谷子转录因子SiNF-YA5通过ABA非依赖途径提高转基因拟南芥耐盐性
黄 锁胡利芹徐东北李微微徐兆师李连城周永斌刁现民贾冠清马有志陈 明,*
1中国农业科学院作物科学研究所 / 农作物基因资源与基因改良国家重大科学工程 / 农业部麦类生物学与遗传育种重点实验室, 北京100081;2西北农林科技大学农学院 / 旱区作物逆境生物学国家重点实验室, 陕西杨凌 712100;3哈尔滨师范大学生命科学与技术学院 / 黑龙江省分子细胞遗传与遗传育种重点实验室, 黑龙江哈尔滨 150025
谷子转录因子SiNF-YA5通过ABA非依赖途径提高转基因拟南芥耐盐性
黄 锁1,**胡利芹1,**徐东北1,2李微微1,3徐兆师1李连城1周永斌1,2刁现民1贾冠清1马有志1陈 明1,*
1中国农业科学院作物科学研究所 / 农作物基因资源与基因改良国家重大科学工程 / 农业部麦类生物学与遗传育种重点实验室, 北京100081;2西北农林科技大学农学院 / 旱区作物逆境生物学国家重点实验室, 陕西杨凌 712100;3哈尔滨师范大学生命科学与技术学院 / 黑龙江省分子细胞遗传与遗传育种重点实验室, 黑龙江哈尔滨 150025
核转录因子Y (nuclear transcription factor Y, NF-Y)类转录因子在植物生长发育和非生物胁迫响应基因表达调控中发挥重要的作用, NF-Y由3种亚基(NF-YA、NF-YB、NF-YC)组成。本研究从抗逆性强的谷子品种龙谷25中克隆1个新的NF-Y类转录因子基因SiNF-YA5。该基因序列为924 bp, 编码307个氨基酸, 分子量为33.76 kD, 等电点为9.19。SiNF-YA5在149~210位氨基酸之间含有CBF保守结构域。亚细胞定位分析表明, SiNF-YA5定位于细胞膜和细胞核。基因功能分析显示, 在不同浓度高盐处理下, 和野生型拟南芥(WT)相比SiNF-YA5转基因拟南芥种子萌发率更高; 苗期SiNF-YA5转基因拟南芥根表面积和植株鲜重显著高于WT, 证明过表达SiNF-YA5基因可以显著提高植物耐盐性。基因表达分析结果显示, 在SiNF-YA5转基因拟南芥中参与盐胁迫响应的基因NHX1和LEA7的表达量明显高于WT。另一方面, SiNF-YA5转基因拟南芥与WT相比对于ABA的敏感性差异不显著, 以上结果证明SiNF-YA5主要通过ABA非依赖途径提高转基因植物对高盐胁迫的耐性。
谷子; NF-Y类转录因子; 高盐胁迫; ABA非依赖途径
干旱、盐碱、低温等非生物胁迫严重影响作物的生长发育及产量[1]。植物在长期的进化过程中逐渐形成一套复杂的逆境应答机制, 以抵御不良环境对植物的损害[2-3]。大量研究证明, 植物细胞在染色体水平、转录水平以及转录后水平精确调控一系列胁迫应答基因的表达, 其中, 一些功能蛋白包括胚胎发育后期丰富蛋白(LEA蛋白)、渗透调节蛋白、离子区域化和水通道蛋白、脯氨酸及果聚糖合成酶及甜菜碱合成酶等直接发挥功能增强植物细胞抗逆性[4]。