Singh R K, Gregorio G B. 2008. Assessment of rice genotypes for salt tolerance using microsatellite markers associated with the saltol QTL., 7(6): 730–736.
Moradi F, Ismail A M, Gregorio G B, Egdane J A. 2003. Salinity tolerance of rice during reproductive development and association with tolerance at the seedling stage., 8: 105–116.
Moradi F, Ismail A M. 2007. Responses of photosynthesis, chlorophyll fluorescence and ROS-scavenging systems to salt stress during seedling and reproductive stages in rice., 99(6): 1161–1173.
Morales S G, Trejo-Téllez L I, Gómez Merino F C, Caldana C, Espinosa-Victoria D, Herrera Cabrera B E. 2012. Growth, photosynthetic activity, and potassium and sodium concentration in rice plants under salt stress., 34(3): 317–324.
Munns R, Fisher D B, Tonnet M L. 1986. Na+and Cl-transport in the phloem from leaves of NaCl-treated barley., 13(6): 757–766.
Munns R, James R A, Lauchli A. 2006. Approaches to increasing the salt tolerance of wheat and other cereals., 57(5): 1025–1043.
Munns R, Rawson H M. 1999. Effect of salinity on salt accumulation and reproductive development in the apical meristem of wheat and barley., 26(5): 459–464.
Munns R, Tester M. 2008. Mechanisms of salinity tolerance., 59: 651–681.
Oyiga B C, Sharma R C, Shen J, Baum M, Ogbonnaya F C, Léon J, Ballvora A. 2016. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach., 202(6): 472–485.
Oyiga B C, Sharma R C, Baum M, Ogbonnaya F C, Léon J, Ballvora A. 2017. Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat: GWAS reveals gene loci for salt tolerance in wheat., 41(1): 1–17.
Palao C D C, de La Viña C B, Gregorio G B, Singh R K. 2013. A new phenotyping technique for salinity tolerance at the reproductive stage in rice., 50(3): 199–207.
Parida A K, Das A B. 2005. Salt tolerance and salinity effects on plants: A review., 60(3): 324–349.
Paupière M J, van Heusden A W, Bovy A G. 2014. The metabolic basis of pollen thermo-tolerance: Perspectives for breeding., 4(4): 889–920.
Platten J D, Egdane J A, Ismail A M. 2013. Salinity tolerance, Na+exclusion and allele mining of HKT1;5 inand; many sources, many genes, one mechanism?, 13: 32.
Qureshi A S, Al-Falahi A A. 2015. Extent, characterization and causes of soil salinity in central and southern Iraq and possible reclamation strategies., 5(1): 84–94.
Rao P S, Mishra B, Gupta S R, Rathore A. 2008. Reproductive stage tolerance to salinity and alkalinity stresses in rice genotypes., 127(3): 256–261.
Rao P S, Mishra B, Gupta S R. 2013. Effects of soil salinity and alkalinity on grain quality of tolerant, semi-tolerant and sensitive rice genotypes., 20(4): 284–291.
Ravikiran K T, Krishnamurthy S L, Warraich A S, Sharma P C. 2018. Diversity and haplotypes of genotypes for seedling stage tolerance analyzed through morpho-physiological and SSR markers., 220: 10–18.
Roy S J, Negrão S, Tester M. 2014. Salt resistant crop plants., 26: 115–124.
Saddiq M S, Afzal I, Basra S M A, Ali Z, Ibrahim A M H. 2017. Sodium exclusion is a reliable trait for the improvement of salinity tolerance in bread wheat., 64(2): 272–284.
Saeedipour S. 2014a. Effects of salinity stress on growth, chlorophyll content and ion accumulation in tworice (L.) cultivars differing in salinity tolerance., 4(4): 33–40.
Saeedipour S. 2014b. The effect of salinity stress on ions distribution in panicle, flag leaf and leaf sheaths of two rice (L.) genotypes differing in salt tolerance., 4(10): 269–275.
Sanoubar R, Cellini A, Veroni A M, Spinelli F, Masia A, Vittori Antisari L, Orsini F, Gianquinto G. 2016. Salinity thresholds and genotypic variability of cabbage (L.) grown under saline stress., 96(1): 319–330.
