Duplication and diversif ication of insulin genes in ray-f inned f ish
David M.Irwin
1 Department of Laboratory Medicine and Pathobiology,University of Toronto,Toronto Ontario M5S 1A8,Canada
2 Banting and Best Diabetes Centre,University of Toronto,Toronto Ontario M5S 1A8,Canada
ABSTRACT Insulin is a key hormone for the regulation of metabolism in vertebrates.Insulin is produced by pancreatic islet cells in response to elevated glucose levels and leads to the uptake of glucose by tissues such as liver and adipose tissue to store energy.Insulin also has additional functions in regulating development.Previous work has shown that the proglucagon gene,which encodes hormones counter regulating insulin,is duplicated in teleost f ish,and that the peptide hormones encoded by these genes have diversif ied in function.Isought to determine whether similar processes have occurred to insulin genes in these species.Searches of f ish genomes revealed an unexpected diversity of insulin genes.A triplication of the insulin gene occurred at the origin of teleost f ish,however one of these three genes,insc,has been lost in most teleost f ish lineages.The two other insulin genes,insa and insb,have been retained but show differing levels of selective constraint suggesting that they might have diversif ied in function.Intriguingly,a duplicate copy of the insa gene,which I named insab,is found in many f ish.The coding sequence encoded by insab genes is under weak selective constraint,with its predicted protein sequences losing their potential to be processed into a two-peptide hormone.However,these sequences have retained perfectly conserved cystine residues,suggesting that they maintain insulin’s three-dimensional structure and therefore might modulate the processing and secretion of insulin produced by the other genes.
Keywords:Insulin;Teleost f ish;Gene duplication;Adaptive evolution;Gene loss
INTRODUCTION
Fish have been important contributors to our understanding of human biology and disease(Lieschke&Currie,2007;Mac Rae & Peterson,2015),especially in endocrinology(Conlon,2000a).In vertebrates,insulin is a hormone produced by the beta-cells of pancreatic islets in response to increased levels of blood glucose,which induces the uptake of glucose by tissues such as liver and adipose tissue for storage,and thus is a key regulator of glucose metabolism(Röder et al.,2016). Def iciencies in insulin production and/or signaling leads to diabetes in humans and other animals(Röder et al.,2016;Weiss,2009).Many aspects of insulin function have been learned from extensive studies in diverse species of f ish(Caruso&Sheridan,2011;Conlon,2000b;Polakof etal.,2012).Fish have also been explored in the development of potential treatments for diabetes.Xenotransplantation of fish islets,using both wild-type and humanized insulin,has been considered due to the relative ease of isolation of islets from pancreatic tissue in fish(e.g.,tilapia,Oreochromis niloticus)compared to mammalian sources(Wright et al.,2014).Insulin,however,in addition to its role in the regulation of metabolism,also has other roles in vertebrate biology,including some in development(Chan&Steiner,2000;Hernández-Sánchez et al.,2006).
Insulin is a polypeptide hormone composed of two peptides,A-and B-chains of about 20 and 30 amino acids,respectively,which are held together by disulf ide bridges(Conlon,2001;Steiner et al.,1985,2009;Weiss,2009).Insulin sequences have been characterized,mostly at the protein level,in a large number of vertebrate and f ish species(Caruso&Sheridan,2011;Conlon,2000b,2001).Typically,vertebrate species have a single copy of the insulin gene in their genomes(Nishi&Nanjo,2011;Steiner et al.,1985),however,multiple insulin genes have been characterized in some species,such as rodents(Long etal.,2013;Soares etal.,1985;Wentworth etal.,1986)and some f ish(Caruso et al.,2008;Caruso&Sheridan,2011,2014;Irwin,2004). Within f ish,it appears that only one of these insulin genes,which produces the hormone that regulates blood glucose levels,is expressed in the pancreas(Caruso&Sheridan,2011;Irwin,2004),with the exception of the multiple insulin genes produced by the very recent genome duplication in salmonid f ish(Caruso et al.,2008;Caruso&Sheridan,2014).While the origin of the two insulin genes found in some f ish(e.g.,zebraf ish)was shown to be early in teleost f ish evolution,whether this ins2 gene has been retained in the genomes of diverse f ish is unclear(Caruso&Sheridan,2011;Irwin,2004).The f ish ins2 gene has been poorly characterized.The initial identif ication of this gene in zebraf ish(Danio rerio)and takifugu(Takifugu rubripes)only showed expression in the embryo of zebraf ish(Irwin,2004).A subsequence study of the expression of the two insulin genes in zebraf ish suggested that ins2 potentially has a function as a growth and neurotrophic factor during development(Papasani et al.,2006).More recently,studies of the tilapia(O.niloticus)ins2 gene showed that it had negligible expression in the pancreas,thus likely is not a major contributor to the regulation of glucose metabolism and would not need to be silenced to allow the xenotransplantation of tilapia islets as a treatment for diabetes in humans(Hrytsenko et al.,2016).
Changes in the function of insulin can be paralleled by changes in the function of glucagon,the hormone that counters the action of insulin(Seino et al.,1988).Recently it has been shown that duplicated proglucagon genes are widespread in teleost f ish,and that the functions of the proglucagon-derived peptides encoded by these genes have diversif ied(Irwin&Mojsov,2018). Here I have taken advantage of the large number of f ish genomes that have been characterized in the past few years(Ravi&Venkatesh,2018)to determine the distribution of insulin genes in the genomes of ray-f inned f ish.Analysis of these sequences allows us to begin to understand the origin of these genes,including the measurement of the selective forces acting upon them,which might provide evidence for differences in function.Surprisingly,I found a greater diversity in the numbers of insulin genes than expected,with the identif ication of a third insulin paralog,as well as lineage-specif ic duplicates of some of these paralogs. My analyses suggest that all three f ish insulin paralogs encode functional protein products.
