两个线形三核配合物的晶体结构及性质
王丽雯 尚启高 周晶晶 周 红 潘志权 程清蓉
(武汉工程大学,武汉 430074)
Polynuclear transition complexes containing nitrogen and phenolic oxygen donor atoms are of considerable interest in inorganic and biomimetic chemistry due to their potential application in catalysis,their biological relevance,and potentially interesting magnetic properties[1-4].The extensively investigated “metallosalen-type” coordination compounds have long been employed as catalysts for the selective epoxidation of olefins and the oxidation of hydrocarbons and in recent years for DNA cleavage[5-8].
Cobalt complexes derived from Schiff bases have attracted an extensive interest in the area of coordination chemistry due to their intriguing structural features and various applications covering catalysis in organic synthesis,photochemistry,electrochemistry and important biological applications[9-12].Ever since the structural determination of Ni3(dpa)4Cl2in 1991[13],when it was realized that the anion of 2,2′-dipyridylamine (dpa)can support the formation of linear chains of three nickel atoms,studies of extended metal atom chains(EMACs)have attracted considerable attention[14].Cotton group reported the linear trinuclear complexes and studied the magnetic susceptibility measurements[15].
In our recent work we have synthesized a series ofdinuclearcomplexesand studied theirDNA cleavage abilities and magnetism[16-23].As part of on going studies on the polynuclear complex,two linear trinuclear nickel complex and cobalt complex were reported.In order to investigate the synergistic effect between metal ions and ligand,and the magnetic interaction among metal ions,their DNA cleavage abilitiesand magnetismswerealsostudied.The structure diagram of two complexes are showed in the Scheme 1.

Scheme 1 Structure diagram of the complexes
1 Experimental
1.1 Materials and measurement
All solvents and chemicals were of analytical grade and used as received,except that ethanol was purified to absolute by the general method.IR spectra were recorded on a Vector 22 FT-IR spectrophotometer using KBr disks.Elemental analyses were performed on a Perkin-Elmer 240 analyzer.Cyclic voltammograms were performed on a CHI model 750B electrochemical analyzer in DMF solution containing tetra(n-butyl)ammonium perchlorate(TBAP)as the supporting electrolyte.A three-electrodecellwas used,which was equipped with a glassy carbonworking electrode,a platinum wire as the counter electrode and an Ag/AgCl electrode as the reference electrode.Scanning rates were in the range of 20~200 mV·s-1.The solution was deaerated for 15 min before measurements.The half-wave potentials were calculated approximately from(Epa+Epc)/2,and the measured error was 2 mV.Magnetic susceptibility of a crystallinepowdered sample was measured on a SQUID-based sample magnetic meter in the temperature range of 2.0~300 K,and the diamagnetic corrections were made according to Pascals constants.UV-Vis spectra were recorded on an UV-2450 spectrophotometer.Circular dichroic spectra of DNA were obtained by using a Jasco J-810 spectropolarimeter.Electrospray mass spectrum(ES-MS)was determined on a Finnigan LCQ ES-MS mass spectrograph using methanol as a mobile phase with a sample concentration of 1.0 mmol·L-1.
1.2 Preparations of the complexes
1.2.1 Preparation of[CoⅢ2CoⅡL2(μ-OAc)4](1)
To the mixture of methanol(1 mL)containing 5-fluorosalicylaldehyde (0.141 g,1 mmol),2-hydroxy-1,3-propane diamine (0.045 g,0.5 mmol)was slowly added.The mixture was stirred for 6 h,then the yellow resolution was obtained.After adding the mixture of methanol(6 mL)containing Co(CH3COO)2·4H2O(0.249 g,1 mmol),The mixture was stirred for 8 h,and the color changed to dark red.Then the filtrate was slowly evaporated in air to give red single crystals.Yield:0.169 g,39%.Anal.Calcd.for C42H40O14N4F4Co3(%):C,46.81;H,3.74;N,5.19.Found(%):C,46.84;H,3.71;N,5.16.IR(KBr,cm-1):3 050,2 911(C-H),1 640(C=N),1 080,625(ClO4-).ESI-MS(m/z):897.92 for[Co3L2(OAc)]+(Supporting information).