另外, 一些调控蛋白包括感应和传导胁迫信号的蛋白激酶(例如 CDPK, cadium dependent phospholylation kinase; MAPK, mitogen-activated protein kinases)以及参与调控基因表达的转录因子(包括Bzip, basic leucine zipper类; NAC, nascent polypeptide-associated complex类; DREB, dehydration responsive element binding protein类; NF-Y, nuclear transcription factor Y类等)在植物胁迫应答过程中也发挥重要的基因表达调控作用[5]。近年来, 研究证明NF-Y类、NAC类、DREB类等转录因子参与多种逆境信号转导途径[6]。ABA (abscisic acid)是调控植物非生物胁迫响应的主要激素[7]。植物调控非生物胁迫响应的信号途径主要被分为 ABA依赖性和ABA非依赖性信号途径[2-3]。DREB1A转录因子主要参与 ABA非依赖的逆境信号途径, 过表达DREB1A提高植物对低温的抗性[8]; 而 MYBR/C (MYB/C recognition site)和ABRE (ABA responsive element)作为ABA依赖性信号途径中的一类重要的顺式作用元件, 能够调节干旱响应基因的表达[9],过表达MYC类转录因子AtMYC2和MYB类转录因子AtMYB2基因可以提高植物对ABA的敏感性和对脱水逆境的耐受性[10]。
植物 NF-Y类转录因子是一类重要的逆境调节因子, 由NF-YA、NF-YB和NF-YC亚基组成。在行使功能时, NF-YB亚基和NF-YC亚基先在细胞质中形成异源二聚体, 然后迁移到细胞核中和NF-YA亚基结合形成有活性的异源三聚体[11-13]。NF-Y异源三聚体具有保守的 CCAAT-box位点结合特性, 而CCAAT-box顺式调控元件存在于真核生物约25%基因的启动子区域, 说明 NF-Y转录因子在调控真核生物细胞内基因表达方面具有重要作用[14-16]。NF-Y转录因子普遍存在于拟南芥[17]、水稻[18]、玉米[19]、大豆[20]、小麦[21-22]等作物中, 参与细胞增殖[23]、叶绿体形成[24]、胚胎发育、种子成熟[25]、光合作用[26-27]、固氮成分的合成[28-29]、花发育[30]等生长发育过程。除此之外, 一些NF-YA类转录因子在ABA信号途径中发挥重要作用。基因芯片分析结果显示, 诱导启动NF-YA2、NFYA3、NF-YA7和NF-YA10基因的表达可以下调PYR1/PYL/RCAR、PP2C和SnRK2等ABA信号途径相关基因的转录[31]。除此之外, 一些NF-YA类基因的拟南芥突变体和过表达植株在萌发期或者干旱条件下也表现与ABA有关的表型。拟南芥AtNF-YA5基因通过依赖ABA信号途径调控干旱胁迫[32], 并且在萌发期, NF-YA5突变体也表现出对ABA高度敏感[33]。定量分析显示, 过表达NF-YA1、NF-YA2、NF-YA3、NF-YA7、NF-YA9和NF-YA10均可导致拟南芥对ABA高度敏感[25]。这些研究结果都说明NF-Y转录因子调控植物抗逆反应依赖ABA信号途径, 关于NF-Y转录因子通过ABA非依赖途径调控植物抗逆反应未见报道。