MATERIALS AND METHODS
Data collection
Genome sequences databases maintained by the National Center for Biotechnology Information(NCBI:https://www.ncbi.nlm.nih.gov/genome/gdv/)and Ensembl(http://www.ensembl.org)were searched in March/April 2018,for sequences that were predicted to encode proteins similar to proinsulin.Initial searches used the tBLASTn algorithm(Gertz et al.,2006)using previously characterized zebraf ish(Danio rerio)proinsulin protein sequences(Irwin,2004;Milewskiet al.,1988)as queries. Subsequent tBLASTn,BLASTn,and BLASTp searches used our putative proinsulin sequences as queries.Genome sequences of 55 fish(52 ray-finned(Superclass Actinopterygii),2 cartilaginous(Superclass Chondrichthyes),and one lobe-finned(Class Sarcopterygii))were examined,with 52 of these contained in the NCBIGenome Data Viewer,and three(Atlantic cod,Gadus morhua;three-spined stickleback,Gasterosteus aculeatus;and the spotted green pufferfish,Tetraodon nigroviridis)only in the Ensembl database(9 were in both NCBI and Ensembl).A list of species with genomes used here,and their phylogenetic relationships,is presented in Figure 1.All sequences with E-scores below 0.001 were examined.As an initial step to assess homology and orthology,reciprocal BLASTp searches of the zebrafish proteome were conducted to examine the similarity of the putative proinsulin sequences.Sequences that were not more obviously similar to insulin-like growth factor-I(igf1)or insulin-like growth factor-II(igf2)were used for the following analyses.Here I suggest a standardized set of names for f ish ins genes,based on the analysis described below,with insa,insb,and insc representing the three ins paralogs that originated early in teleost f ish evolution,and insaa and insab representing duplicates of the insa gene shared by multiple species.Arabic numerals are used for recent lineage-specif ic duplicates and do not indicate necessarily indicate orthology.
Phylogenetic analysis
Prior to phylogenetic analysis, proinsulin (ins) coding sequences were aligned using the MAFFT algorithm(Katoh et al.,2001)as implemented at the Guidance2 server(http://guidance.tau.ac.il/ver2/; Penn et al.,2010).Phylogenetic relationships were established using both maximum likelihood and Bayesian methods. PhyML 3.0(http://www.atgc-montpellier.fr/phyml/;Guindon et al.,2010)was used to generate maximum likelihood trees,with 500 bootstrap replications,where the Smart Model Selection(SMS)(Lefort et al.,2017)option was used to identify the best fitting evolutionary model. Bayesian trees were constructed using MrBayes 3.2.6(Huelsenbeck et al.,2001;Ronquist et al.,2012),with 5 000 000 generations and four simultaneous Metropolis-coupled Monte Carlo Markov chains sampled every 100 generations under the same evolutionary model used for maximum likelihood.The first 25%of the trees were discarded as burn-in with the remaining samples used to generate consensus trees.Insulin coding sequences from cartilaginous(Callorhinchus milli(Elephant shark)and Rhincodon typus(Whale shark))and lobe-finned fish(Latimeria chalumnae(Coelacanth))were used as outgroups to root the trees for all insulin coding sequences.Subsequent analyses,using subsets of the sequence data,were conducted with similar approaches,and used sequences identified in our initial analyses as outgroups.
Evolutionary analysis
Genes adjacent to insulin genes in fish genomes were identified by examining genomic contigs in the NCBI and Ensembl databases using methods that I have previously used(Irwin,2002,2012;Irwin&Mojsov,2018).Briefly,the identity and orientation of genes neighboring insulin genes were identified from the genomic databases.The orthology of the genes adjacent to insulin genes was assessed(or conf irmed if annotated)using BLASTp searches of the zebraf ish and human proteomes(Kuraku&Meyer,2012).

Figure 1 Diversity and evolution of f ish insulin genesSummary of the evolution of insulin genes in ray f inned f ish.Phylogenetic relationships of species with genome sequences is based on Betancur-R et al.(2017),with modif ications in an area that is not conf idently resolved to minimize the number of gene duplications or deletions.Some phylogenetic relationships are not resolved.Branches are not proportional to evolutionary time.Classif ication of f ish is shown to the right.Solid diamonds indicate genome duplications.The numbers of different types of insulin gene found in each genome is listed,with numbers in brackets indicating incomplete sequences.Gene additions or gene losses are indicated by plus(+)and minus(-)before an a,b,or c,representing the insa,insb,and insc paralogs,respectively.The insa,insb,and insc paralogs originated near the genome duplication in the ancestor of teleosts and are indicated as+a,+b,and+c to refer to the two gene duplications.The duplication to generate the insaa and insab genes is indicated by+aa/ab.+ins indicated the gene duplication on the Lepisosteus oculatus lineage.+ψins,+ψa,and+ψb indicates retroposition events that generated processed pseudogenes.
The strength of selective pressure acting upon the insulin coding sequences can be measured by comparing the relative rates of nonsynonymous(dN)and synonymous substitutions(dS).Sequences under stronger selective pressure for protein function will have lower nonsynonymous to synonymous(dN/dS)ratios.dN/dSrate ratios were obtained from analyses using RELAX(Wertheim et al.,2014)as implemented on the Datamonkey Adaptive Evolution server(http://datamonkey.org/;Weaver et al.,2018),which also tested for intensif ication or relaxation of the levels of selection on tested lineages.Evidence for positive selection on branches of the phylogenetic tree was tested using aBSREL(Smith et al.,2015)on the Datamonkey Adaptive Evolution server(Weaver et al.,2018).
