青藏高原濒危植物唐古特大黄的遗传多样性
王爱兰,李维卫
1 鲁东大学生命科学学院, 烟台 264025 2 鲁东大学学报编辑部, 烟台 264025
青藏高原濒危植物唐古特大黄的遗传多样性
王爱兰1,李维卫2,*
1 鲁东大学生命科学学院, 烟台 264025 2 鲁东大学学报编辑部, 烟台 264025
唐古特大黄(Rheumtanguticum)是中国传统的中藏药材,近几年由于生境的严重破坏,已濒临灭绝,并被列入濒危植物名单。为了探索唐古特大黄物种濒危的原因并保护其野生资源,本研究采集了9个居群87个个体的唐古特大黄样本,基于该物种的叶绿体基因trnS-G序列对其进行了遗传多样性研究。结果表明,唐古特大黄物种具有较高的遗传多样性水平(Ht=0.694),其中95.97%的遗传分化来自于居群间(GST=0.960),4.03%的遗传分化来自于居群内(Hs=0.028)。AMOVA分析也显示唐古特大黄居群间基因流较小(Nm=0.01),存在较高的遗传分化(FST=0.9631)。唐古特大黄较高的遗传多样性水平可能与该物种较长的进化史和生活史有关,居群间较高的遗传分化可能与高山地区特殊的地理环境和人类活动有关。根据研究结果,建议对唐古特大黄所有野生居群进行就地保护,同时收集种质资源开展异地繁殖工作,以保护物种的遗传多样性,维持其进化潜力。
唐古特大黄;trnS-G序列;遗传多样性;基因流
青藏高原是世界上海拔最高、面积最大的高原,具有丰富的物种多样性,是全球25个生态热点之一,有超过12000种植物生长在青藏高原及其邻近地区[1- 2]。由于高海拔和干旱等极端环境的存在,导致该地区生态系统对环境变化及人类活动非常敏感[3],已有1000多种植物被列为濒危物种[4]。过去,青藏高原地区居民依靠野生植物资源获得收入,导致野生资源过分采挖,由于环境恶劣,致使这些物种很难恢复,从而引起一系列生态问题。
唐古特大黄(RheumtanguticumMaxim. ex Balf.)是蓼科(Polygonaceae)大黄属植物,主要分布在青藏高原地区,其根入药,具有通便、抗炎等功效,是传统的中藏药材[5]。由于青藏高原地区的野生资源品质较高,出口到日本、韩国和其他地区[6]。近几年由于经济利益的驱使导致人类过度采挖,使得唐古特大黄野生分布区急剧减少,因而被列入濒危物种名单[4]。
根据实地调查,发现大黄野生资源已经非常稀少,对其野生居群的保护迫在眉睫。居群遗传多样性研究对濒危物种的保护具有重要意义[7- 9],相关研究工作也已逐步开展,如Chen等[10]、Hu等[11]和Wang等[12]分别利用SSR、ISSR和基因片段trnL-F对唐古特大黄进行了遗传多样性的分析,迄今为止,尚未见利用trnS-G序列研究其遗传多样性的报道。叶绿体基因片段trnS-G序列因其具有较快的进化速率和较高的遗传变异等特点,已被广泛应用到物种遗传多样性和种内种间基因交流的研究中[13-14]。因此,研究选择trnS-G序列对唐古特大黄物种多样性水平及居群间的基因流进行了研究。本研究重点探讨以下问题:1)唐古特大黄的遗传多样性水平;2)唐古特大黄居群的遗传结构;3)通过多样性水平调查,为该物种的遗传保护提出合理建议。
1 材料与方法
1.1 实验材料
