2014年长沙市禽类场所环境H9N2亚型禽流感病毒分子流行特征分析
张如胜,姚 栋,叶 文,陈静芳,黄 政,刘晓蕾,陈田木,欧新华,孙边成
2014年长沙市禽类场所环境H9N2亚型禽流感病毒分子流行特征分析
张如胜,姚 栋,叶 文,陈静芳,黄 政,刘晓蕾,陈田木,欧新华,孙边成
目的 了解2014年长沙市禽类场所环境中H9N2亚型禽流感病毒(Avian influenza virus,AIV)血凝素(Hemagglutinin, HA)、神经氨酸酶(Neuraminidase, NA)和非结构蛋白(ron-structural, NS)基因的分子进化特征,为人感染H9N2亚型AIV防控提供实验室科学证据支持。方法 对2014年长沙市禽类场所中采集的501份环境标本(禽类饮水263份,污水221份,其他标本17份)利用real-time PCR方法进行A型、H5、H7和H9亚型流感病毒核酸检测,然后对单独H9阳性标本利用HA和NA基因通用引物进行RT-PCR扩增和核苷酸测序,病毒HA、NA及NS基因测序结果在线BLAST分析,利用Bioedit和Mega5软件进行氨基酸比对和进化树构建。结果 从501份环境标本中检出A型AIV核酸阳性标本350份,H9亚型核酸阳性标本191份,占总标本数量的38.12%,H5亚型核酸阳性标本177份(35.33%),H7亚型核酸阳性11份(2.20%),H5和H9亚型混合核酸阳性标本68份(13.57%)。部分单独H9亚型核酸阳性标本经RT-PCR和核苷酸测序鉴定出阳性H9N2亚型病毒核酸标本23份,分子进化分析表明2014年长沙市禽类场所环境中的绝大部分H9N2亚型AIV HA、NA、NS基因来源于A/Chicken/Shanghai/F/98 (F98)类似株病毒,属于新的S基因型,HA蛋白受体结合位点(Receptor binding site,RBS)第235位(对应H3流感第226位)氨基酸为Leu(L),表现为特异性结合人流感病毒受体特征,病毒HA、NA及NS蛋白关键分子位点表现为低致病性的分子特征。结论 2014年长沙市禽类场所环境中H9亚型AIV核酸阳性率最高,H9N2亚型AIV的HA、NA和NS基因表现为低致病性的分子特征,但具有容易感染人类的特征,需要进一步加强监测。
禽类场所;环境;禽流感病毒;H9N2亚型;基因;进化分析
禽流感病毒(Avian influenza virus,AIV)属于正粘病毒科A型流感病毒属。根据流感病毒表面糖蛋白血凝素(Hemagglutinin, HA)和神经氨酸酶(Neuraminidase, NA)抗原性的不同,A型流感病毒可分为18个HA亚型(H1-H18)和11个NA亚型(N1-N11)[1-2]。AIV除感染禽外,还可感染人、猪、马、水貂和海洋哺乳动物。近年来,部分AIV亚型突破种间屏障,造成人类感染,如H5N1、H5N2、H6N1、H9N2、H7N7、H7N2、H7N3、H10N7、H7N9、H10N8和H5N6亚型AIV[3-13]。截至2017年2月22日,全球共报告22例人感染H9N2亚型AIV病例[14],其中21例发生在中国大陆和香港地区(广东12例,香港5例,湖南2例,安徽2例),1例发生在孟加拉国,病例年龄7月-86岁不等,13例为女性,9例为男性,以儿童为主,其中5岁以下儿童占68.1%(15例),病例临床表现以呼吸道症状为主,大多表现为轻症,无死亡病例;流行病学调查显示病例大多有禽类接触史。研究[12,15]表明中国大陆首次报道的人感染新型重配H7N9和H10N8亚型AIV病毒,其内部基因均来自于H9N2亚型AIV。
AIV的致病性与病毒的基因特征相关。AIV基因组由8个基因节段(PB2: RNA polymerase basic subunit 2;PB1: RNA polymerase basic subunit 1;PA: RNA polymerase acidic subunit;HA: haemagglutinin;NP: nucleoprotein;NA: neuraminidase;M: matrix gene;NS: non-structural gene)组成,其中节段4编码的HA蛋白,具有抗原性,除了能对A型流感病毒HA亚型进行区分外,还在感染宿主和增加致病力方面起重要作用,其HA1和HA2连接肽处编码碱性氨基酸的多少与病毒的致病性相关[16],且HA蛋白编码氨基酸第226-228位点(H3亚型流感病毒氨基酸位点)为特异性受体结合区域,如为Gln-Ser-Gly(QSG)则表现为特异性结合AIV受体-唾液酸A-2,3半乳糖(SA-2,3-Gal),Leu-Ser-Ser(LSS)则表现为对人流感病毒受体-唾液酸A-2,6半乳糖(SA-2,6-Gal)亲和[17-18]。节段6编码的NA蛋白氨基酸[19]出现69-73位点缺失则表明该病毒在小鼠体内的毒力明显增加。节段8编码NS1和NS2蛋白,其中NS1蛋白编码氨基酸的第42位出现P42S突变及其末端结构编码氨基酸的第218-230位出现缺失,分别与病毒在小鼠体内的毒力增强和减轻相关[20-21]。
黄一伟等[22]报道了1例H9N2亚型AIV轻症病例,该病例AIV病毒分离株基因特征为禽源,与禽类市场环境中分离到的H9N2亚型AIV病毒高度相似(98.5~99.8%),其感染来源为与病例有流行病学关联的活禽市场。近年来研究[23-25]也表明活禽市场为人感染H5N1、H7N9和H10N8亚型AIV的重要感染来源,研究[26]显示长沙市禽类场所环境中H5N1亚型禽流感病毒核酸阳性率较高,存在传播AIV的风险。为此,本研究对2014年长沙市禽类场所环境中监测到的H9N2亚型AIV HA、NA及NS基因进行测序和基因特性分析,对阳性基因中与致病性相关的氨基酸变异和分子遗传(Phylogenetic)等分子特征进行探讨,为人感染H9N2亚型AIV疫情防控提供科学数据支持。
1 材料与方法
1.1 仪器及试剂 7300 Real-time RT-PCR System购于美国AB Applied Biosystems公司,MycyclerTMthermal cycler购于美国BIO-RAD公司,Centrifuge 5430R购于德国eppendorf公司,PowerpacTM Universal购于美国BIO-RAD公司,SuperScript○RIII One-Step RT-PCR System with Platinum○RTaq购于美国Life Technologies公司,SuperScript○RIII Platinum○ROne-Step Quantitative RT-PCR System购于美国Life Technologies公司,Rneasy○RMini Kit购于德国QIAGEN公司。