Prediction of protein processing sites and alignment of peptide sequences
Signal peptidase cleavage sites in the proinsulin protein sequences were predicted using SignalP 4.1(http://www.cbs.dtu.dk/services/SignalP/;Petersen et al.,2011).I used two prediction programs to identify potential prohormone protease processing sites in the proinsulin sequences,NeuroPred(http://neuroproteomics.scs.illinois.edu/neuropred.htm;Southey et al.,2006)and ProP 1.0(http://www.cbs.dtu.dk/services/ProP/;Duckert et al.,2004),with“general PC prediction”selected.To visual the conservation of the processing sites and insulin peptide sequences,the proinsulin protein sequences were aligned using Clustal Omega(https://www.ebi.ac.uk/Tools/msa/clustalo/;Sievers et al.,2011). Consensus proinsulin Aand B-chain peptide sequences were generated from the alignments using WebLogo 3(http://weblogo.threeplusone.com/;Crooks et al.,2004).
RESULTS
Numbers of insulin genes in f ish genomes
Searches of genome databases maintained by NCBI and Ensembl resulted in the identif ication of 168 insulin genes from the genomes of 55 species examined,with 141 of these predicted to be intact full-length coding sequences(Figure 1 and Supplementary Table S1).While only single copy insulin genes were found in the genomes of cartilaginous and lobe-f inned f ish,multiple genes were found in the genomes of most ray-f inned f ish. Within ray-f inned f ish,the number of identif ied insulin genes ranged from 1(Sclerophages fromosus)to 8(Sinocyclocheilus anshuiensis).As expected,f ish species that experienced recent genome duplications,i.e.,carp(Cyprinus carpio;Xu et al.,2014),members of the genus Sinocyclocheilus(Yang et al.,2016),and salmonids(Oncorhynchus kisutch,O.mykiss,Salmo salar,and Salvelinus alpinus;Lien et al.,2016)have the largest numbers of insulin genes. Phylogenetic analysis(see below)suggested the presence of three insulin paralogs in teleost f ish:insa,insb,and insc.Of the 162 insulin genes identif ied in the 51 species of teleost f ish(infraclass Teleostei),92 were classif ied as insa(which includes insaa and insab genes),57 as insb,and 13 as insc,with 11,12,and 3 of the sequences found to be incomplete,respectively,for these three paralogs(Figure 1 and Supplementary Table S1).All 51 species of teleost f ish possessed at least one insa gene,with 45 having insb genes,and 10 with insc genes(Figure 1 and Supplementary Table S1).The 6 species of teleost fish where I failed to identify insb genes are distributed across the accepted phylogeny of fish(Figure 1;Betancur-R et al.,2017;Near et al.,2012),suggesting they were lost independently several times(or are missing from these genome assemblies).In contrast,almost all of the species lacking insc genes share a common ancestor(Figure 1,clade Euteleostei),suggesting a single gene loss event explains the absence of insc genes in most teleost fish.The coding sequences of the insulin(ins)genes used in our analysis described below are presented in Supplementary Figure S1.
Phylogeny of f ish insulin genes
To better understand the origin and evolution of the multiple insulin (ins)genes found in the genomes of ray-f inned f ish,their phylogenetic relationships,rooted using ins gene sequences from cartilaginous and lobe-f inned f ish,were inferred using maximum likelihood(Figure 2)and Bayesian(Supplementary Figure S2)methods.Both analyses generated similar phylogenies with independent duplications of the insulin gene on the spotted gar(Lepisosteus oculatus;infraorder Holostei)and teleost(infraorder Teleostei)f ish lineages.While duplicated insa and insb genes were previously identif ied in teleost f ish(Caruso&Sheridan,2014;Hrytsenko et al.,2016;Irwin,2004),here If ind that these species have a third insulin paralog,which Inamed insc.The species relationships within each paralog of insulin genes,given its limited power at resolving species relationships,is in general agreement with the accepted phylogeny of teleost f ish(Betancur-R et al.,2017;Near et al.,2012).This observation suggests that the triplication of the insulin gene occurred in an early teleost.The relationships among the insa,insb,and insc paralogs is poorly resolved in our phylogenetic analyses,with the maximum likelihood analysis suggesting a closer relationship between insa and insb,while the Bayesian analysis suggesting that insb and insc are most closely related.
Most teleost species that possess insb and insc genes have a single copy of each gene,except those species that have experienced additional genome duplications(i.e.,Cyprinus carpio,salmonids(O.mykiss,O.kisutch,Salmo salar,and Salvelinus aplinus),and the genera Sinocyclocheilus)).
In contrast,a large number(30 of 51)of teleosts have multiple insa genes(Figure 1 and Supplementary Table S1).While some of these appear to be due to lineage-specif ic genome duplications(i.e.,Cyprinus carpio,salmonids,and the genera Sinocyclocheilus)or single-gene duplications(e.g.,Labrus bergylta and Larimichthys crocea),many are due to a duplication event that occurred on an early Percomorpha lineage(Figures 1,2,and Supplementary Figure S2).The genomes of many Percomorpha f ish contain two(or more)insa genes that form monophyletic clades that are largely in accord with the accepted species phylogeny(Betancur-R et al.,2017;Near et al.,2012).This observation suggests that a duplication of the insa gene,generating insaa and insab genes,occurred in an early lineage of Percomorpha f ish(see Figure 1). In species that are inferred to be descendants of this Percomorpha-specif ic ins gene duplication,all but Nothobranchius furzeri possess an insaa gene,however,a larger number of species,such as tilapia(Oreochromis niloticus),some cichlids (e.g.,Pundamilia nyererei and Maylandia zebra),and species of the genus Poecilia do not have an insab gene(Figures 1,2 and Supplementary Figure S2 and Table S1).This suggests thatthe insab gene was lost in parallel in a number of species of Percomorpha(see Figure 1).