本研究采用了唐古特大黄9个居群87个样本,所有样本均分布在青藏高原地区(图1),采样地点基本覆盖唐古特大黄现存野生居群的分布地区,样本信息见表1。采样时,样本间隔根据居群大小确定在3—5m之间,采集的叶片用硅胶干燥后保存备用。
1.2 DNA提取,扩增及测序
采用改良的CTAB法[15]从干燥的唐古特大黄叶片中提取基因组DNA。利用Hamilton[16]设计的‘S’ (5′-GCCGCTTTAGTCCACTCAGC- 3′)和 ‘G’ (5′-GAACGAATCACACTTTTACCAC- 3′)引物进行PCR扩增,扩增体系为:25 μL体系,包括40 ng模板DNA,17.5 μL无菌水,2.5 μL 10×Taq 酶缓冲液,0.5 μL 10 mmol/L dNTPs,5 μmol/L引物各1.25 μL,1 U DNA Taq酶。扩增程序为:94 ℃预变性4.0 min;94 ℃变性1.0 min,51℃退火50 s,72 ℃延伸1.75 min,循环38次;最后72 ℃延伸8 min。PCR产物用1.0%的琼脂糖凝胶进行电泳检测,采用纯化试剂盒进行纯化(购自上海生工),然后送至北京六合华大基因公司进行核苷酸序列分析。

图1 唐古特大黄样本分布图Fig.1 The samples distributions of R. tanguticum
1.3 数据分析
用Clustal X软件[17]进行序列比对并进行手工校正。在进行单倍型分析时,插入缺失和碱基替换作相同处理。利用DnaSP5.1软件[18]计算居群的单倍型多态性(Hd)、核苷酸多态性(pi)和物种水平居群间的平均基因流(Nm)。采用PERMUT软件[19]计算居群内平均遗传多样性(Hs)、总遗传多样性(Ht)和居群遗传分化系数(GST)。应用Arlequin3.1软件进行分子变异分析(AMOVA)[20],并进行1000次随机抽样的显著性检验。采用TFPGA软件对遗传距离矩阵和地理距离矩阵进行mantel检测,并进行1000次随机置换的显著性检验。另外,采用MEGA 6.0的邻接法NJ(Neighbor-Joining)进行聚类分析,建立无根系统发育树[21]。采用NETWORK 4.6.1软件[22]中的中点连接法进行种内谱系的单倍型网络分析,并用溯祖理论来推测单倍型间的亲缘关系。
2 结果
2.1 CpDNA序列变异和单倍型分布
87个样本获得的trnS-G序列长度在944—954 bp之间,校正后总长度为965 bp。共检测到23个核苷酸变异位点,其中10个是碱基缺失或插入造成的突变位点,13个为核苷酸置换位点(表2)。

表1 用于实验的珊瑚菜材料及其来源
基于这些核酸突变,在9个居群的87个样本中共发现4种单倍型H1—H4(表3,图2),这些单倍型的代表序列均提交到GenBank数据库(表3)。H1单倍型是Pop4居群特有单倍型,包含7个样本;H2单倍型分布在Pop3和Pop8两个居群,共16个样本;H3是分布最广泛且最丰富的单倍型,分布在5个居群(Pop1、Pop2、Pop5、Pop7和Pop9),共48个样本,占采样总数的55.17%;H4单倍型出现在Pop3和Pop6 这两个居群,分别包括7个和9个样本(表3)。

表2 唐古特大黄4个单倍型变异位点信息
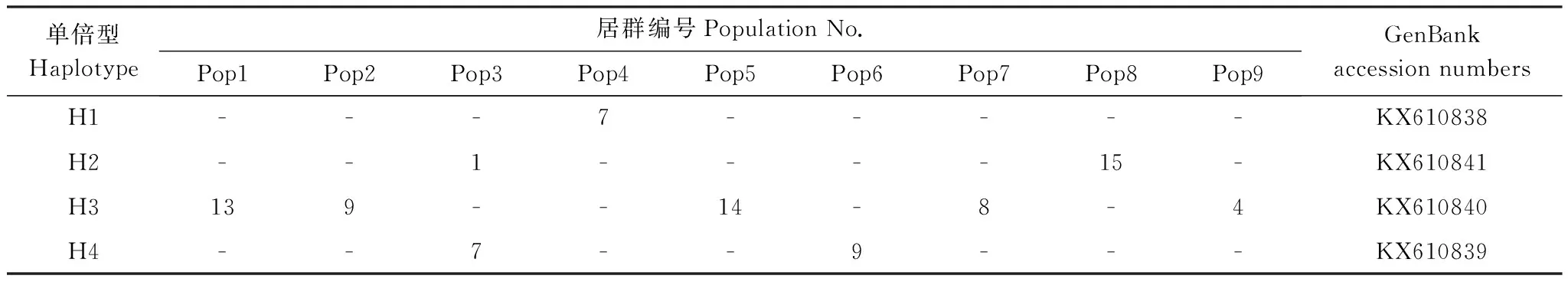
表3 各居群的单倍型分布数目
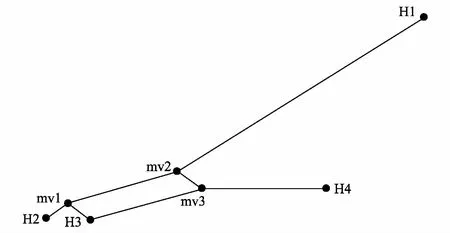