1.2 禽类场所环境标本H9亚型AIV核酸检测 按照中国疾病预防控制中心下发的《职业暴露人群血清学和环境高致病性禽流感监测方案》(2011年版)要求,采集2014年长沙市辖区禽类场所环境标本501份(禽类饮水263份、禽类污水221份、其他标本17份),利用real-time RT-PCR方法进行A型及H5、H7、H9亚型流感病毒核酸检测,核酸提取、检测步骤及结果判断按照《全国流感监测方案(2010年版)》进行[27-28],检测用real-time RT-PCR引物和探针序列来源于中国国家流感中心(Chinese National Influenza Center,CNIC)[29-30],引物及探针由上海Invitrogen公司合成。
1.3 H9N2亚型AIV HA、NA及NS基因扩增及核苷酸序列测定 将在2014年长沙市禽类场所环境中监测到的H9亚型AIV阳性标本(禽类污水、禽类饮水共23份,排除H5和H7亚型AIV核酸阳性)利用Rneasy○RMini Kit (德国QIAGEN 公司)进行RNA核酸提取(提取步骤参照试剂盒说明书进行),提取的RNA利用SuperScript○RIII One-Step RT-PCR System with Platinum○RTaq RT-PCR(Life Technologies 公司)对HA和NA基因片段分别进行RT-PCR核酸扩增,反应体积(25 μL):RNase Free Water 6.0 μL;2×PCR反应缓冲液12.5 μL;Platinum@Taq酶混合液1.0 μL;20 μmol/L引物(Bm-HA-1、Bm-NS-890R; Ba-NA-1、Ba-NA-1413R,引物序列来源于文献[31],见表1)各0.25 μL;RNA模板5.0 μL。HA和NA基因预期扩增片段长度分别为1 778 bp和1 413 bp,其中HA基因扩增能同时扩增出NS片段,预期片段长度为890 bp。扩增反应条件:60 ℃ 60 min;94 ℃ 2 min;再按94 ℃ 15 s,58 ℃ 30 s,68 ℃ 7 min 循环40 次,68 ℃ 7 min。分别取扩增后的HA和NA基因片段RT-PCR产物10 μL,在琼脂糖凝胶中90 v 40 min电泳后成像,观察有无预期目的片段。如有预期目的片段,将PCR产物TA克隆后进行核苷酸序列测定,TA克隆测序由上海Life Technologies 公司利用ABI PRISMTM 3730XL DNA Analyzer测序仪和BigDye○RTerminator v3.1 Cycle Sequencing Kit测序试剂完成核苷酸序列测定,正、反两方向测序,测通。
1.4 环境中H9N2亚型AIV HA、NA及NS基因进化分析 长沙市禽类场所环境中H9N2亚型AIV病毒HA、NA及NS基因测序结果利用BioEdit软件拼接后提交至美国国立生物信息中心(National Center for Biotechnology Information,NCBI),利用在线基本局部相似性搜索工具(Basic local alignment search tool, BLAST)进行同源性分析(http://BLAST.ncbi.nlm.nih.gov/BLAST.cgi)。从NCBI 流感病毒资源库(Influenza Virus Resource,http://www.ncbi.nlm.nih.gov/genomes/FLU/Database/nph-select.cgi)下载长沙市禽类场所环境中H9N2亚型AIV、国内H9N2亚型AIV及G1(A/Quail/Hong Kong/G1/97)、Y280(A/duck/Hong Kong/Y280/97)、Y439(A/duck/Hong Kong/Y439/1997)、F98(A/Chicken/Shanghai/F/98)和BJ/1/94(A/Chicken/Beijing/1/94)等谱系代表株HA、NA及NS基因片段核苷酸序列,利用MEGA 5软件的Neighbor-joining方法和Tamura-Nei 模式构建进化树。
表1 H9N2亚型禽流感病毒测序引物序列
Tab.1 Sequences of sequencing primers for H9N2 virus

PrimerSeguence(5'→3')Bm-HA-1TATTCGTCTCAGGGAGCAAAAG-CAGGGGBm-NS-890RATATCGTCTCGTATTAGTAGAAA-CAAGGGTGTTTTBa-NA-1TATTGGTCTCAGGGAGCAAAAG-CAGGAGTBa-NA-1413RATATGGTCTCGTATTAGTAGAAA-CAAGGAGTTTTTT
1.5 环境中H9N2亚型AIV HA、NA及NS基因关键位点分析 选择并从NCBI流感病毒资源库中下载人感染H9N2亚型病毒湖南株A/Lengshuitan/11197/2013 (LST/11197)、H9N2亚型AIV各谱系代表株(G1、Y280、Y439、F98、BJ/1/94)和H5N1亚型AIV A/duck/Guangxi/xa/2001(GX/xa)HA、NA及NS基因片段编码氨基酸序列,利用BioEdit软件将上述AIV和本研究中的环境H9N2亚型AIV进行多重比对、分析各基因关键位点分子特征。
2 结 果
2.1 禽类场所环境标本各基因相关检测结果 对2014年长沙市禽类场所环境中采集到的501份禽类饮水(263份)、污水(221份)和其他(17份)标本进行AIV病毒核酸检测,共检出A型病毒核酸阳性标本350份,其中主要A型病毒核酸亚型为H9和H5, H9亚型核酸阳性标本191份,占总标本的38.12%, H5亚型核酸阳性标本177份(35.33%),H7亚型核酸阳性11份(2.20%),H5和H9亚型混合核酸阳性标本68份(13.57%);263份禽类饮水标本中H9和H5亚型核酸检出率分别为50.19%和32.70%,221份污水标本中H9和H5亚型核酸检出率分别为25.34%和39.37%,具体结果见表2。