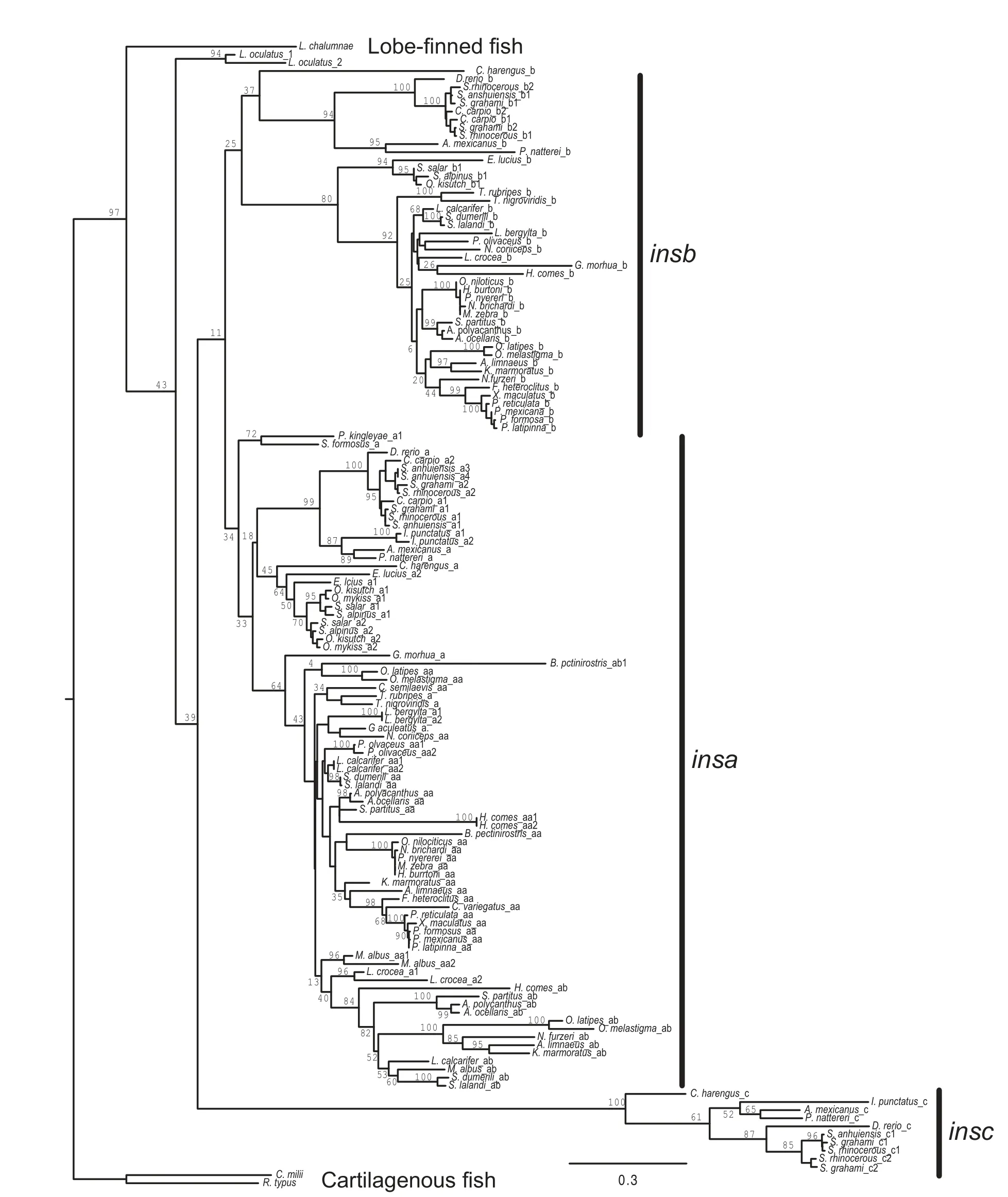
Figure 2 Phylogeny of f ish insulin genesPhylogenetic relationship of insulin coding sequences was inferred using maximum likelihood with the best f itting evolutional model GTR+G+I(GTR with gamma distribution and invariant sites)and bootstrapped with 500 replications.A similar phylogeny was generated using Bayesian methods(Supplementary Figure S2).Percent bootstrap support for lineages are shown.The tree in Newick format,with all branch lengths and support values,is presented in Supplementary Figure S3.The phylogeny was rooted with sequences from cartilaginous(Callorhinisus milli and Rhincodon typus)and lobe-f inned(Latimeria chalumnae)f ish.Full species names are listed in Figure 1.insa,insb,and insc indicate the three clades of insulin genes found in teleost f ish.
Duplications of f ish insulin genes
To aid in the assessment of the orthology of insulin genes,and to gain insight into the gene duplication events,Iidentif ied genes f lanking the insulin genes in the genomic sequences of 55 species of f ish(Supplementary Table S2,examples given in Figure 3).A genome duplication occurred in the ancestor of teleost f ish,the teleost f ish-specif ic genome duplication(Glasauer&Neuhauss,2014;Meyer&Van de Peer,2005).Our phylogenetic analyses(Figure 2 and Supplementary S2)suggests that the three insulin paralogs found in teleost f ish(insa,insb,and insc)originated on this lineage. If duplicate genes originated through a genome duplication,then one might expect similar genomic neighborhoods for these genes(i.e.,similar genes found adjacent to different ins genes),as observed for the duplicated proglucagon(gcg)and glucagon receptor(gcgr)genes of teleost f ish(Irwin,2014;Irwin&Mojsov,2018). For the three insulin paralogs examined here,only very limited similarity was seen in the genomic neighborhoods(longer genomic regions were examined,with immediate neighborhoods summarized in Supplementary Table S2). A gene similar to tenm2(teneurin transmembrane protein 2)was found adjacent to insa genes in Exox lucius,O.kisutch,O.mykiss,Paramormyrops kingsleyae,Salmo salar,and Sclerophages formosus,and insb genes in Pygocentrus nattereri,Salvelinus alpinus,and Sinocyclocheilus rhinocerous(Supplementary Table S2).This might suggest that the insa and insb genes originated via the teleost-f ish specif ic genome duplication.However,as the linkage between the tenm2 and ins genes was only observed in a smallnumber of species,this would require large number of independentlosses(or genomic rearrangements)of tenm2-like genes to yield the absence of a linked tenm2-like with most ins genes,therefore the evidence for the origin of insa and insb by a genome duplication event is not strong.No genes near insc were similar to those adjacent to insa or insb,thus it more likely originated as a single gene duplication event rather than through a genome duplication. If the insa and insb genes originated via a genome duplication,an addition single-gene duplication would stillbe needed to explain the origin of insc.The gene neighborhood data does not help resolve the relationships among the three ins genes.Intriguingly,the genomic neighborhoods of none of the insulin gene paralogs in teleost fish is similar to the typical vertebrate insulin gene neighborhood,where insulin is flanked by insulin-like growth factor 2(igf2)and tyrosine hydroxylase(th)genes(Patton et al.,1998),although similar neighborhoods were found for the insulin genes in cartilaginous and lobe-finned fish(Supplementary Table S2).This suggests that the insulin genomic neighborhood was rearranged on the ancestral ray-finned fish lineage.