谷子具有抗旱、耐贫瘠等特点, 是研究作物抗逆的理想材料[34]。本研究通过克隆并分析高盐胁迫响应NF-Y类转录因子SiNF-YA5基因的特性和生物学功能, 旨在为 NF-Y类转录因子调控植物抗逆性的信号途径提供证据, 同时也为作物耐盐遗传改良提供新的遗传资源。
1 材料与方法
1.1 试验材料
1.1.1 植物材料 拟南芥野生型(Columbia生态型, WT)由本实验室保存, 谷子品种龙谷 25由中国农业科学院作物科学研究所刁现民课题组提供。
1.1.2 载体和菌株 大肠杆菌、农杆菌GV3101、pBI121载体、GFP载体都由本实验室保存, pZero-Back载体购于北京天根公司。
1.1.3 试剂 限制性内切酶、T4 DNA连接酶购于Promega公司; in-Fusion克隆试剂盒购于TaRaKa公司; RT-PCR试剂盒购于全式金生物技术有限公司:质粒提取试剂盒、RNA提取试剂盒、DNA凝胶回收试剂盒、qRT-PCR试剂盒购于天根公司; 引物合成和测序由奥科生物技术科技有限公司完成; 其他化学药品为国产分析纯试剂。
1.2 谷子SiNF-YA5基因的生物信息学分析
谷子数据来源于Phytozome数据库(http://www. phytozome.net/search.php), 利用 SMART 数据库(http://smart.embl-heidelberg.de/)在线工具分析谷子SiNF-YA5蛋白的保守结构域。
1.3 SiNF-YA5基因的克隆及载体构建
根据谷子基因SiNF-YA5的CDS序列设计基因引物 A5-F1和A5-R1 (表 1), 用 RNA提取试剂盒(TIANGEN, 北京)提取龙谷 25植株总 RNA, 用TransScript II一步法反转录试剂盒(TransGen, 北京)反转录成cDNA, 以cDNA为模板扩增SiNF-YA5, 并将其回收纯化, 采用in-Fusion试剂盒(TaKaRa)将其连接到pZeroBack载体上。以pZeroBack-SiNF-YA5质粒为模板, 引物为 A5-F2和 A5-R2 (表 1)扩增SiNF-YA5, BamH I酶切 GFP表达载体, 利用in-Fusion技术构建载体 16318hGFP-SiNF-YA5。同样以 pZeroBack-SiNF-YA5质粒为模板, 引物为A5-F3和 A5-R3 (表 1)扩增 SiNF-YA5, Sma I酶切pBI121表达载体, 利用 in-Fusion技术构建载体pBI121-SiNF-YA5。
1.4 SiNF-YA5的亚细胞定位
参考 Yoo等[35]的方法制备谷子原生质体, 将融合表达的重组质粒 p16318hGFP-SiNF-YA5和 GFP空载体质粒作为对照分别转化原生质体, 黑暗培养16 h以上, 并在激光共聚焦显微镜(Zeiss LSM700)下观察定位情况。
1.5 拟南芥转化