表2 长沙市禽类场所环境标本AIV real-time RT-PCR检测结果分析
Tab.2 Prevalence of AIV in specimens collected from poultry markets in Changsha City, Hunan Province, China, determined by real-time RT-PCR
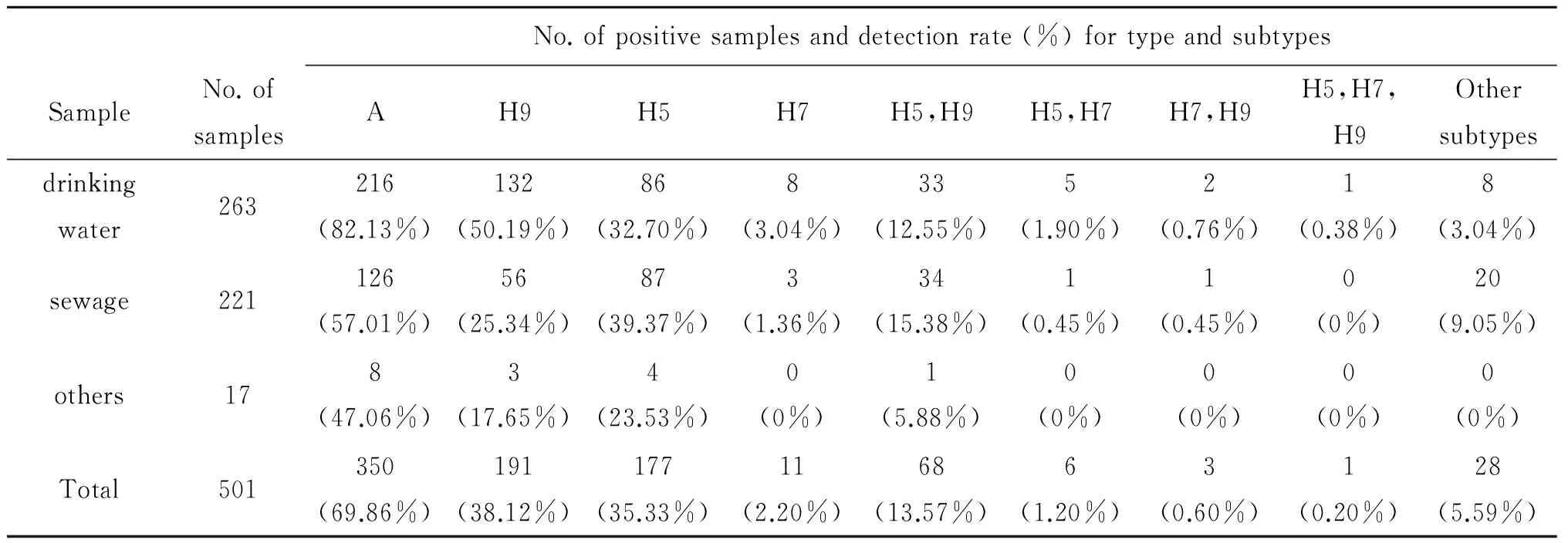
No.ofpositivesamplesanddetectionrate(%)fortypeandsubtypesSampleNo.ofsamplesAH9H5H7H5,H9H5,H7H7,H9H5,H7,H9Othersubtypesdrinkingwater263216(82.13%)132(50.19%)86(32.70%)8(3.04%)33(12.55%)5(1.90%)2(0.76%)1(0.38%)8(3.04%)sewage221126(57.01%)56(25.34%)87(39.37%)3(1.36%)34(15.38%)1(0.45%)1(0.45%)0(0%)20(9.05%)others178(47.06%)3(17.65%)4(23.53%)0(0%)1(5.88%)0(0%)0(0%)0(0%)0(0%)Total501350(69.86%)191(38.12%)177(35.33%)11(2.20%)68(13.57%)6(1.20%)3(0.60%)1(0.20%)28(5.59%)
2.2 H9N2亚型AIV核苷酸序列测定结果 从H9亚型核酸阳性标本中筛选出23份(H5、H7亚型AIV核酸均为阴性),通过HA和NA基因通用引物(其中HA基因扩增引物能够同时扩增出NS基因片段)RT-PCR扩增和TA克隆核苷酸序列测定,成功获得58条基因序列。其中HA和NA基因序列分别为23条,NS序列为12条,各基因序列已上传至NCBI,GenBank登录号为:KX247857-KX247914。其中H9N2亚型AIV HA基因核苷酸序列全长1 683 bp,NA基因核苷酸序列全长1 401 bp,NS基因核苷酸序列全长883 bp。在线BLAST相似性比对分析表明:23条HA基因序列分别与来源于湖南、河南、江西、江苏、北京和青岛等省市环境及鸡H9N2、H9亚型AIV分离株HA基因高度相似(≥98.63%);22条H9N2亚型AIV NA基因序列分别与来源于湖南、江苏、江西、北京、东莞和深圳等省市环境及鸡H9N2亚型AIV分离株NA基因高度相似(≥98.07%), 1条H9N2亚型AIV(A/environment/Changsha/369/2014(H9N2))NA基因序列与来源于蒙古麻鸭H11N2亚型AIV分离株(A/ruddy shelduck/Mongolia/590C2/2009(H11N2))NA基因高度相似(97.94%);7条NS基因序列分别与武汉和温州等地鸡源H9N2亚型AIV分离株NS基因高度相似(≥98.98%),5条NS基因序列分别与安徽、无锡、温州等省市人、鸡和鸭H7N9亚型AIV分离株NS基因高度相似(≥99.09%);HA和NA基因BLAST结果表明上述23份环境标本中存在的病毒为H9N2亚型AIV,具体病毒名称及其BLAST比对结果见表3。
2.3 H9N2亚型AIV进化树构建 H9N2 亚型AIV在世界范围内广泛分布,在遗传进化上可以分为北美和欧亚谱系两大分支,其中欧亚系又进一步衍生出以Y280、G1、Y439、F98以及BJ/1/94等为代表的病毒系进化分支[32-33]。HA基因进化树构建出Y280、G1和Y439三个谱系进化分支,其中本研究中的23株H9N2亚型AIV、国内H9N2亚型AIV HA基因聚集在一个亚分支内,Y280、F98和CK/BJ/1谱系代表株HA基因聚集在另一个亚分支内,两个亚分支共同构成Y280谱系进化分支[22],本研究中的23株H9N2病毒与Y280谱系代表株进化距离近;NA进化树显示本研究中的23株环境H9N2亚型AIV有22株病毒聚集在一个单独进化分支内,与Y280、F98谱系代表株及H9N2亚型人源湖南株LST/11197进化距离近,而本研究中的另外1株病毒CS/369位于Y439谱系;NS基因进化树显示,本研究中的12株H9N2亚型AIV NS基因全部聚集在F98谱系,但分别形成3个亚分支,见图1。
表3 长沙市禽类场所环境23株H9N2亚型病毒BLAST相似性分析
Tab.3 Comparisons of twenty-three H9N2 viruses from Changsha City with isolates in GenBank of highest nucleotide identity (%) (The date of this BLAST search was Feb 27, 2016)

VirusHANucleotideidentityofH9N2virus(%)NANucleotideidentityofH9N2virus(%)NSNucleotideidentityofH9N2virus(%)CS/92A/chicken/Beijing/0331/2013(99.17%)A/chicken/Jiangsu/JS86/2013(99.13%)A/chicken/Wenzhou/YHQL04/2014(98.98%)CS/94A/chicken/Jiangxi/29075/2013(99.58%)A/chicken/Jiangxi/29075/2013(99.71%)A/chicken/Wuhan/JXQL01/2015(99.66%)CS/219A/environment/Hunan/27420/2014(99.94%)A/chicken/Dongguan/1674/2014(99.93%)A/chicken/Wuhan/JXQL01/2015(99.09%)CS/250A/environment/Hunan/28028/2014(99.23%)A/environment/Hunan/28034/2014(99.79%)A/Anhui/DEWH72-09/2013b(99.55%)CS/279A/environment/Hunan/28034/2014(99.58%)A/environment/Hunan/28034/2014(99.64%)A/duck/Wuxi/0405006/2013b(