Orthologous genes often have similar genomic neighborhoods,thus can these neighborhoods can be used to assess orthology when gene sequence similarity is limited(Kurokawa et al.,2005).When the genomic neighborhoods surrounding insb and insc genes were examined(Figure 3 and Supplementary Table S2)shared features were identif ied for each gene. All insb genes are adjacent to a hmmr(hyaluronan mediated motility receptor)-like gene,while all insc genes are f lanked by prr12(proline rich 12)and scaf1(SR-related CTD associated factor 1)-like genes(as shown for the Clupea harengus insb and Danio rerio insc genes in Figure 3),suggesting that their genomic neighborhoods are largely conserved since they originated in the early teleost.In contrast,several different genomic neighborhoods were observed for the insa genes(Supplementary Table S2,examples shown in Figure 3). The most phylogenetically widespread genomic neighborhood has a nipal(magnesium transporter NIPA3-like)-like gene adjacent to the insa gene(Supplementary Table S2)and is found in species representing Osteoglossomorpha(e.g.,Scelerophages formosus,shown in Figure 3),Protacanthopterygii(e.g. Esox lucius and Salmo salar),Paracanthopterygii(e.g.,Gadus morhua),and Percomorpha(e.g.,Takifugu rubripes and Oryzias latipes).The only major group of teleost f ish that does not have an insa gene adjacent to nipal are those of Otomorpha(e.g.,Danio rerio,shown in Figure 3),suggesting a change in the gene order occurred in this group and that the linkage of insa with nipal is ancestral.Within Percomorpha(Supplementary Table S2),the insaa genes are found adjacent to nipal-like genes(as in Scelerophages formosus,Figure 3),while the insab genes are near a sh3pxd2b(SH3 and PX domains 2B)-like genes(e.g.,Haplochromis burtoni,Figure 3).The linkage of the insab genes with sh3pxd2b strengthens the conclusion that all insab genes are orthologous and indicates that insaa is located at the ancestral genomic location and was the source(locus-of-origin)for the insab gene that was inserted into a new genomic locus.

Figure 3 Genomic neighborhoods surrounding f ish insulin genesExamples of genomic neighborhoods found for insulin paralogs in ray-f inned f ish.Genomic neighborhoods are listed in Supplementary Table S2.A:Typical genomic neighborhood for insa and insaa genes as illustrated from Sceleropages formosus.B:Genomic neighborhood for insa genes in Danio rerio and close relatives.C:Typical genomic neighborhood for insab genes as illustrated from Haplochromis burtoni.D:Typical genomic neighborhood for insb genes as illustrated from Clupea harengus.E:Typical genomic neighborhood for insc genes as illustrated from D.rerio.See Supplementary Table S2 for full gene names.
Lineage-specif ic changes in insulin gene number
The gene duplication events described above explain most,but not all,of the insulin genes identif ied(Figure 1 and Supplementary Table S1). Many of the additional genes can be explained by tandem gene duplication,which lead to two insulin genes arranged head-to-tail in the genome(see Supplementary Tables S1,S2)and includes the insa1 and insa2 genes in Parmormyrops kingsleyae and in Larmichthys crocea,the insaa1 and insaa2 genes in Hippocampus comes,and the insab1 and insab2 genes in Boleopththalmus pectinirostris. Both Cyprinus carpio and Sinocyclocheilus rhinocerous have tandemly arranged insb genes(insb3 and insb4 in C.carpio and insb2 and insb3 in S.rhinocerous,Supplementary Table S2)that might have originated prior to the divergence of these two species(although,they would then have to be lost in the two other Sinocyclocheilus species). Sinocyclocheilus anshuiensis has two pairs of tandemly arranged genes,insa1/insa2 and insa3/insa4,which either originated prior to this species genome duplication(and then were lost in the closely related species that share this genome duplication),had parallel tandem duplications,or is an assembly error(the tandemly arranged genes are identical in sequence).The ins1 and ins2 genes of Lepisosteus oculatus might also have originated via a tandem duplication,which was then followed by rearrangements so that these lineage-specif ic gene duplicates are now found in opposite orientations on either side of a hmmr-like gene(see Supplementary Table S2).More complex duplication events,resulting in genes moving to new locations are need to explain the remaining genes,with some being extremely recent(e.g.,the identical insaa1 and insaa2 genes in Labrus bergylta)or more ancient(e.g.,insa1 and insa2 gene of Esox lucius).