Sarhadi E, Bazargani M M, Sajise A G, Abdolahi S, Vispo N A, Arceta M, Nejad G M, Singh R K, Salekdeh G H. 2012. Proteomic analysis of rice anthers under salt stress., 58: 280–287.
Schmidt T, Situ A J, Ulmer T S. 2016. Structural and thermodynamic basis of proline-induced transmembrane complex stabilization., 6: 29809.
Shabala S, Cuin T A. 2008. Potassium transport and plant salt tolerance., 133(4): 651–669.
Shigeoka S, Yokota A, Nakano Y, Kitaoka S. 1979. The effect of illumination on theascorbic acid content in., 43(10): 2053–2058.
Singh R K, Flowers T J. 2010. The physiology and molecular biology of the effects of salinity on rice.: Pessarakli M. Handbook of Plant and Crop Stress. Florida: Taylor & Francis: 899–939.
Suzuki K, Yamaji N, Costa A, Okuma E, Kobayashi N I, Kashiwagi T, Katsuhara M, Wang C, Tanoi K, Murata Y, Schroeder J I, Ma J F, Horie T. 2016. OsHKT1;4-mediated Na+transport in stems contributes to Na+exclusion from leaf blades of rice at the reproductive growth stage upon salt stress., 16: 22.
Szabados L, Savouré A. 2010. Proline: A multifunctional amino acid., 15(2): 89–97.
Taïbi K, Taïbi F, Abderrahim L A, Ennajah A, Belkhodja M, Mulet J M. 2016. Effect of salt stress on growth, chlorophyll content, lipid peroxidation and antioxidant defence systems inL., 105: 306–312.
Wankhade S D, Sanz A. 2013. Chronic mild salinity affects source leaves physiology and productivity parameters of rice plants (L., cv. Taipei 309)., 367: 663–672.
Wassmann R, Hien N X, Hoanh C T, Tuong T P. 2004. Sea level rise affecting theDelta: Water elevation in the flood season and implications for rice production., 66: 89–107.
Wu H H, Shabala L, Zhou M X, Shabala S. 2014. Durum and bread wheat differ in their ability to retain potassium in leaf mesophyll: Implications for salinity stress tolerance., 55(10): 1749–1762.
Wu H H, Zhu M, Shabala L, Zhou M X, Shabala S. 2015. K+retention in leaf mesophyll, an overlooked component of salinity tolerance mechanism: A case study for barley., 57(2): 171–185.
Yang J C, Peng S B, Visperas R M, Sanico A L, Zhu Q S, Gu S L. 2000. Grain filling pattern and cytokinin content in the grains and roots of rice plants., 30(3): 261–270.
Zahra J, Nazim H, Cai S G, Han Y, Wu D Z, Zhang B L, Haider S I, Zhang G P. 2014. The influence of salinity on cell ultrastructures and photosynthetic apparatus of barley genotypes differing in salt stress tolerance., 36(5): 1261–1269.
Zeng L, Shannon M C. 2000. Salinity effects on seedling growth and yield components of rice., 40(4): 996–1003.
Zhu M, Shabala S, Shabala L, Fan Y, Zhou M X. 2016. Evaluating predictive values of various physiological indices for salinity stress tolerance in wheat., 202(2): 115–124.
11 May 2018;
29 October 2018
Maria Elisa B. Gerona (mbgerona@up.edu.ph); Abdelbagi M. Ismail (a.ismail@irri.org)
Copyright © 2019, China National Rice Research Institute. Hosting by Elsevier B V
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/)
Peer review under responsibility of China National Rice Research Institute
http://dx.doi.org/10.1016/j.rsci.2019.05.001
(Managing Editor: Wang Caihong)
- Rice Science的其它文章
- Ustilaginoidea virens: A Fungus Infects Rice Flower and Threats World Rice Production
- Photocatalytic Treatment of Waste Water from Rice Husk Alkaline Hydrolysate
- Fine-Mapping of qTGW1.2a, a Quantitative Trait Locus for 1000-Grain Weight in Rice
- Screening for Spikelet Fertility and Validation of Heat Tolerance in a Large Rice Mutant Population
- Assessment of Genetic Diversity of Drought Tolerant and Susceptible Rice Genotypes Using Microsatellite Markers
- Comparison of Five Endogenous Reference Genes for Specific PCR Detection and Quantification of Rice