图2 NETWORK单倍型网络图 Fig.2 Haplotype relationship constructed by the software NETWORK 图中圆圈代表单倍型,H代表检测到的单倍型,mv代表缺失的单倍型
Network单倍型网络图显示,4个单倍型组成了星型遗传结构(图2)。单倍型H1、H2和H4在网络的边缘,表明可能是新近发生的单倍型;H3单倍型和3个缺失的单倍型在星型网络中心,且H3单倍型出现的频率最高,推测为祖先单倍型。
2.2 遗传多样性分析
DnaSP分析显示,物种水平上,单倍型多样性Hd为0.6218,核苷酸多样性指数pi为0.0049;9个居群的单倍型多样性(Hd)范围在0—0.25之间,核苷酸多样性(Pi)在0—0.0026之间。其中,Pop3居群含有H2和H4两种单倍型,其单倍型多样性和核苷酸多样性最高(Hd=0.25,Pi=0.0026),而其他8个居群因只有一个单倍型(表3),居群单倍型多样性和核苷酸多样性均为0。PERMUT分析显示,居群总的遗传多样性指数(Ht)为0.694,表明了较高的遗传多样性水平;居群内遗传多样性指数(Hs)为0.028,表明居群内遗传多样性水平较低。
2.3 遗传结构分析
由居群总的遗传多样性指数(Ht)和居群内遗传多样性指数(Hs)推算,居群间遗传分化较大,4.03%的变异发生在居群内,而其余95.97%的遗传变异发生在居群间,居群间分化系数(GST)为0.960,也验证了上述结果。AMOVA分析得出居群间遗传分化系数(FST)为0.9631,同样表明遗传变异主要发生在居群间。DnaSP分析显示,居群间基因流(Nm)极小,仅为0.01,表明不同居群间存在有限的基因交流,这也支持了PERMUT和AMOVA的分析结果。
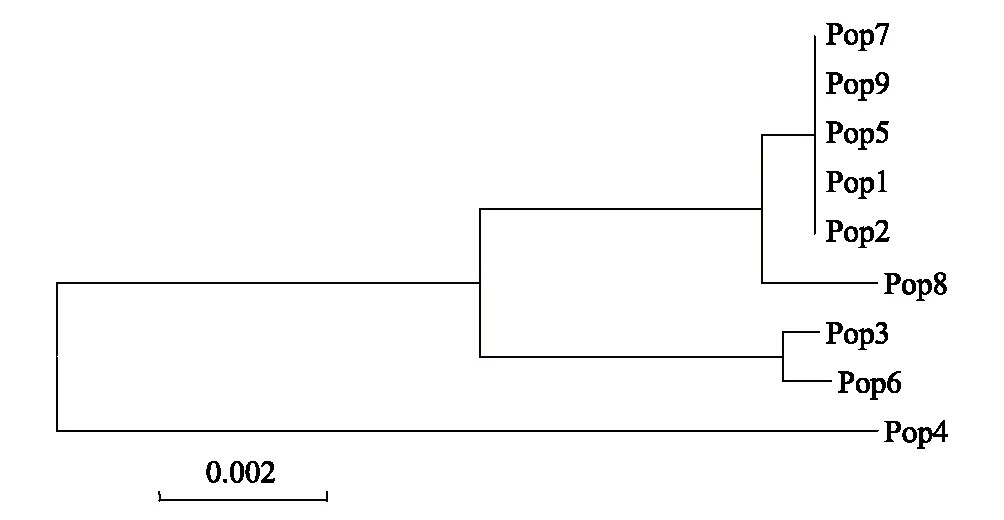
图3 基于遗传距离构建的唐古特大黄居群聚类分析图 Fig.3 NJ dendrogram based on genetic distances of R. tanguticum
基于遗传距离构建的NJ树显示了3个组(图3)。Pop3和Pop6聚为一组,表明了相近的亲缘关系;Pop7、Pop9、Pop5、Pop1和Pop2聚为一个小分支,又与Pop8聚为一个较大的分支,表明相对较近的亲缘关系;Pop4没有与其他居群聚在一起,表明其与其他居群亲缘关系较远。聚类分析表明,地理距离较近的居群并未聚到一起,说明地理距离与遗传距离没有直接的相关关系。Mantel检测结果(r=0.2461,p=0.2060)也验证了这一点,表明居群地理距离与遗传距离存在不显著的正相关关系。
3 讨论
3.1 唐古特大黄的遗传多样性
遗传多样性降低被认为是物种濒危的主要原因[23-26]。但是某些研究发现濒危物种也具有较高的遗传多样性,例如罂粟科的毛黄堇Corydalistomentella[27](Ht=0.804)和伞形科的珊瑚菜Glehnialittoralis[28](Ht=0.2471)。