While duplication of genomic DNA is a frequent mechanism for the origin of duplicate genes,they can also be generated by retroposition of c DNA generated from mRNA transcripts(Long et al.,2013).Indeed,the well-characterized duplicate ins1 gene in mice was generated from an incompletely processed mRNA transcript(Shiao et al.,2008;Soares et al.,1985).Typically,insulin genes contain two coding exons that must be spliced together to create an intact coding sequence. Here I identif ied 6 insulin genes that are likely generated by reverse-transcribed mRNAs,as these genomic sequences do not have an intron interrupting the coding sequence(see Supplementary Figure S4 for an example).The 6 retro-processed insulin genes can be explained by three integration events:(1)between ankrd6(ankyrin repeat domain 6)and lyrm2(LYRmotif containing 2)-like genes on the Lepisosteus oculatus lineage to generateψins;(2)within an apbb3(amyloid beta precursor protein binding family 8 member 3)-like gene on the Monopterus albus lineage to generate ψinsb;and(3)within a psd3(PH and SEC7 domain containing protein 3)-like gene on the ancestral lineage for salmonids(Oncorhynchus mykiss,O.kisutch,Salmo salar,and Salvelinus aplinus)and the Northern pike(Esox lucius)that generated
ψinsa in O.kisutch,S.salar,and S.alpinus(but was not found in O.mykiss)and an intact coding sequence(insa2)in Esox lucius(Figure 1 and Supplementary Figure S4 and Table S2).The maintenance of an intact open reading frame in Exox lucius insa2(Supplementary Figure S4)suggests that this retropositioned sequence,like the mouse ins1 gene(Soares et al.,1985),is still functional.However,this sequence has lost its coding potentialin salmonid f ish and is a pseudogene(Figure 1 and Supplementary Table S1).Additional processed genes might also exist,as several incomplete genes(e.g.,insb4 from Cyprinus carpio and insa3 from Paramormyrops kingsleyae)were identif ied(see Supplementary Table S1)thatwere located at unique location of the genome(see Supplementary Table S2)consistent with being inserted processed c DNAs,but the sequences were similar to either exon 1 or 2(and not both),thus cannot be distinguished from a sequence generated by an DNA-mediated incomplete gene duplication event.
Gene for insulin a(insa)is evolving under greater evolutionary constraint
Our genomic and phylogenetic analyses of f ish genome sequences demonstrated that teleost f ish have three paralogous insulin genes.In contrast,most other vertebrate species only have one(Chan&Steiner,2000;Conlon,2000b). The increased number of insulin genes in teleost f ish raises the possibility that they have been diversif ied to: (1)subfunctionalize distinct functions of insulin,(2)neofunctionalize to acquire novel functions,or(3)lose function(pseudogenize).As a f irst step to explore possible changes in the biological roles of these distinct insulin genes Iassessed the selective pressures acting upon the sequences. This can be done by comparing the rates of nonsynonymous(dN)to synonymous(dS)substitutions(Yang&Bielawski,2000).If the protein encoded by a gene has lost its function,then it would be expected that there would be no selection against nonsynonymous substitutions,and that rates of nonsynonymous and synonymous substitutions would be the same rate(i.e.,dN/dS=1,neutral evolution). If the protein encoded by these genes still had a function,then selection should act against a subset of nonsynonymous substitutions(the deleterious mutations)and be lower than the synonymous rate(i.e.,dN/dS<1,purifying selection).Rarely,one might see dN/dS>1,which would indicate positive selection for change in amino acid sequence(Yang&Bielawski,2000).If protein coding sequences of genes are being maintained for different functions,with different parts of the sequence being important for these functions,then one might see differing levels of selective constraint.
I used the program RELAX(Wertheim et al.,2014)to assess the levels of selective pressure(dN/dS)for each of the three insulin gene paralogs,and to determine whether the selective pressure was intensif ied or relaxed compared to the other insulin genes,with sequences from a non-teleost ray-f inned f ish,Lepisosteus oculatus,used as outgroup.When all sequences were analyzed,insa genes showed the stronger levels of purifying selection(dN/dS=0.266 6,0.3332,and 0.3258 for insa,insb,and insc,respectively;Table 1). If I restricted these analyses to either species that have all three paralogs,or species that only have a single copy of these paralogs,to attempt to minimize species-specif ic effects(changes seen in species that only have some of the genes)or due to differences in gene number,the difference in the selective constraints acting on insa vs.insb and insc became even more pronounced(Table 1).The results from RELAX also indicated that selection intensif ication,compared to the other insulin sequences,occurred on insa,while selection relaxation occurred for insb(Table 1).No signif icant change in selection intensity was seen for insc vs.all other insulin sequences.Similar results,with insa showing greater constraint,were seen in comparisons of insa and insb genes in species that had both of these genes(Table 1).These results suggest that the insa paralog is under greater purifying selective constraint than either the insb or insc paralog,but also importantly demonstrate that both insb and insc are continuing to experience purifying selection. Thus,it can be concluded that the protein coding sequence of insa is under the greatest level of selective constraint,however,the coding sequences of both insb and insc are also experiencing selective constraints consistent with their protein products having essential biological functions.

Table 1 Differences in the selective pressure(d N/d S)acting on insulin paralogs in teleosts
Adaptive evolution of insulin genes
While the protein products of the three insulin genes are being maintained for function,they might not be the same function.To examine whether any of the sequences might have gained new functions I tested for evidence of positive selection on lineages using aBSREL(Smith et al.,2015).Only one lineage showed signif icant evidence for episodic diversifying selection,the ancestral lineage for the insc genes.Inspection of the phylogenetic tree generated from all of the insulin coding sequences suggest that the ancestral branch for the insc genes is longer than for the insa and insb genes(Figure 2 and Supplementary Figure S2),consistent with a greater number of nonsynonymous substitutions driven by positive selection.No evidence for positive selection was found on the ancestral lineages for the insa or insb genes.These results might suggest that insc has acquired a new function(neofunctionalization)driven by positive selection,while insa and insb have been retained due to subfunctionalization of insulin functions between the two genes.