1.2.2 Preparation of[Ni3L2(μ-OAc)2(CH3OH)2]·2H2O(2)
Complex 2 was prepared by the same method as described above except that Ni(CH3COO)2·4H2O was used instead of Co(CH3COO)2·4H2O.Yield:0.182 g,42%.Anal.Calcd.for C40H46O14N4F4Ni3(%):C 45.37,H 4.38,N 5.29.Found(%):C 45.68,H 3.92,N 6.01.IR(KBr,cm-1):3 049,2 910(C-H),1 641(C=N),1 083,624(ClO4-).ESI-MS(m/z):839.17 for[Ni3L2(OH)]+(Supporting information).
1.2.3 Crystal structure determination
Crystalsweremeasured on aBrukerAXS SMART diffractometer (Mo Kα,λ=0.071 073 nm).Data reduction and cell refinement were performed by SMART and SAINT[24].The structures were solved by direct methods(Bruker SHELXTL)and refined on F2by full-matrix least squares(Bruker SHELXTL)using all unique data[25].The non-hydrogen atoms in the structure were treated as anisotropic.All the hydrogen atoms(except the ones bound to water molecules)were placed in calculated positions with fixed isotropic thermal parameters and included in structures factor calculations in the final stage of full-matrix leastsquares refinement.The hydrogen atoms of water moleculeswere located bydifferencemapsand constrained to ride on their parent O atoms.The crystallographic data and the details about the data collection are presented in Table 1.
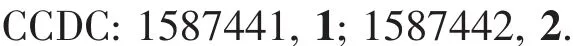

Table 1 Crystal data and structure refinements for 1 and 2
2 Results and discussion
2.1 Crystal structure of complexes
The crystal structures of the complexes are shown in Fig.1 and Fig.2,together with the atom numbering scheme.And selected bond lengths (nm)and angles(°)relevant to the metal coordination spheres of the complexes are listed in Table 2.
The molecular structure of 1 consists of two Coバions and one Coギion,two ligand units and four OAc-anions.Co1 and Co3 ions are hexa-coordinated with a distorted octahedron geometry and have similarcoordinated environment.Co1/Co3 ion resides in the N2O2sites of the Schiff-base ligand,and the basal plane is formed by two imine nitrogen atoms arising from the Schiff-base ligand,and two phenoxo oxygen atoms.The apical position is occupied by the oxygen atom arising from OAc-anion(Co1-O7 0.191 5(7)nm,Co1-O9 0.192 0(6)nm;Co3-O11 0.189 7(2)nm,Co3-O14 0.187 2(6)nm).The Co1/Co3-N and Co1/Co3-O distances in the basal plane are 0.190 1~0.193 4 nm.The values of the O1-Co1-O3 and N1-Co1-N2 angles are 82.2(2)°and 94.9(3)°,and the values of the O4-Co3-O6 and N3-Co3-N4 angles are 83.7(2)°and 94.8(3)°.
Co1,Co2 and Co3 ions are linked by OAc-anions and phenoxo oxygen atoms,and the value of the Co1…Co2…Co3 angle is 155.49(4)°.

Table 2 Selected bond lengths(nm)and angles(°)for complexes 1 and 2

Fig.1 Perspective view of complex 1 with ellipsoids drawn at 30%probability
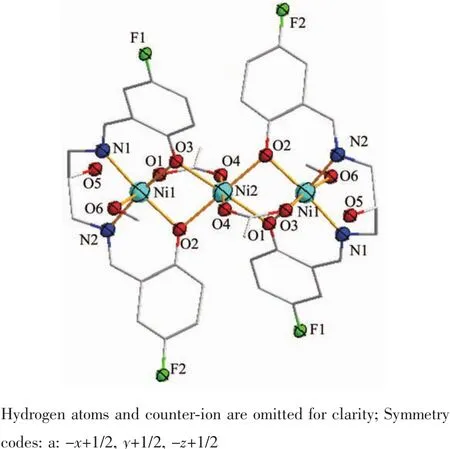
Fig.2 Perspective view of complex 2 with ellipsoids drawn at 30%probability
The molecular structure of 2 consists of three Niギions,two ligand units,two methanol molecules and two OAc-anions.The whole molecule is high symmetrical with Ni2 as symcenter.Ni1 ions are hexacoordinated with a distorted octahedron geometry,in which the apical position is occupied by the oxygen atoms arising from the methanol molecule(Ni1-O6 0.214 5(2)nm)and OAc-anion(Ni1-O3 0.203 7(2)nm),respectively.The basal plane is formed by two imine nitrogen atoms arising from the Schiff-base ligand,and two phenoxo oxygen atoms.Ni2 ion also has a distorted octahedron geometry,in which the apical position is occupied by the oxygen atoms arising from OAc-anion(Ni2-O4 0.201 7(2)nm).The basal plane is formed by four phenoxo oxygen atoms,and all the Ni2-O distances in the basal plane are 0.201 7(2)nm.The value of the Ni1…Ni2…Ni1 angle is 180.00°,so all the Ni ions are in a line.