本研究基于叶绿体基因trnS-G序列构建了9个野生唐古特大黄居群的遗传结构,结果显示该物种具有较高的遗传多样性,这一结果与SSR(He=0.515)、ISSR(He=0.2689)和trnL-F(Ht=0.632)的研究结果[10-12]一致。物种遗传多样性应该是长期形成的遗传分化,唐古特大黄较高的遗传多样性可能与该物种的进化史和生活史有关。大黄属物种分化时间发生在360万—679.6万年前[29-30],唐古特大黄经历了漫长的进化过程,保存了丰富的基因库;且该物种为多年生草本植物,具有较长的生活史和复杂的异交繁育系统,决定了该物种具有较高的基因扩散和交流能力,确保了较高的遗传多样性[31]。
3.2 唐古特大黄遗传结构
唐古特大黄居群间存在强烈的遗传分化。Buso等[32]认为,当GST>0.25时,表明居群间存在极强的遗传变异。本研究中,唐古特大黄居群GST为0.960,表明唐古特大黄居群间存在强烈的遗传分化,其中95.97%的遗传分化来源于居群间,4.03%的遗传分化来自居群内。AMOVA分析与DnaSP、PERMUT分析均取得了一致的结果,均表明遗传分化主要来自于居群间。产生这种遗传结构的原因可能与其特殊的高山生态环境有关。唐古特大黄主要生长在青藏高原地区,青藏高原的高山和深谷可能阻碍了花粉和种子的传播,从而降低了居群间的基因流,导致居群间遗传分化加大。唐古特大黄居群间较低的基因流(Nm=0.01)也证实了这一推论。另外,在经济利益的驱使下,人类对唐古特大黄野生资源过度采挖,也可能导致部分居群消失,造成某些等位基因的缺失,打破了居群间连续的基因交流,最终导致居群遗传分化加剧。单倍型网络图分析结果验证了这一点,4个单倍型的简约网络图并未确定所有祖源单倍型,而是表明存在3个缺失的古老单倍型。
聚类分析结果表明,9个居群分为3组,居群间的遗传距离与地理距离无显著的正相关关系,地理相距较近的居群并没有聚到一起,如西藏地区的居群4和居群9以及青海地区的6个居群分别聚到不同的分支(图3),推测现存居群可能起源于不同的冰期避难所,支持唐古特大黄经历了冰期的退缩和间冰期的扩张,在间冰期扩张过程中发生了遗传漂变,部分单倍型丢失,居群保留下来的单倍型由于来源不同,造成居群间的遗传分化较大。遗憾的是根据本研究结果,尚无法得知唐古特大黄的避难所所在,增加数据量对该植物开展分子亲缘地理学研究,可能会更好地揭示青藏高原地区第四纪冰期以来该植物居群变化的历史过程。
3.3 唐古特大黄遗传多样性保护
遗传变异是物种进化和发展的基础,因此居群遗传多样性的保护是濒危物种保护的基本要求。Hamrick和Bhagwat等指出,若某一居群遗传多样性占总遗传多样性比例达到99%,则建议该居群单独保护以维持其较高的多样性[24,33],否则应当对所有居群加以保护。基于以上调查结果,发现9个居群中遗传多样性最高的为Pop3(Hd=0.25,Pi=0.0026),占总遗传多样性比例为53.06%,未达到单独保护的标准。另外,唐古特大黄遗传多样性主要存在于居群间(GST=0.960,FST=0.9631),而居群内的遗传多样性较低(Hs=0.028),居群呈片段化存在,居群间的基因交流受阻(Nm=0.01),这种格局极易造成居群的退化甚至灭绝。综合以上因素,提出如下建议:1)对唐古特大黄所有野生居群进行就地保护,包括一些遗传多样性程度不高,但拥有特殊等位基因的种群,维持居群数量,确保居群间的基因交流,维持物种的进化潜力;为了避免遗传多样性高的小居群走向灭绝,如果就地保护不利的情况下建议迁地保护;2)采集和保存种子建立种质资源库,开展异地繁育等保护措施。以上这些保护策略将有利于增加唐古特大黄居群数量,并逐步扩大其分布范围。
4 结论