99.09%)CS/334A/environment/Hunan/27420/2014(98.87%)A/chicken/Jiangsu/YZLH3/2013(99.07%)NosequenceCS/341A/chicken/Beijing/1115/2013(99.23%)A/chicken/Jiangxi/20482/2013(99.14%)A/chicken/Wuhan/JXQL01/2015(99.66%)CS/347A/environment/Hunan/28034/2014(99.47%)A/environment/Hunan/28034/2014(99.71%)NosequenceCS/360A/environment/Hunan/27420/2014(99.52%)A/chicken/Beijing/0512/2013(99.00%)NosequenceCS/366A/environment/Hunan/27420/2014(99.47%)A/environment/Hunan/28034/2014(99.57%)NosequenceCS/368A/environment/Hunan/27420/2014(99.58%)A/environment/Hunan/28034/2014(99.50%)NosequenceCS/369A/environment/Hunan/27420/2014(99.47%)A/ruddyshelduck/Mongolia/590C2/2009a(97.94%)NosequenceCS/372A/chicken/Qingdao/013/2014(98.63%)A/chicken/Jiangxi/20478/2013(99.29%)NosequenceCS/400A/chicken/Henan/SH01/2015(99.52%)A/chicken/Dongguan/1674/2014(99.64%)NosequenceCS/408A/environment/Hunan/27420/2014(99.23%)(A/chicken/Dongguan/1674/2014(99.50%)NosequenceCS/418A/environment/Hunan/27420/2014(99.70%)(A/chicken/Dongguan/1674/2014(99.07%)A/chicken/Wenzhou/RAQL18/2015b(99.09%)CS/450A/chicken/Jiangsu/J3156/2014(99.05%)A/chicken/Jiangxi/20482/2013(99.21%)A/chicken/Wuhan/JXQL01/2015(99.21%)CS/451A/environment/Hunan/27420/2014(99.41%)A/chicken/Dongguan/1674/2014(99.36%)NosequenceCS/462A/chicken/Henan/SH01/2015(99.34%)A/chicken/Dongguan/1674/2014(99.57%)A/chicken/Wuhan/JXQL01/2015(99.21%)CS/478A/chicken/Henan/SH01/2015(99.28%)A/chicken/Jiangxi/29117/2013(98.93%)A/chicken/Wuhan/JXQL01/2015(99.21%)CS/481A/environment/Hunan/27420/2014(99.41%)A/environment/Hunan/28034/2014(99.29%)NosequenceCS/484A/chicken/Jiangsu/J3294/2014(99.35%)A/chicken/Shenzhen/1949/2013(98.07%)A/duck/Wuxi/0405006/2013b(99.21%)CS/490A/environment/Hunan/27420/2014(99.17%)A/chicken/Jiangxi/29117/2013(98.86%)(A/duck/Wuxi/0405006/2013b(99.32%)
Note: CS/92: A/environment/Changsha/92/2014; CS/94: A/environment/Changsha/94/2014; CS/219: A/environment/Changsha/219/2014; CS/250: A/environment/Changsha/250/2014; CS/279: A/environment/Changsha/279/2014; CS/334: A/environment/Changsha/334/2014; CS/341: A/environment/Changsha/341/2014; CS/347: A/environment/Changsha/347/2014; CS/360: A/environment/Changsha/360/2014; CS/366: A/environment/Changsha/366/2014; CS/368: A/environment/Changsha/368/2014; CS/369: A/environment/Changsha/369/2014; CS/372: A/environment/Changsha/372/2014; CS/400: A/environment/Changsha/400/2014; CS/408: A/environment/Changsha/408/2014; CS/418: A/environment/Changsha/418/2014; CS/450: A/environment/Changsha/450/2014; CS/451: A/environment/Changsha/451/2014; CS/462: A/environment/Changsha/462/2014; CS/478: A/environment/Changsha/478/2014; CS/481: A/environment/Changsha/481/2014; CS/484: A/environment/Changsha/484/2014; CS/490: A/environment/Changsha/490/2014; a: H11N2; b: H7N9.