“因为我亲口告诉过他们。”她笑得有点僵硬,从牙缝里挤出这几个字。她的牙齿上面很整齐,下面却歪歪斜斜。她看着我说:“在诚实派,我们都敞开心扉说出自己的感受。很多人告诉过我他们不喜欢我,但也有很多人没说,谁在乎啊?”
2.2 Spectroscopic analysis of DNA-binding activity
Electronic absorption spectroscopy is one of the most useful methods for DNA-binding studies of metal complexes.The absorption spectra of complex 1 in the absence and presence of calf thymus DNA at different concentrations(8~175 μmol·L-1)are shown in Fig.3.The spectrum of complex 1 shows a very strong absorption at 385 nm,which is attributed to a metalto-ligand charge transfer(MLCT)[26-27].The band shows a hypochromism of 23.4%after adding DNA.The finding supports the hypothesis of a DNA intercalating interaction of the complex[28].The results suggest that complex 1 interacts with DNA most likely through a stacking interaction between the aromatic chromophore and the base pairs of DNA.After intercalating the base pairs of DNA,the π*orbital of the intercalated ligand can couple with the π orbitals of the base pairs,decreasing the π-π*transition energy and resulting in bathochromism.
In order to quantitatively investigate the binding strength of the complex with CT-DNA,the intrinsic binding constant Kbwas obtained by monitoring the changes in absorbance at 385 nm for the complex as increasing concentration of CT-DNA using the equation,cDNA/Eap=cDNA/E+1/(KbE),where Eap=εa-εf,E=εb-εf;εa,εfand εbcorrespond to Aobs/(ccomplexl),the extinction coefficients for the free complex and the complex in the fully bound form,respectively.As shown in Fig.3b,the value of Kbwas obtained from the ratio of slope to the intercept from the plot of cDNA/Eapvs cDNA[29].The Kbvalue is 2.43×105L·mol-1.

Fig.3 (a)UV-visible spectra of complex 1 in the presence of different concentrations of CT-DNA;(b)Plot of cDNA/Eapversus cDNAfor absorption titration of CT-DNA with complex 1
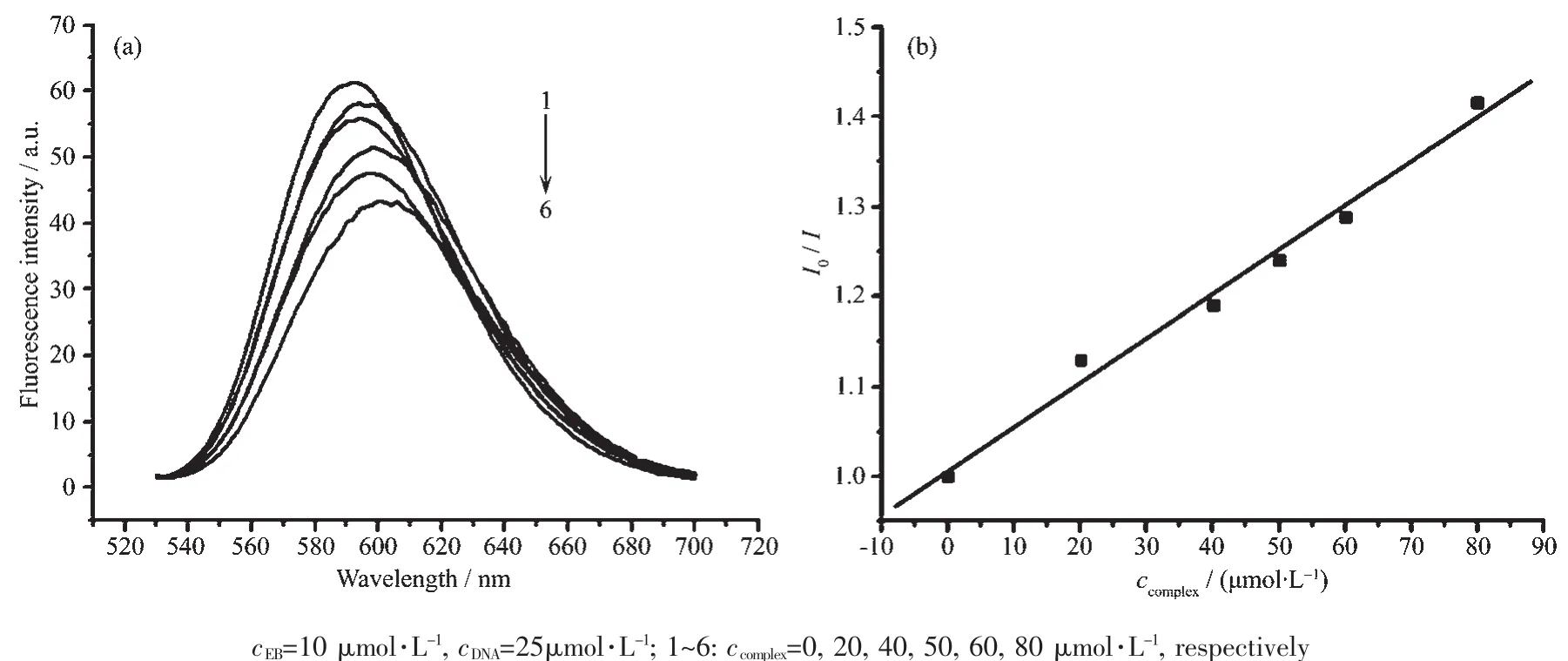
Fig.4 (a)Emission spectra of EB bound to DNA in the presence of complex 1 of different concentration;(b)Stern-Volmer quenching plot of EB bound to DNA by complex 1