本研究基于叶绿体基因片段trnS-G 的序列变异对9个唐古特大黄野生居群的遗传结构进行了分析,结果表明该物种具有较高的遗传多样性,遗传分化主要来自于居群间,居群间的基因流较低。产生这种遗传结构的原因可能与唐古特大黄特殊的高山生境和人类活动的干扰有关。为保护唐古特大黄的遗传多样性,建议对其所有的野生居群进行就地保护,并采集和保存种子建立异地种质资源库,开展异地繁育等保护工作。
致谢:感谢刘建全教授在野外考察和材料收集过程中给予的指导和帮助。
[1] 施雅风, 李吉均, 李炳元. 青藏高原晚新生代隆升与环境变化. 广州: 广东科学技术出版社, 1998.
[2] Myers N, Mittermeier R A, Mittermeier C G, da Fonseca G A B, Kent J. Biodiversity hotspots for conservation priorities. Nature, 2000, 403(6772): 853- 858.
[3] Zhang X S, Yang D A, Zhou G S, Liu C Y, Zhang J. Model expectation of impacts of global climate change on biomes of the Tibetan Plateau//Omasa K, Kai K, Taoda H, Uchijima Z, Yoshino M, eds. Climate Change and Plants in East Asia. Tokyo: Springer-Verlag, 1996.
[4] 傅立国. 中国植物红皮书——稀有濒危植物(一). 北京: 科学出版社, 1991.
[5] Yang M H, Zhang D M, Zheng J H, Liu J Q. Pollen morphology and its systematic and ecological significance inRheum(Polygonaceae) from China. Nordic Journal of Botany, 2001, 21(4): 411- 418.
[6] 杨永昌. 藏药志. 青海: 青海人民出版社, 1991.
[7] Hamrick J L, Godt M J W. Allozyme diversity in plant species//Brown A H D, Clegg M T, Kahler A L, Weir B S, eds. Plant Population Genetics, Breeding and Genetic Resources. Sunderland: Sinauer Associates, 1990: 43- 63.
[8] Gitzendanner M A, Soltis P S. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany, 2000, 87(6): 783- 792.
[9] Cole C T. Genetic variation in rare and common plants. Annual Review of Ecology, Evolution, and Systematics, 2003, 34(1): 213- 237.