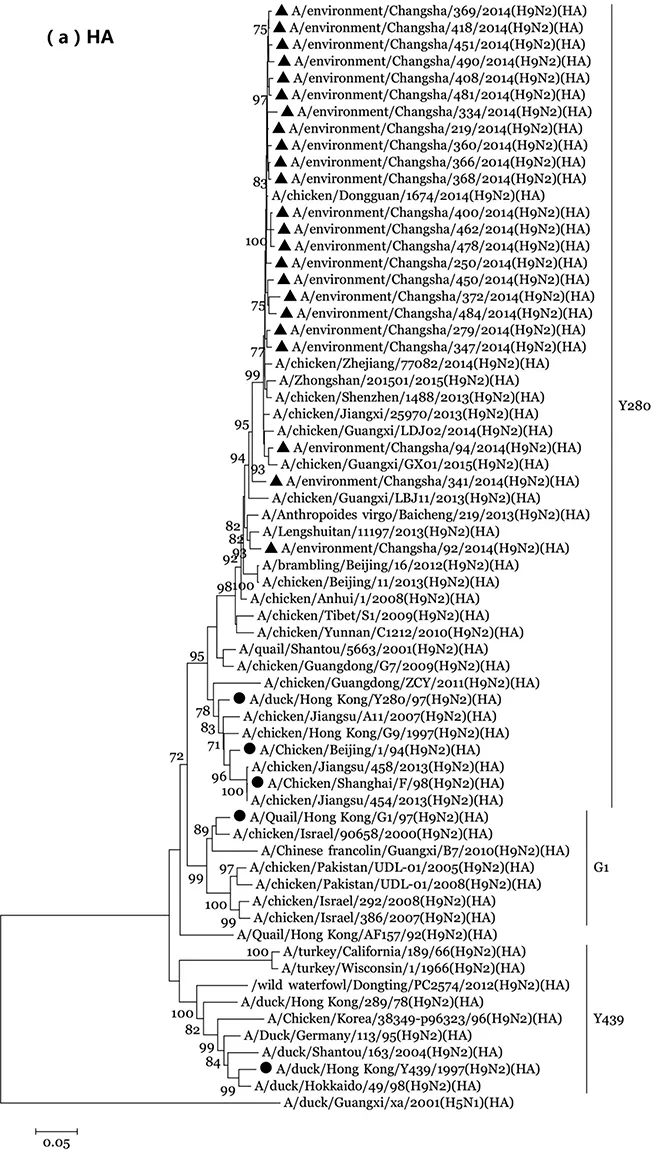


The sequence of the H9N2 virus in our study is marked by ▲, and the sequences of representative H9N2. Viruses of G1, Y280, Y439, BJ/1/94 and F98 lineages from GenBank are marked by ●.
>图1 2014年长沙市禽类场所环境H9N2亚型AIV HA(a)、NA(b)及NS(c)基因进化分析
Fig.1 Phylogenetic analysis of the HA (a), NA(b) and NS(c) genes of the influenza H9N2 viruses isolated from poultry markets in Changsha, China in 2014
2.3.1 H9N2亚型AIV HA、NA及NS基因关键位点分析 本研究中的23株H9N2亚型AIV HA基因HA1和HA2 蛋白连接肽(335-341 位点)出现2个碱性氨基酸(SRSSRGL,其中R为碱性氨基酸),与其他H9N2亚型AIV谱系代表株HA基因致病特征相似,对禽表现为低致病性特征,与H5N1亚型高致病性AIV具有多个碱性氨基酸的高致病性分子特征不同[16],见表4。
本研究中的23株H9N2亚型AIV HA 蛋白第235~237位(对应H3型流感病毒编码氨基酸第226-228位)氨基酸为LMG,表明其受体为人源流感病毒特异性受体[17],与人源H9N2亚型AIV湖南株LST/11197及其他不同谱系H9N2亚型AIV代表株HA基因特征相似,具有容易感染人的受体特征,见表4。
本研究中的1株H9N2亚型AIV CS/369 NA基因编码氨基酸第63~64位点为NR氨基酸,与G1、Y439和BJ/1/94谱系代表株病毒NA基因特征相似,但本研究中的另外22株H9N2亚型AIV NA基因编码氨基酸第63-64位点出现缺失,与Y280和F98谱系代表株病毒相似,见表4。
本研究中的12株H9N2亚型AIV与其他不同谱系代表株NS1蛋白全长编码217个氨基酸,相比H5N1亚型AIV GX/xa,表4中的H9N2亚型AIV第80-84位氨基酸(TIASV)均未发生缺失现象,但本研究中的12株H9N2亚型AIV及1株人源H9N2亚型AIV湖南株LST/11197缺失第218-230位氨基酸,导致了病毒致死性相关PL基序(ESEV/EPEV)的缺失,见表4。本研究中的12株H9N2亚型AIV NS1编码蛋白第92位氨基酸均为D,未出现D92E突变,不同于G1谱系代表株NS1蛋白出现D92E氨基酸突变,见表4。
表4 长沙市H9N2亚型AIV与参考株病毒HA、NA及NS1蛋白关键位点特征分析
Tab.4 Analysis of the important amino acids of the HA, NA and NS1 proteins of H9N2 AIV strain from Changsha City with reference strains

VirusHANANS1Cleavagesite335-341Receptorbindingsite235-23763-64Totalofno.ofaaDeletionofaa80-8492CS/92SRSSRGLLMGDeletion217NODCS/94SRSSRGLLMGDeletion217NODCS/219SRSSRGLLMGDeletion217NODCS/250SRSSRGLLMGDeletion217NODCS/279SRSSRGLLMGDeletion217NODCS/334SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/341SRSSRGLLMGDeletion217NODCS/347SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/360SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/366SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/368SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/369SRSSRGLLMGNRNosequenceNosequenceNosequenceCS/372SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/400SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/408SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/418SRSSRGLLMGDeletion217NODCS/450SRSSRGLLMGDeletion217NODCS/451SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/462SRSSRGLLMGDeletion217NODCS/478SRSSRGLLMGDeletion217NODCS/481SRSSRGLLMGDeletionNosequenceNosequenceNosequenceCS/484SRSSRGLLMGDeletion217NODCS/490SRSSRGLLMGDeletion217NODLST/11197SRSSRGLLMGDeletion217NODG1ARSSRGLLQGNR218NOEY280ARSSRGLLQGDeletion218NODY439AASNRGLQQGNR218NODF98ARSSRGLQQGDeletion217NODBJ/1/94ARSSRGLQQGNR217NODGX/xaRERRRKRGLQSGIR225YESD
Note: Substitutions of particular concern are shown in bold.