It is well known that mixed solutions of DNA and EB show very strongly enhanced fluorescence emission[30]due to intercalation of EB to DNA.The emission spectra of EB bound to DNA in the absence and presence of the complex are given in Fig.4.It can be noted that the fluorescence intensity of the EBDNA solutions decreases with the addition of the complex obviously.The resultssuggestthatthe complex can replace the EB and bind to the DNA molecule.
According to the classical Stern-Volmer equation I0/I=1+Kccomplex,in which I0and I are the fluorescence intensities in the absence and presence of complex,respectively;K is the linear Stern-Volmer quenching constant and ccomplexis the concentration of the complex,the relative binding propensity of the complex to CTDNA can be determined[29].The fluorescence intensity at(597±4)nm(λex=520 nm)of EB in the bound form was plotted against the compound concentration.The quenching constant K of the complex is obtained by the slopes of the plot of I0/I vs ccomplexin Fig.4b.The calculated quenching constant K is 4.92×103L·mol-1,which is smaller than the values of classical intercalative mode[28,31].
2.3 Electrochemical studies
The electrochemical properties of complex 1 were studied by cyclic voltammetry in 50 mmol·L-1Tris-HCl/50 mmol·L-1NaCl buffer solution(pH=7.2)using tetrabutyl ammonium perchlorate(TBAP)as supporting electrolyte and a sweep range of-1.0~-0.4 V.The cyclic voltammograms of complex 1 in the absence and in the presence of CT-DNA are shown in Fig.5.
Fig.5 Cyclic voltammograms of complex 1 in the absence(a)and presence(b)of CT-DNA
In the absence of DNA,the CV curve of complex 1 has a cathodic peak at-0.834 V and an anodic peak at-0.729 V.The separation between the cathodic and anodic peak potential,ΔE=0.105 V,indicates an irreversible process[32].Addition of CT-DNA to complex 1 results in a decrease in the cathodic and anodic peak current,which could be explained by a reduction in the apparent diffusion coefficient of the complex on binding to DNA[33].These results again indicate that an intercalative interaction occurred between complex 1 and DNA[34].