[10] Chen F J, Wang A L, Chen K M, Wan D S, Liu J Q. Genetic diversity and population structure of the endangered and medically importantRheumtanguticum(Polygonaceae) revealed by SSR markers. Biochemical Systematics and Ecology, 2009, 37(5): 613- 621.
[11] Hu Y P, Wang L, Xie X L, Yang J, Li Y, Zhang H G. Genetic diversity of wild populations ofRheumtanguticumendemic to China as revealed by ISSR analysis. Biochemical Systematics and Ecology, 2010, 38(3): 264- 274.
[12] Wang A L, Li W W. Genetic diversity ofRheumtanguticum(Polygonaceae), an endangered species on Qinghai-Tibetan Plateau. Biochemical Systematics and Ecology, 2016, 69(1): 132- 137.
[13] Sharma A, Poudel R C, Li A R, Xu J C, Guan K Y. Genetic diversity ofRhododendrondelavayivar.delavayi(C. B. Clarke) Ridley inferred from nuclear and chloroplast DNA: implications for the conservation of fragmented populations. Plant Systematics and Evolution, 2014, 300(8): 1853- 1866.
[14] Lu Z Q, Chen P, Bai X T, Xu J M, He X D, Niu Z M, Wan D S. Initial diversification, glacial survival, and continuous range expansion ofGentianastraminea(Gentianaceae) in the Qinghai-Tibet Plateau. Biochemical Systematics and Ecology, 2015, 62(1119): 219- 228.
[15] Doyle J J, Doyle J L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin, 1987, 19: 11- 15.
[16] Hamilton M B. Four primer pairs for the amplification of chloroplast intergenic regions with intraspecific variation. Molecular Ecology, 1999, 8(3): 521- 523.
[17] Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 1997, 25(24): 4876- 4882.
[18] Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 2009, 25(11): 1451- 1452.
[19] Pons O, Petit R J. Measuring and testing genetic differentiation with orderedversusunordered alleles. Genetics, 1996, 144(3): 1237- 1245.
[20] Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics, 2007,2005(1), 1: 47- 50.
[21] Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 2013, 30(12): 2725- 2729.
[22] Bandelt H J, Forster P, Röhl L. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 1999, 16(1): 37- 48.
[23] Barrett S C H, Kohn J R. Genetic and evolutionary consequences of small population size in plants: implications for conservation//Falk D A, Holsinger K E, eds. Genetics and Conservation of Rare Plants. Oxford: Oxford University Press, 1991: 3- 30.
[24] Hamrick J L, Godt M J W. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society B: Biological Sciences, 1996, 351(1345): 1291- 1298.
[25] 肖猛, 李群, 郭亮, 唐琳, 王丽, 陈放. 四川西部濒危植物桃儿七遗传多样性的RAPD分析. 生态学报, 2015, 35(5): 1488- 1495.
[26] Ross A A, Travers S E. The genetic consequences of rarity in the western prairie fringed orchid (Platantherapraeclara). Conservation Genetics, 2016, 17(1): 69- 76.
[27] Zhang Z X, Niu H Y, Guo X, Wang D, Eaton W D. Genetic diversity and genetic structure ofCorydalistomentellaFranch. (Papaveraceae), an endangered herb species from Central China. Biochemical Systematics and Ecology, 2015, 63(346): 27- 33.
[28] Wang A L, Zhang P, Liu X, Liang J G, Li W W. Genetic structure and diversity ofGlehnialittoralis, an endangered medicinal plant in China. Biochemical Systematics and Ecology, 2016, 66: 265- 271.
[29] Wang A L, Yang M H, Liu J Q. Molecular phylogeny, recent radiation and evolution of gross morphology of the rhubarb genusRheum(Polygonaceae) inferred from chloroplast DNAtrnL-F sequences. Annals of Botany, 2005, 96(3): 489- 498.
[30] Sun Y S, Wang A L, Wan D S, Wang Q, Liu J Q. Rapid radiation ofRheum(Polygonaceae) and parallel evolution of morphological traits. Molecular Phylogenetics and Evolution, 2012, 63(1): 150- 158.