3 讨 论
H9N2亚型AIV在中国及东亚地区鸡群中广泛流行,其中Y280谱系病毒在中国中南部流行最为广泛[34-35],H9N2病毒同时也是活禽市场中最常见的AIV,研究[22]显示与人感染H9N2亚型AIV病例有流行病学关联的湖南省永州市活禽市场中H9亚型AIV核酸阳性率最高(18%),其他研究[36]显示广西地区活禽市场低致病性AIV具有相似的阳性率10.8%(336/3 121)。同时人感染H7N9、H10N8、H5N6、H9N2等[22-25]亚型AIV疫情流行病学调查已证实禽类场所,特别是活禽市场已经成为人感染AIV的重要来源,CNIC每年都在开展禽类场所AIV日常监测工作,监测禽类场所环境中的不同亚型AIV污染情况,本研究结果显示2014年长沙市禽类场所环境中H9亚型AIV核酸阳性率最高(38.12%),高于H5亚型AIV核酸阳性率(35.33%),并且高于其他研究报道[23,37],原因可能与标本种类有关,本研究中的标本种类主要为禽类饮水和污水,这两类标本分别用于禽类的饮用水和禽类宰杀后的清洗用水,聚集了禽类口腔和内脏器官的AIV,造成了AIV核酸检出率的升高。
研究对2007-2013年中国东部地区分离的H9N2病毒基因进化分析显示出现六个不同的基因型,其中的一个新基因型(S),以F98谱系代表株病毒为骨架,通过与G1谱系代表株病毒的PB2和M基因重配而成[37]。HA及NA进化树分析显示本研究中的H9N2亚型AIV与国内2013年以后分离的部分H9N2亚型AIV HA、NA基因聚集在一个单独的亚分支内,但与Y280及F98谱系代表株同属于一个大的进化分支;由于本研究中的H9N2病毒CS/369与H11N2亚型AIV分离株(A/ruddy shelduck/Mongolia/590C2/2009)NA基因核苷酸序列高度相似(97.94%),导致CS/369病毒与Y439谱系代表株进化距离近,属于Y439谱系;NS基因进化树显示本研究中的12株H9N2亚型AIV病毒NS基因全部聚集在F98谱系,但分别形成3个亚分支,与本研究中的NS基因分别来源于H9N2和H7N9亚型AIV相关。进化表明长沙市禽类场所环境中的绝大部分H9N2亚型AIV HA、NA、NS基因来源于F98 类似株病毒,属于新的S基因型,并且随着时间的推移,已经开始进化形成新的分支,需要引起关注。另外HA及NA基因进化分析还显示本研究中的环境H9N2亚型AIV与人源H9N2亚型AIV湖南株LST/11197位于同一分支,且进化距离近,进一步证实了活禽市场是人感染H9N2亚型AIV的重要来源[22]。
流感病毒在感染宿主细胞时,HA蛋白需要蛋白酶将其裂解为HA1和HA2,然后才能吸附到宿主细胞膜上。H7N9、H10N8和H9N2等低致病性AIV在HA1和HA2连接肽区域只有一个碱性氨基酸,只能被局限在有R蛋白酶表达的粘膜表面和组织中进行复制,感染后症状较轻。而H5N1,H5N6等高致病性AIV在HA1和HA2连接肽区域有4个以上碱性氨基酸,故能被广泛分布在全身各组织和器官的蛋白酶所识别并裂解,造成全身感染,故流感病毒HA蛋白的HA1和HA2连接肽位点碱性氨基酸数目的多少与病毒的致病性密切相关。本研究中的2014年长沙市禽类场所环境中H9N2亚型AIV HA基因HA1和HA2蛋白连接肽位点出现2个碱性氨基酸,与其他H9N2亚型AIV谱系代表株HA基因致病特征相似,对禽表现为低致病性基因特征[16]。流感病毒HA蛋白还起识别宿主细胞受体的作用,其第226~228位氨基酸为QSG,表明其对AIV受体-SA-2,3-Gal亲和,如为LSS序列,则对人流感病毒受体-SA-2,6-Gal亲和[38]。研究报道近年来国内流行的H9N2病毒HA蛋白RBS区域几乎均突变为L226,并且多数具有与SA-2,6-Gal结合的能力[39],有研究还证实含L226的H9N2亚型AIV能够在人的气管上皮细胞中进行有效的复制[40]。本研究中的H9N2亚型AIV HA基因RBS区域为L226,表现为人源流感病毒受体,具有与SA-2,6-Gal结合的能力,容易造成人类感染。目前报道的22例人感染H9N2亚型AIV病例绝大多数表现为轻症,表明更多的H9N2亚型AIV轻症和无症状携带者尚未被发现,这种温和的感染使得H9N2亚型AIV能够进一步在人体内适应并有可能与其他人流感病毒发生基因重组而具备造成流感流行的潜力。
本研究中的绝大部分H9N2亚型AIV NA基因蛋白第63-64位氨基酸出现缺失,仅1株H9N2亚型AIV CS/369第63-64位氨基酸未发生缺失,表现为NR,证实了H9N2亚型AIV NA基因来源的多样性。研究表明H9N2亚型AIV NS1蛋白编码氨基酸发生D92E突变,具有提高H9N2亚型AIV感染人类的能力,本研究中的12株H9N2病毒的NS1蛋白未发生D92E突变,且缺失了与病毒致死性相关的PL基序(ESEV/EPEV), 该缺失可以减轻病毒对小鼠的致病能力[21],与本地区环境中的其他H9N2亚型AIV NS1蛋白编码氨基酸相似,具有低致病性流感病毒的分子特征[41]。然而,研究显示不同谱系的H9亚型病毒的内部基因在哺乳动物和人体细胞模型中都具备复制能力[42],表明H9N2亚型AIV的内部基因仍然有潜力为产生新的流行株病毒提供部分甚至整套内部基因,譬如新型H7N9和H5N1亚型病毒。
目前,国内不断新增人感染H9N2亚型AIV病例报告,且禽类市场活禽销售模式未改变、禽类场所环境中H9N2亚型AIV污染状况未进一步减轻的情况下,需要密切监测禽类场所环境中H9N2亚型AIV病毒进化状况,及时评估、预警H9N2亚型AIV感染人类的潜在风险。
[1] Tong S, Zhu X, Li Y, et al. New world bats harbor diverse influenza A viruses[J]. PLoS Pathog, 2013, 9(10): e1003657. DOI: 10.1371/journal.ppat.1003657
[2] Tong S, Li Y, Rivailler P, et al. A distinct lineage of influenza A virus from bats[J]. Proc Natl Acad Sci U S A, 2012, 109(11): 4269-4274. DOI: 10.1073/pnas.1116200109
[3] Pan M, Gao R, Lv Q, et al. Human infection with a novel, highly pathogenic avian influenza A (H5N6) virus: Virological and clinical findings[J]. J Infect, 2016, 72(1): 52-59. DOI: 10.1016/j.jinf.2015.06.009
[4] Koopmans M, Wilbrink B, Conyn M, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands[J]. Lancet, 2004, 363(9409): 587-593. DOI: 10.1016/S0140-6736(04)15589-X
[5] Hirst M, Astell CR, Griffith M, et al. Novel avian influenza H7N3 strain outbreak, British Columbia[J]. Emerg Infect Dis, 2004, 10(12): 2192-2195. DOI: 10.3201/eid1012.040743
[6] Ogata T, Yamazaki Y, Okabe N, et al. Human H5N2 avian influenza infection in Japan and the factors associated with high H5N2-neutralizing antibody titer[J]. J Epidemiol, 2008, 18(4): 160-166. DOI: 10.2188/jea.JE2007446
[7] Cheng VC, Chan JF, Wen X, et al. Infection of immunocompromised patients by avian H9N2 influenza A virus[J]. J Infect, 2011, 62(5): 394-399. DOI:10.1016/j.jinf.2011.02.007
[8] To KK, Ng KH, Que TL, et al. Avian influenza A H5N1 virus: a continuous threat to humans[J]. Emerg Microbes Infect, 2012, 1(9): e25. DOI: 10.1038/emi.2012.24
[9] Ostrowsky B, Huang A, Terry W, et al. Low pathogenic avian influenza A (H7N2) virus infection in immunocompromised adult, New York, USA, 2003[J]. Emerg Infect Dis, 2012, 18(7): 1128-1131. DOI: 10.3201/eid1807.111913
[10] Arzey GG, Kirkland PD, Arzey KE, et al. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia[J]. Emerg Infect Dis, 2012, 18(5): 814-816. DOI: 10.3201/eid1805.111852
[11] Wei SH, Yang JR, Wu HS, et al. Human infection with avian influenza A H6N1 virus: an epidemiological analysis[J]. Lancet Respir Med, 2013, 1(10): 771-778. DOI: 10.1016/S2213-2600(13)70221-2.