2.4 Viscosity study

Fig.6 Effects of increasing amounts of complex 1 on the relative viscosities of CT-DNA at(23±0.1)℃
2.5 DNA cleavage
In order to evaluate the function of complex 1 to incise pBR322 DNA,the cleavage reaction of supercoiled pBR322 DNA was monitored by agarose gelelectrophoresis.The cleavage products were subjected to gel electrophoretic separation and the gels were analyzed after ethidium bromide staining.As shown in Fig.7,with increasing concentrations of the complex,the amount of FormⅠ(supercoiled form)of DNA diminished obviously,whereas FormⅡ(nicked form)and FormⅢ (linear form)increased.This suggests that complex 1 can cleave the DNA effectively.
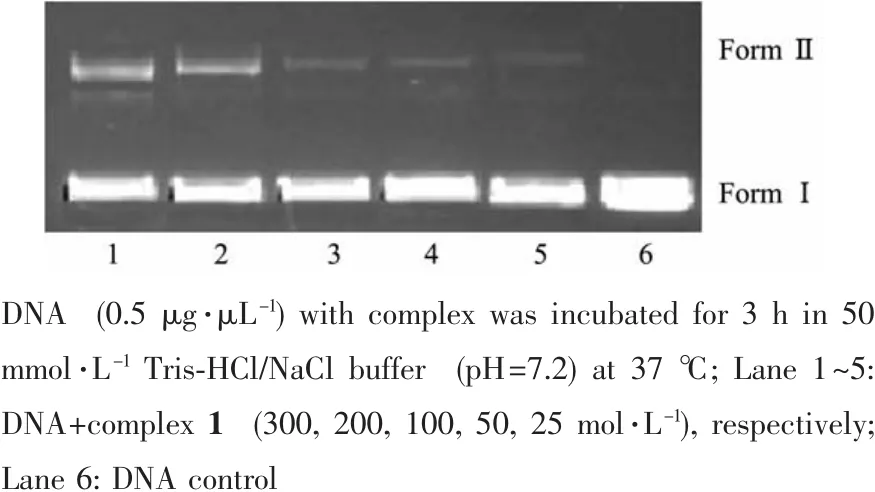
Fig.7 Agarose gel electrophoresis of pBR322 plasmid DNA in the presence of different concentrations of complex 1
In order to further clarify the DNA cleavage mechanism,some scavenging agents,hydroxyl radical scavengers (5 mmol·L-1DMSO and 5 mmol·L-1EtOH),superoxide scavenger (5 mmol·L-1KI)and singlet oxygen scavenger (5 mmol·L-1NaN3),were added in the incubation solution.No obvious change of DNA cleavage activity of the complex is observed in the presence or absence of the scavenging agents(Fig.8).Therefore,DNA cleavage promoted by this complex might not occur by an oxidative pathway but rather by a hydrolytic pathway and/or intercalation.This result is consistence with the deduction from UVVis and fluorescence spectroscopy studies.

Fig.8 Gel electrophoresis diagram showing the cleavage of pBR322 DNA (0.5 μg·μL-1)by complex 1 at different scavenging agents in 50 mmol·L-1Tris-HCl/NaCl buffer(pH=7.2)and 37℃for 3 h
2.6 Magnetism
The magnetic susceptibilities of complexes 1 and 2 were measured on a SQUID magnetometer within the temperature range of 2.0~300 K.The temperature dependence of the magnetic susceptibilities of 1 and 2,in the form of χmand χmT,are shown in Fig.9 and 10.The χmT product is 3.01 emu·K·mol-1for 1 at room temperature,which is obvious smaller than that of[Co3(pytrz)6(H2O)6](NO3)6[38].The χmT value decreases slowly upon cooling till about 100 K,then decreases rapidly and reaches 2.73 emu·K·mol-1at 2 K.The magnetic behavior reveals the antiferromagnetic interactions among three Co centers in 1.The χmand χmT versus T plot of 2 is very different from that of 1.The χmT value is 0.46 emu·K·mol-1at room temperature,which is obvious smaller than that of[Ni3(dpa)4(CH3CN)2](PF6)2[15].The χmT value decreases rapidly upon cooling till about 100 K,then decreases slowly and reaches 0.01 emu·K·mol-1at 2 K,which indicates antiferromagnetic interactions among three Niギcenters.