Relaxed selection on the insab duplicate of the insulin a(insa)gene
An intriguing observation from our analysis of the selective constrains acting upon insa,insb,and insc paralogs was that the calculated value for the selective constraint acting on insa varied considerably depending upon which sequences were used for the analysis(Table 1).The ratio of the nonsynonymous to synonymous rates was higher when all insa sequences were used than if sequences were used only from species that had all three paralogs or had only single copies of the insa and insb paralogs(Table 1). These observations suggest that inclusion of insa sequences from species that have multiple insa sequences yields higher estimates of the dN/dSratio due to some of these sequences having lower levels of sequence constraint.As the duplication to generate the insaa and insab genes is a major source of the multiple insa genes in teleost f ish(see Figures 1,2 and Supplementary Figure S2 and Table S1),I compared the selective constraints acting upon insaa and insab genes(Table 2). The insaa coding sequences were found to be under selective constrains(Table 2)similar to those of other insa sequences,and especially those from species that had single copy insa,while the insab sequences displayed the lowest levels of constrains seen for any f ish ins sequence(Tables 1,2).Test for intensif ication or relaxation of selective constraint showed that insaa was under signif icant intensif ication of selective constraint,while insab was signif icant relaxation of selective constraint was demonstrated for insab(Table 2).These results suggest that insaa retains the function of insa,which would be consistent with itbeing atthe locus of origin,while insab is evolving with far less constraint,to the extent thatit has been loston a number of lineages(e.g.,Oreochromis niloticus,Pundamilia nyererei,M.zebra,and species of the genus Poecilia,see Figure 1).This might suggest that insab is not essential.

Table 2 Differences in the selective pressure(d N/d S)acting on insaa and insab genes
Processing of proinsulin sequences
To be functional,the protein product encoded by insulin genes need to be proteolytically processed and secreted to generate two-peptide chain insulin molecules(Steiner et al.,1996,2009). Our analysis was initially only focused on sequences that showed similarity to previously characterized proinsulin sequences and had intact coding sequences,i.e.,had an initiation codon,a termination codon and intact open reading frame with greater similarity to proinsulin than to other insulin-like sequences(e.g.,insulin-like growth factors). To determine whether the encoded protein sequences could be secreted and properly processed Isearched for potential signal peptidases and proprotein processing sites using programs that predict these sites(Duckert et al.,2004;Petersen et al.,2011;Southey et al.,2006).
The coding sequences of intact proinsulin open reading frames from all ray-f inned f ish identif ied here are predicted to have functional signal peptides(Table 3 and Supplementary Table S3 and Figure S6),thus should be able to be secreted.In addition,allof the proinsulin protein sequences encoded by the insa,insab,insb,and insc genes have potential prohormone protease cleavage sites that could yield typical two-chain insulin hormone molecules(Table 3 and Supplementary Table S3 and Figure S6).In contrast,however,only one of the 14 proteins encoded by insab genes is predicted to potentially produce a two-chain insulin(Table 3).For the insab sequences,only 2 have potential B-chain/C-peptide processing site and 9 have potential C-peptide/A-chain processing sites(Table 3 and Supplementary Table S3 and Figure S6),indicating that neither site is conserved.Both NeuroPred(Southey et al.,2006)and ProP(Duckert et al.,2004)predicted similar sites,but ProP also provided a list of other potential processing sites that did not score well enough to be predicted sites(see Supplementary Table S3).The alternative potential sites identified by ProP in the insab proteins still would not generate 2-chain insulin molecules similar to functionally characterized insulin molecules(Steiner et al.,1996,2009)as they would generate very short B-chains,often missing residues essential for function.
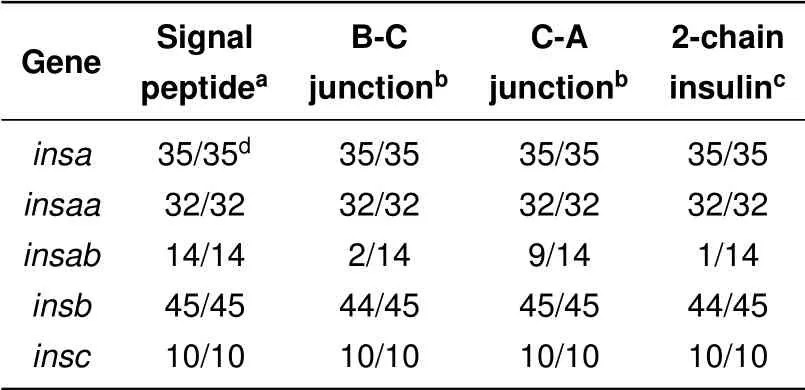
Table 3 Secretion and processing of f ish proinsulin proteins
The hormone insulin is a two-chain peptide molecule held together by disulf ide bridges(Steiner et al.,1996,2009).
To conf irm that two-chain molecules could be produced from the proinsulin protein sequences Igenerated consensus sequences for the A-and B-chains for the different types of insulins(Figure 4).The lengths of the predicted A-and B-chains were similar between the different types of insulins,with more variation,atboth the N-and C-termini,within proteins encoded by a type of gene than between types of genes(Figure 4 and Supplementary Figure S6). When cysteine residues were examined one of the two insc sequences from Sinocyclocheilus grahami(insc2)had a cystine replaced by an arginine residue.It is possible that the S.graham insc2 gene does not encode a functional insulin,but this is compensated by the presence of an insc1 gene that encodes an insulin that can be secreted processed and has all six cysteine residues.Surprisingly,when I examined A-and B-chain sequences homologous to those of other insulins in the protein encoded by insab genes,all of the cysteine residues were perfectly conserved,however much of the rest of the sequence showed greater variation than within other types of insulins(Figure 4 and Supplementary Figure S6).This raises the possibility that insab could still fold in a similar fashion as typical insulin,thus be transported to insulin secretory granules,but then not be processed into an active hormone(Liu et al.,2015).