[12] Chen H, Yuan H, Gao R, et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study[J]. Lancet, 2014, 383(9918): 714-721. DOI: 10.1016/S0140-6736(14)60111-2
[13] Zhang R, Chen T, Ou X, et al. Clinical, epidemiological and virological characteristics of the first detected human case of avian influenza A(H5N6) virus[J]. Infect Genet Evol, 2016, 40: 236-242. DOI: 10.1016/j.meegid.2016.03.010
[14] Freidl GS, Meijer A, de Bruin E, et al. Influenza at the animal-human interface: a review of the literature for virological evidence of human infection with swine or avian influenza viruses other than A(H5N1)[J]. Euro Surveill, 2014, 19(19): 8-26. DOI: 10.2807/1560-7917
[15] Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7N9) virus[J]. N Engl J Med, 2013, 368(20): 1888-1897. DOI: 10.1056/NEJMoa1304459
[16] Senne DA, Panigrahy B, Kawaoka Y, et al. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential[J]. Avian Dis, 1996, 40(2): 425-437. DOI: 10.2307/1592241
[17] Srinivasan K, Raman R, Jayaraman A, et al. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin[J]. PLoS One, 2013, 8(2): e49597. DOI: 10.1371/journal.pone.0049597
[18] Shu YL,Lan Y,Wen LY, et al.Analysis of human H5N1 virus hemagglutinin gene isolated from the mainland of China[J]. Chin J Exp Clin Virol, 2006, 20(2): 8-10. DOI: 10.3760/cma.j.issn.1003-9279.2006.02.003 (in Chinese)
舒跃龙,蓝雨,温乐英,等.我国分离人H5N1禽流感病毒血凝素基因特性的研究[J]. 中华实验和临床病毒学杂志,2006,20(2):8-10.
[19] Mckimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors[J]. J Virol, 1998, 72(3): 2456-2462.
[20] Jiao P, Tian G, Li Y, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice[J]. J Virol, 2008, 82(3): 1146-1154. DOI: 10.1128/JVI.01698-07
[21] Jackson D, Hossain MJ, Hickman D, et al. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity[J]. Proc Natl Acad Sci U S A, 2008, 105(11): 4381-4386. DOI: 10.1073/pnas.0800482105
[22] Huang Y, Li X, Zhang H, et al. Human infection with an avian influenza A (H9N2) virus in the middle region of China[J]. J Med Virol, 2015, 87(10): 1641-1648. DOI: 10.1002/jmv.24231
[23] Wan XF, Dong L, Lan Y, et al. Indications that live poultry markets are a major source of human H5N1 influenza virus infection in China[J]. J Virol, 2011, 85(24): 13432-13438. DOI: 10.1128/JVI.05266-11
[24] Shi J, Deng G, Liu P, et al. Isolation and characterization of H7N9 viruses from live poultry markets-Implication of the source of current H7N9 infection in humans[J]. Chin Sci Bull, 2013, (16): 1857-1863. DOI: 10.1007/s11434-013-5873-4
[25] Zhang T, Bi Y, Tian H, et al. Human infection with influenza virus A(H10N8) from live poultry markets, China, 2014[J]. Emerg Infect Dis, 2014, 20(12): 2076-2079. DOI: 10.3201/eid2012.140911
[26] Zhang RS, Ou XH, Song KY, et al. Risk related to the transmission of H5N1 subtype avian influenza virus in the environment of poultry markets in Changsha, China[J]. Chin J Epidemiol, 2012, 33(8): 768-773. DOI: 10.3760/cma.j.issn.0254-6450.2012.08.003 (in Chinese)
张如胜,欧新华,宋克云, 等. 长沙市家禽市场环境中H5N1亚型禽流感病毒传播风险研究[J]. 中华流行病学杂志, 2012, 33(8): 768-773.
[27] Zhang RS, Sun BC, Yao D, et al. Evolution of the HA, NA and NS genes of H5N1 avian influenza viruses from sewage in live bird markets in Changsha, 2014[J]. Chin J Zoonoses, 2017, 33(1): 85-88, 80. DOI: 10.3969/j.issn.1002-2694.2017.01.016 (in Chinese)
张如胜, 孙边成, 姚栋, 等. 2014年长沙市活禽市场污水中H5N1亚型禽流感病毒HA、NA及NS基因进化分析[J]. 中国人兽共患病学报, 2017, 33(1): 85-88, 80.
[28] Ministry of Health of the People’s Republic of China. National influenza surveillance program (2010)[EB/OL]. (2010-09-10)[2017-02-23]. http://www.moh.gov.cn/jkj/s3577/201009/3fa356d0f4834d408fde6c12891a6482.shtml. (in Chinese)
中华人民共和国卫生部:全国流感监测方案(2010年版)[EB/OL]. (2010-09-10)[2017-02-23]. http://www.moh.gov.cn/jkj/s3577/201009/3fa356d0f4834d408fde6c12891a6482.shtml.
[29] WHO Global Influenza Surveillance Network. Manual for the laboratory diagnosis and virological surveillance of influenza[EB/OL]. (2013-09-15)[2017-02-23]. http://whqlibdoc.who.int/publications/2011/9789241548090_eng.pdf.