参考 Clough等[36]方法进行 SiNF-YA5基因的遗传转化, 将收获的T0代种子种于含卡那霉素(50 mg L-1)的MS0培养基上, 筛选、扩繁获得T3代的纯合转基因株系 OE1、OE2和 OE3, 进一步分析其功能。
1.6 RNA提取及SiNF-YA5的表达谱分析
将谷子幼苗在营养土中正常生长(温度22℃、相对湿度65%、光照周期16 h光照/8 h黑暗) 3周后分别移至干旱(6% PEG-6000)、ABA (100 μmol L-1)、NaCl (100 mmol L-1)、低氮(0.2 mmol L-1)的水培营养液中胁迫处理, 于处理后0、1、6和24 h分别取样,用 RNA提取试剂盒(TIANGEN)提取谷子植株总RNA。另外取NaCl (MS0+125 mmol L-1)处理的拟南芥转基因和 WT植株, 提取总 RNA, -80℃保存备用。分别用4种胁迫下的谷子总RNA和NaCl处理下的拟南芥转基因和WT植株RNA反转录产物作为模板, 以 TransScriptII一步法反转录试剂盒(TransGen, 北京)反转录成 cDNA, 以 SYBR Green染料法, 在 ABI Prism 7500上进行实时荧光定量PCR。RT-PCR反应体系含: 2×SuperReal PreMix Plus (含荧光染料)(TIANGEN) 12.5 μL、10 μmol L-1正向引物和反向引物各0.5 μL、50×ROX Reference DyeΔ 0.5 μL、RNase-free ddH2O 9.5 μL。反应条件为95℃预变性10 min; 95℃变性15 s, 60℃退火20 s, 72℃延伸30 s, 并收集荧光信号, 35个循环, 用2-ΔΔCt法计算该基因表达量。谷子SiNF-YA5基因qRT-PCR引物为 A5-F4和 A5-R4, 内参基因(Si001873m.g)引物为 SiActin-F和 SiActin-R (表 1); 拟南芥下游基因qRT-PCR引物为NHX1-F和NHX1-R、LEA7-F和LEA7-R, 内参基因(AT3G15260)引物为AtActin-F和AtActin-R (表1)。
1.7 转SiNF-YA5基因拟南芥的抗盐性分析
将WT和转SiNF-YA5基因株系OE1、OE2、OE3的种子经70%酒精处理3 min, 无菌水清洗3次, 每次1 min左右; 用0.5%~0.8%的次氯酸钠处理15 min,无菌水清洗3次, 每次1 min; 4℃春化3 d, 再将种子分别点播至MS0、MS0+75 mmol L-1NaCl、MS0+100 mmol L-1NaCl和MS0+125 mmol L-1NaCl的培养基上, 每种材料64粒种子, 重复3次。在22℃、相对湿度65%、光照周期16 h光照/8 h黑暗条件下萌发种子, 统计萌发率, 从第1天开始统计, 连续统计4 d;同时, 将MS0培养基上正常生长7 d的拟南芥幼苗转移至MS0、MS0+100 mmol L-1NaCl、MS0+125 mmol L-1NaCl和MS0+150 mmol L-1NaCl培养基上, 垂直培养7 d, 统计不同浓度处理下SiNF-YA5过表达株系和 WT植株鲜重, 使用根系扫描仪(WINRHIZO proLA2400)分析根长, 试验重复 3次, 运用方差分析软件分析转基因株系与野生型之间的差异。