DISCUSSION
Searches of diverse f ish genomes,including in the extensively characterized zebraf ish(Danio rerio)genome,unexpectedly revealed the existence of three insulin gene paralogs(Figure 1 and Supplementary Figure S1 and Table S1).Previously,two paralogous insulin genes,insa and insb,had been found in the zebraf ish genome that have distinct expression patterns(Irwin,2004;Papasani et al.,2006).Here Ifound insa genes in the genomes of all teleost f ish(Figure 1 and Supplementary Figure S1 and Table S1),which encode the previously isolated insulin hormone sequences(Caruso&Sheridan,2011;Conlon,2000b)that regulate glucose metabolism.Given the central role of insulin in the regulation of metabolism(Caruso&Sheridan,2011;Röder et al.,2016;Weiss,2009),it is not surprising thatthis gene was found in allf ish.The presence of a high selective constraint(Tables 1,2)on the protein sequences encoded by insa and insaa genes supports the conclusion that this sequence is essential.A surprising discovery was the presence of a duplicate of the insa gene,the insaa and insab genes,in a large number of teleost f ish(Figure 1 and Supplementary Table S1).The protein coding sequence encoded by insab genes have the highest nonsynonymous substitution rate(Table 2),and thus least selective constraint,yet the consensus sequences of the potential A-and B-chains of insulin predicted from these genes have perfectly conserved cystine residues(Figure 4).This suggests that these genes might encode proinsulin-like proteins that could properly fold but would not be properly processed in secretory granules to produce active insulin molecules(Liu et al.,2015).Thus,insab proteins might compete with insaa proteins for processing enzymes and modulate the release of functional insulin molecules in these species.

Figure 4 Consensus f ish insulin peptide sequencesConsensus sequences for the A-chain(A)and B-chain peptides(B)for insulin encoded by insa,insaa,insab,insb,and insc genes.Consensus sequences were generated from aligned predicted protein sequences(see Supplementary Figure S5)of genes with complete open reading frames(Larimichthys crocea insa2 was removed as it may not be functional).Hight of the residue is proportional to representation among the compared sequences,while residue width represents proportion of sequences without a gap(thin residues are gaps in many sequences).
The insb gene was found in the genomes of almostallteleost f ish species examined,demonstrating that it is conserved in these f ish(Figure 1 and Supplementary Figure S1 and Table S1).The selective forces acting on insb are weaker than those acting on insa(Table 1)suggesting that these two genes have different functions.Previous work indicates that insb is not appreciably expressed in the pancreas,and instead is more predominantly expressed in early development(Hrytsenko et al.,2016;Irwin,2004;Papasani et al.,2006).The analysis of selective constraints and these expression results are consistent with speculation that insb is not primarily involved in the regulation of blood glucose level,but instead has a role in development(Hrytsenko et al.,2016;Papasani et al.,2006).Given the conservation of the insb gene across teleost f ish,it likely acquired this role soon after its origin and has been conserved.
Unexpectedly Ifound an insc paralog,and it was present in the well characterized zebraf ish(D.rerio)genome(Figure 1 and Supplementary Figure S1 and Table S1).Upon closer examination,the insc gene in both the Ensembl(gene ID:ENSDARG00000096862)and NCBI(GenBank accession No.:100534937)zebraf ish genomes fail to predict an intact open reading frame,with the intact insc coding sequence only coming from the linked mRNA sequence(XM_009299616.3)in the NCBIdatabase(see Supplementary Table S1).Icould f ind no raw sequence data(e.g.,ESTs)that supports the existence of an intact open reading frame for zebraf ish insc,as all mRNA derived sequences(GenBank accession Nos.EH533807.1 and EH507658.1,forward and reverse sequences of the same EST clone)also contained the deletion found in the genomic sequence.This suggests that the zebraf ish insc gene does not encode an intact protein product.However,our analysis of the selective forces acting upon the insc coding sequences indicate that the nonsynonymous rate of substitutions(dN)is much lower than the rate of synonymous substitutions(dS)consistent with selection to maintain a protein coding sequence(Table 1)in most,if not all,species that have intact insc genes.The insc gene,in contrast to insa and insb,is restricted to the genomes of only a few lineages,with this gene been lost from the genome of most teleost f ish(Figure 1 and Supplementary Table S1).This distribution of the gene among f ish suggests that it does not encode a function that is essential in most species,and its loss might have been compensated by the presence of paralogs of insulin in the f ish genomes.Expression of insc,based on a single ESTclone(with sequences from both ends)that was identif ied in zebraf ish,is found in the“gut and internal organs”of adults,thus potentially it could be expressed in the pancreas and be replaced by insa.Further studies on the expression of insc in other f ish are needed to identify its role in physiology.
CONCLUSION
Searches of ray-f inned f ish genomes have demonstrated that they contain more insulin genes than previously appreciated and suggest that the roles of these genes have diversif ied.The diversif ication of the functions of insulin parallels the diversif ication of the proglucagon-encoded peptides found in f ish(Irwin&Mojsov,2018). Similarly,parallel changes in the biological functions of insulin and proglucagon-derived peptides have been observed in hystricomorph rodents(i.e.,guinea pig and relatives),where changes in sequence and action of insulin have compensating changes in glucagon(Seino et al.,1988).
COMPETING INTERESTS
The author declare that they have no competing interests.
AUTHORS’CONTRIBUTIONS
D.M.I.designed the study,conducted the analyses,and wrote the manuscript.
- Zoological Research的其它文章
- Specific function and modulation of teleost monocytes/macrophages:polarization and phagocytosis
- Anew species of Crocodile Newt,genus Tylototriton(Amphibia,Caudata,Salamandridae)from the mountains of Kachin State,northern Myanmar
- A new species of the endemic Himalayan genus Liurana(Anura,Ceratobatrachidae)from southeastern Tibet,China,with comments on the distribution,reproductive biology,and conservation of the genus
- Effects of C-terminal amidation and heptapeptide ring on the biological activities and advanced structure of amurin-9KY,a novel antimicrobial peptide identif ied from the brown frog,Rana kunyuensis
- Purification and characterization of a novel anti-coagulant from the leech Hirudinaria manillensis
- Passive eye movements induced by electromagnetic force(EMF)in rats