[30] World Health Organization. WHO information for molecular diagnosis of influenza virus-update[EB/OL]. (2014-03-31)[2016-06-05]. http://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis_influenza_virus_humans_update_201403.pdf?ua=1.
[31] Hoffmann E, Stech J, Guan Y, et al. Universal primer set for the full-length amplification of all influenza A viruses[J]. Arch Virol, 2001, 146(12): 2275-2289. DOI: 10.1007/s007050170002
[32] Xu KM, Smith GJ, Bahl J, et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005[J]. J Virol, 2007, 81(19): 10389-10401. DOI: 10.1128/JVI.00979-07
[33] Lu JH, Liu XF, Shao WX, et al. Phylogenetic analysis of eight genes of H9N2 subtype influenza virus: a mainland China strain possessing early isolates’ genes that have been circulating[J]. Virus Genes, 2005, 31(2): 163-169. DOI: 10.1007/s11262-005-1790-1
[34] Ji K, Jiang W M, Liu S, et al. Characterization of the hemagglutinin gene of subtype H9 avian influenza viruses isolated in 2007-2009 in China[J]. J Virol Methods, 2010, 163(2): 186-189. DOI: 10.1016/j.jviromet.2009.09.01
[35] Chu YC, Cheung CL, Hung LC, et al. Continuing evolution of H9N2 influenza viruses endemic in poultry in southern China[J]. Influenza Other Respir Viruses, 2011, 5 (Suppl 1): 68-71.
[36] Peng Y, Xie ZX, Liu JB, et al. Epidemiological surveillance of low pathogenic avian influenza virus (LPAIV) from poultry in Guangxi Province, Southern China[J]. PLoS One, 2013, 8(10): e77132. DOI: 10.1371/journal.pone.0077132
[37] Gu M, Chen H, Li Q, et al. Enzootic genotype S of H9N2 avian influenza viruses donates internal genes to emerging zoonotic influenza viruses in China[J]. Vet Microbiol, 2014, 174(3/4): 309-315. DOI: 10.1016/j.vetmic.2014.09.029
[38] Stevens J, Blixt O, Tumpey TM, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus[J]. Science, 2006, 312(5772): 404-410. DOI: 10.1126/science.1124513
[39] Jiang W, Liu S, Hou G, et al. Chinese and global distribution of H9 subtype avian influenza viruses[J]. PLoS One, 2012, 7(12): e52671. DOI: 10.1371/journal.pone.0052671
[40] Vines A, Wells K, Matrosovich M, et al. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction[J]. J Virol, 1998, 72(9): 7626-7631.
[41] Zhang RS, Ou X , Song KY, et al. Genetic analysis on the NS gene of H9N2 avian influenza virus isolated from sewage in poultry market[J]. Chin Prev Med, 2013, 14(03): 205-208. (in Chinese)
张如胜,欧新华,宋克云,等. 长沙市家禽市场污水来源H9N2亚型禽流感病毒NS基因进化分析[J]. 中国预防医学杂志, 2013,14(03): 205-208.
[42] The SJCEIRS H9 Working Group. Assessing the fitness of distinct clades of influenza A (H9N2) viruses[J]. Emerg Microbes Infect, 2013, 2(11): e75. DOI: 10.1038/emi.2013.75
Molecular epidemiological characteristics of avian influenza A(H9N2) viruses from environment in poultry markets in Changsha, China, 2014
ZHANG Ru-sheng, YAO Dong, YE Wen, CHEN Jing-fang, HUANG Zheng, LIU Xiao-lei, CHEN Tian-mu, OU Xin-hua, SUN Bian-cheng
(ChangshaCenterforDiseaseControlandPrevention,Changsha410004,China)
We analyzed the evolutional and molecular characteristics of Hemagglutinin(HA), Neuraminidase(NA) and non-structural(NS) genes of avian influenza A(H9n2) viruses from environment in poultry markets in Changsha, China, 2014, providing laboratory data for prevention and control of human infection with avian influenza A(H9N2) virus. Five hundred and one specimens (263 poultry drinking water specimens, 226 poultry sewage specimens and 17 others specimens) were collected from environment in poultry markets in Changsha, 2014, and real-time RT-PCR was used for influenza A typing and subtyping (H5, H7 and H9) detection. HA and NA universal primer sets for conventional RT-PCR and sequencing were used for the positivity of single H9. The sequence homology of HA, NA and NS genes of the viruses were analyzed with the online Basic Local Alignment Search Tool (BLAST). The ClustalW multiple alignments of amino acids and the phylogenetic trees for HA, NA and NS genes were constructed using the BioEdit and MEGA 5 software, respectively. Results showed that among 501 environmental samples, 350 samples were positive for influenza A virus, 191 (38.12%) for H9 subtype, 177 (35.33%) for H5 subtype, 11 (2.20%) for H7 subtype and 68 (13.57%) for H5 and H9 subtypes co-detection. Twenty-three H9N2 subtype AIV were confirmed by conventional RT-PCR and sequencing from the samples of the positivity of single H9. Phylogenetic analysis revealed that most of HA, NA and NS genes of the H9N2 subtype AIV isolated in Changsha City had gene constellations of genotype S,and these virues might have acquired their HA, NA and NS from A/Chicken/Shanghai/F/1998-like (H9N2). L235 (correspond to H3 numbering 226) of the HA protein of the receptor binding site (RBS) were found in these H9N2 viruses, and the characteristics was shown to be associated with increased affinity of HA to the glycan-receptors of human influenza virus, and the low pathogenicity molecular characteristics of HA, NA and NS genes were showed in these viruses. The positive rate of nucleic acid of the H9 subtype of avian influenza virus from environment was the highest in poultry markets in Changsha, 2014, and molecular characteristics of the HA, NA and NS of these H9N2 subtype AIV showed low pathogenicity, but that may facilitate human infection. So, the prevalence and genetic evolution of this virus should be closely monitored.
poultry markets; environment; avian influenza virus; H9N2 subtype; gene; phylogenetic analysis Supported by the Hunan Provincial Health Medicine Research Project (No. B2015-153) Corresponding author: Sun Bian-cheng, Email: sunbiancheng2013@163.com
10.3969/j.issn.1002-2694.2017.03.005
孙边成,Email: sunbiancheng2013@163.com
长沙市疾病预防控制中心,长沙 410004
R373.1+3
A
1002-2694(2017)03-0212-10
2017-02-09 编辑:林丹
湖南省卫生计生委2015年度科研计划项目(No.B2015-153)