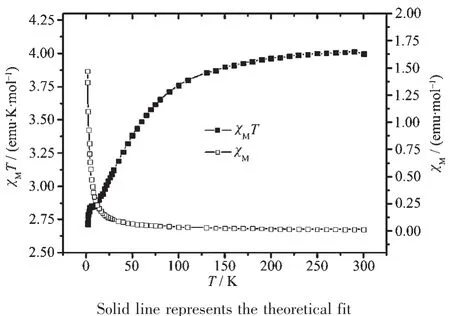
Fig.9 Magnetic susceptibility data for 1
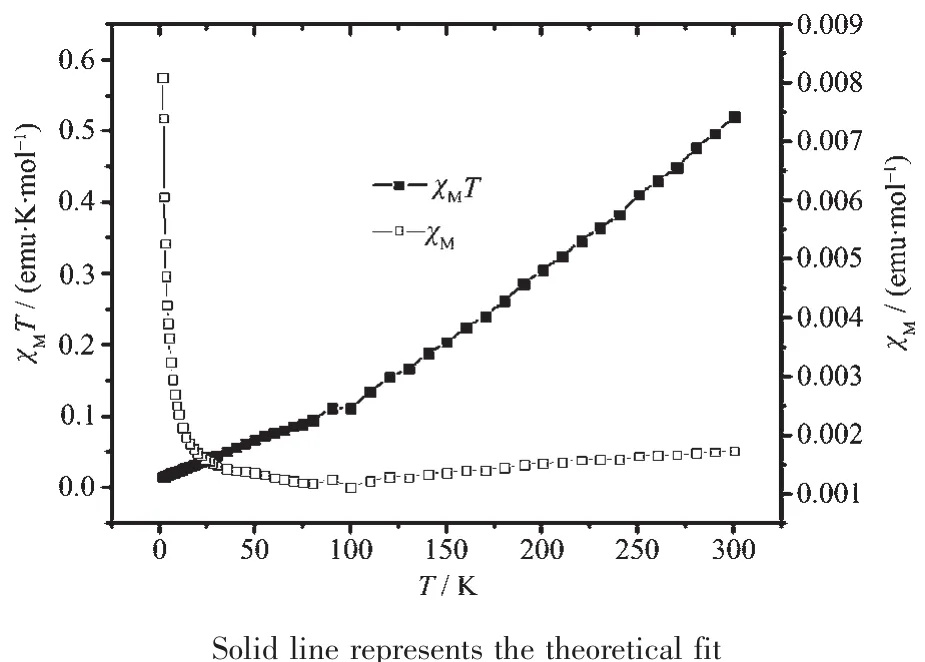
Fig.10 Magnetic susceptibility data for 2

The magnetic susceptibility data was first treated on the basis of Eq.(1).All the symbols in Eq.(1)have their usual meaning.In this expression,2J(in the spin Hamiltonian H=2JS^1S^2)is the singlet-triplet splitting or exchange integral,N is Avogadro′s number, β is the Bohr magneton,k is the Boltzmann constant,T is the absolute temperature,g is Lande g factor,z is the number of nearest-neighboring Co centers(z=3),q is the ratio of the paramagnetic impurity and j′accounts for the presence of magnetic interactions between neighboring Co centers.An excellent set of fit parameters are g=2.0,J=-212.91 cm-1,ρ=1.29 for complex 1,which is similarity with the trinuclear Co complex[38],and g=2.2,J=-189.64 cm-1,ρ=0.003 5 for complex 2.The g value for 2 is larger than 2.0 indicating that orbital contribution from Ni2+,which is similar to those in other Ni36+compounds[15,39],but the J value is significantly smaller than that of the trinuclear Ni complexes(-242 cm-1)[15].
3 Conclusions
In summary,we have prepared and structurally characterized two linear trinuclear complexes.DNA-binding activity studies showed that that complex 1 interacts with DNA by intercalative interaction,and the DNA cleavage mechanism is by hydrolytic pathway.The variable-temperature magnetisms of complexes 1 and 2 show that there are antiferromagnetic couplings among the three metal centers.