[31] Loveless M D, Hamrick J L. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics, 1984, 15(1): 65- 95.
[32] Buso G S C, Rangel P H, Ferreira M E. Analysis of genetic variability of South American wild rice populations (Oryzaglumaepatula) with isozymes and RAPD markers. Molecular Ecology, 1998, 7(1): 107- 117.
[33] Bhagwat R M, Banu S, Dholakia B B, Kadoo N Y, Lagu M D, Gupta V S. Evaluation of genetic variability inSymplocoslaurinaWall. from two biodiversity hotspots of India. Plant Systematics and Evolution, 2014, 300(10): 2239- 2247.
Geneticdiversityofanendangeredspecies,Rheumtanguticum(Polygonaceae),ontheQinghai-TibetanPlateau
WANG Ailan1, LI Weiwei2,*
1SchoolofLifeandSciences,LudongUniversity,Yantai264025,China2EditorialOfficeofLudongUniversityJournal,LudongUniversity,Yantai264025,China
Rheumtanguticum(Polygonaceae), an endangered traditional Chinese and Tibetan medicines, is distributed in the Tibetan Plateau. To investigate endangered reasons and protect the wild resources, we collected 87 individuals fromnine populations ofR.tanguticum, and estimated the genetic diversity of this speciesbased on the cpDNAtrnS-G region.The results showed that there was high genetic diversity within this species. The length of the totaltrnS-G region of the 87 individuals ranged from 944 to 954bp, and covered 965 nucleotide positions after being aligned. Variations were detected in 23 nucleotides, including 13 nucleotide substitutions and 10 indel sites. On the basis of these nucleotide mutations, a total of four nucleotide haplotypes (H1—H4) were identified. The development of a parsimony network showed that the four haplotypes made up a shallow star genetic tree. Haplotypes H1, H2, and H4 were located at the tips of the network, representing recent haplotypes.Haplotype H3 and three missing haplotypes were in the center of the star network.Moreover,since haplotype H3 had the highest frequency, it was identified as an ancestral haplotype.The total diversity (Ht) among the individuals collected was 0.694, indicating a high genetic diversity. In addition, a high level of population differentiation was observed (GST=0.960), as determined by PERMUT analysis, showing 95.97% genetic differentiation between populations, whereas the remainder of the total genetic variation (4.03%) was attributed to within-population diversity (Hs=0.028). AMOVA analysis (FST=0.9631) also indicated that the genetic diversity among populations was greater than that within populations. The value for gene flow was fairly low (Nm=0.01), indicating a limited gene flow between populations, which is consistent with the findings of PERMUT and AMOVA analysis.The NJ tree of populations based on genetic distance revealed three supported groups. However, populations with close geographic distance did not cluster together, indicating that there was no direct relationship between the geographic distances and genetic distances of the nine populations, which was confirmed by the Mantel test(r=0.2461,p=0.2060). The high genetic diversity of the examined populations is probably associated with the long evolutionary history and life history ofR.tanguticum.The large genetic differentiation between populations might be related tothe alpine geographical environment and human activity. Therefore, we suggest that more wild populations with high genetic diversity should be protected by an in-situ conservation program at sites distant from human activity. We also recommend the collection and conservation of seeds from populations with rich genetic diversity for the ex-situ conservation of genetic stocks and their propagation. These conservation measures will certainly provide the basic resources required to increase the number of populations of this species and extend its range of distribution.
Rheumtanguticum;trnS-G region; genetic diversity; gene flow
国家自然科学基金项目(31000104)
2016- 08- 09; < class="emphasis_bold">网络出版日期
日期:2017- 07- 11
*通讯作者Corresponding author.E-mail: lwwal@163.com
10.5846/stxb201608091635
王爱兰,李维卫.青藏高原濒危植物唐古特大黄的遗传多样性.生态学报,2017,37(21):7251- 7257.
Wang A L, Li W W.Genetic diversity of an endangered species,Rheumtanguticum(Polygonaceae), on the Qinghai-Tibetan Plateau.Acta Ecologica Sinica,2017,37(21):7251- 7257.