Supporting information is available at http://www.wjhxxb.cn
[1]Tangoulis V,Raptopoulou C P,Paschalidou S,et al.Angew.Chem.,Int.Ed.,1997,36:1083-1085
[2]Mukherjee A,Nethaji M,Chakravarty A R.Angew.Chem.,Int.Ed.,2004,43:87-90
[3]Nishida Y,Kida S.J.Chem.Soc.Dalton Trans.,1986,12:2633-2640
[4]Hilms E,Elias H,Paulus H.J.Chem,Soc.Dalton Trans.,1986,10:2169-2172
[5]Breslow R,Zhang X J,Xu R,et al.J.Am.Chem.Soc.,1996,118:11678-11679
[6]Adam W,Fell R T,Stegmann V R,et al.J.Am.Chem.Soc.,1998,120:708-714
[7]Routier S,Bernier J L,Waring M J,et al.J.Org.Chem.,1996,61:2326-2331
[8]Lamour E,Routier S,Bernier J L,et al.J.Am.Chem.Soc.,1999,121:1862-1869
[9]Maleev V I,North M,Larionov V A,et al.Adv.Synth.Catal.,2014,356:1803-1810
[10]Kunkely H,Vogle A.J.Photochem.Photobiol.A:Chem.,2001,138:51-54
[11]Honda T,Kojima T,Fukuzumi S.J.Am.Chem.Soc.,2012,134:4196-4206
[12]Salehi M,Amirnasr M,Meghdadi S,et al.Polyhedron,2014,81:90-97
[13]Aduldecha S,Hathaway B.J.Chem.Soc.Dalton Trans.,1991,4:993-998
[14]Chien C H,Chang J C,Yeh C Y,et al.Dalton Trans.,2006,17:2106-2113
[15]Cotton F A,Murillo C A,Wang Q S.Inorg.Chem.Commun.,2007,10:1088-1090
[16]Cheng Q R,Zhang F Q,Zhou H,et al.J.Coord.Chem.,2015,68(11):1997-2005
[17]Cheng Q R,Yu L,Li P,et al.Transition Met.Chem.,2015,40:789-797
[18]Cheng Q R,You D,Hu H,et al.J.Coord.Chem.,2014,67(5):910-920
[19]Cheng Q R,Li P,Zhou H,et al.J.Coord.Chem.,2014,67(9):1584-1595
[20]Cheng Q R,Zhou H,Pan Z Q.J.Mol.Struct.,2014,1071:255-262
[21]Cheng Q R,Zhou H,Pan Z Q.Polyhedron,2014,81:668-674
[22]CHENG Qing-Rong(程清蓉),ZHANG Ya(张雅),ZHOU Hong(周红),et al.Chinese J.Inorg.Chem.(无机化学学报),2014,33(11):2591-2500
[23]Cheng Q R,Zhou H,Pan Z Q.Transition Met.Chem.,2012,37:407-414
[24]SMART and SAINT,Area Detector Control and Integration Software,Siemens Analytical X-ray Systems Inc,Madison,WI,1996.
[25]Sheldrick G M.SHELXTL Version 5.1,Software Reference Manual,Bruker AXS Inc.,Madison,WI,1997.
[26]Neves A,Brito M A de,Vencato I,et al.Inorg.Chem.,1996,35:2360-2368
[27]Gaber B P,Miskowski V,Spiro T G.J.Am.Chem.Soc.,1974,96:6868-6873
[28]Tian J L,Feng L,Gu W,et al.J.Inorg.Biochem.,2007,101:196-202
[29]Wolfe A,Shimer G H,Meehan T.Biochemistry,1987,26:6392-6396
[30]Meyer-Almes F J,Porschke D.Biochemistry,1993,32:4246-4253
[31]Qian J,Gu W,Liu H,et al.Dalton Trans.,2007:1060-1066
[32]Zhang J A,Pan M,Yang R,et al.Polyhedron,2010,29:590-591
[33]Ibrahim M S.Anal.Chim.Acta,2001,443:65-71
[34]Michael T C,Marisol R,Alle J B.J.Am.Chem.Soc.,1989,111(24):8901-8911
[35]Satyanarayana S,Dabrowiak J C,Chaires J B.Biochemistry,1993,32:2573-2584
[36]Sun J,Shuo S,An Y,et al.Polyhedron,2008,27:2845-2850
[37]Liu J G,Ye B H,Li H,et al.J.Inorg.Biochem.,1999,76:265-271
[38]Ding B,Yang E C,Zhao X J,et al.J.Coord.Chem.,2008,23(61):3793-3799
[39]Clérac R,Cotton F A,Dunbar K R,et al.Inorg.Chem.,1999,38:2655-2657