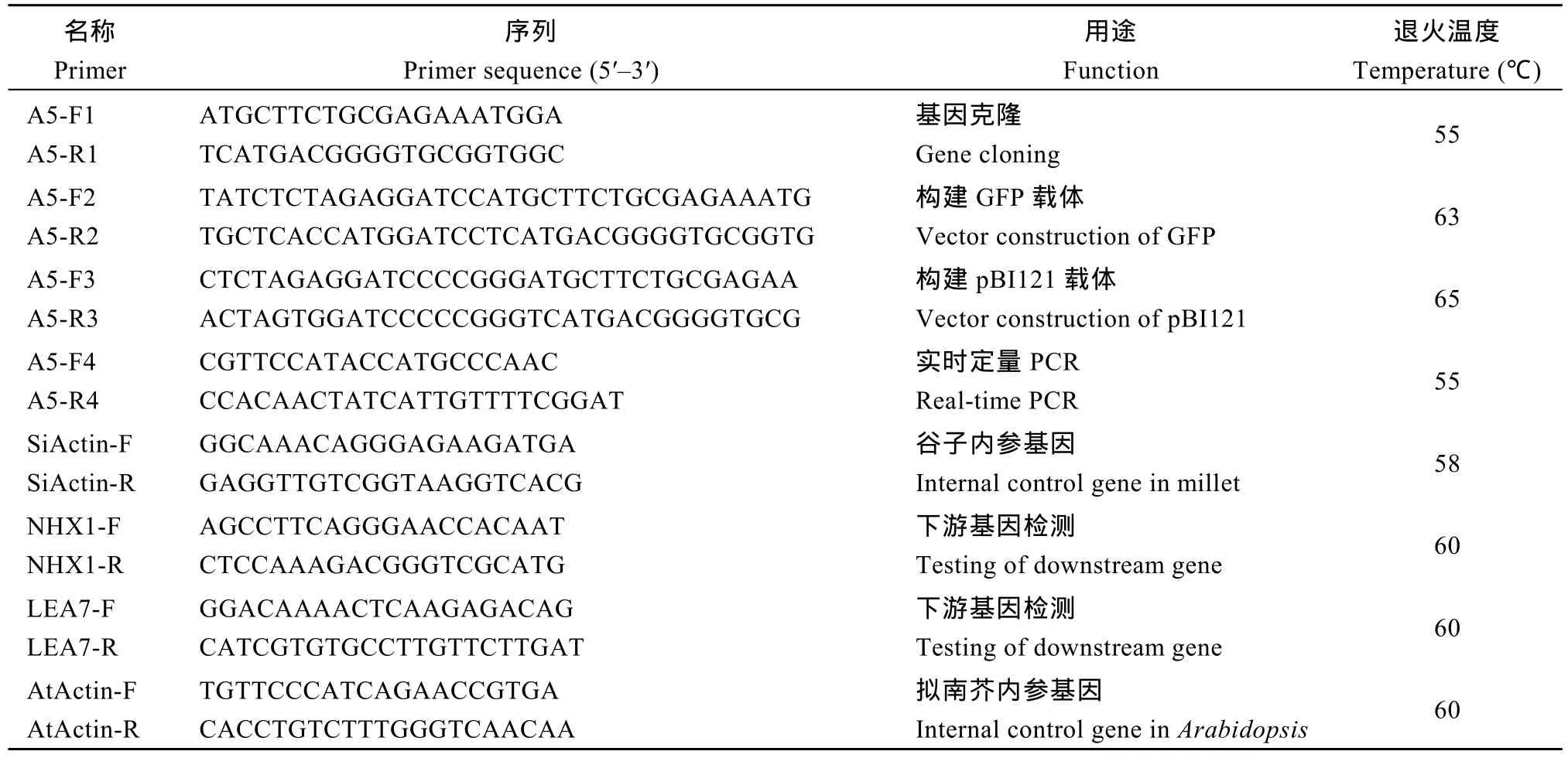
表1 SiNF-YA5基因克隆和Real-time PCR分析所用引物Table 1 Primers used for gene cloning and real-time PCR analysis
1.8 转SiNF-YA5基因拟南芥对ABA敏感性分析
方法同 1.7。萌发试验的 ABA处理浓度为 0.5 μmol L-1和1 μmol L-1, 苗期敏感性试验的ABA处理浓度为30 μmol L-1和40 μmol L-1。
2 结果与分析
2.1 SiNF-YA5基因的特性分析
前期工作对谷子干旱胁迫转录组分析发现 1个在干旱处理下表达上调的 NF-YA类转录因子基因SiNF-YA5。在谷子基因组数据库(http://www. phytozome.net/)搜索 SiNF-YA5全长序列, 发现SiNF-YA5基因编码序列为924 bp, 有6个外显子, 5个内含子, 编码307个氨基酸, 分子量为33.76 kD。SiNF-YA5在149~210位氨基酸之间含有CBF保守域, 属于CCAAT结合蛋白家族。
2.2 SiNF-YA5基因的表达模式分析
利用qRT-PCR分别检测结果(图1), 在NaCl处理下, SiNF-YA5的表达量逐渐上升并在24 h达到最大, 表达量是处理前的 13.0倍; 在 PEG处理下, SiNF-YA5的表达量逐渐上升, 在24 h达最高值, 表达量提高了4.0倍; 在低氮处理下, SiNF-YA5的表达量也呈上升趋势, 在12 h达最高值, 表达量提升了6.0倍左右。在ABA处理下, SiNF-YA5的表达量在1 h有所上升, 但相比处理前仅提高1.5倍。
2.3 SiNF-YA5蛋白亚细胞定位分析
将融合表达的重组质粒p16318hGFP-SiNF-YA5和GFP空载体质粒作为对照分别转化制备的原生质体, 激光共聚焦显微镜下观察结果显示, 对照 GFP蛋白在细胞核、细胞质、细胞膜中均有表达; 而转入 16318hGFP-SiNF-YA5融合载体的原生质体在细胞核和细胞膜上都能观察到绿色荧光信号, 表明SiNF-YA5定位在细胞膜和细胞核中(图2)。
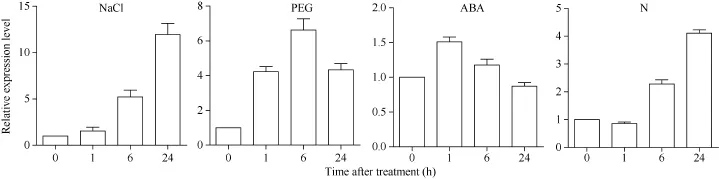
图1 SiNF-YA5在不同处理下的表达模式Fig. 1 Expression patterns of the SiNF-YA5 gene under various treatments
2.4 高盐条件下SiNF-YA5转基因拟南芥种子萌发率分析
从第1天观察WT和SiNF-YA5转基因拟南芥株系OE-1、OE-2和OE-3, 连续4 d统计萌发率。结果显示, 在MS0培养基中的SiNF-YA5转基因拟南芥和WT种子萌发率基本保持一致, 在24 h以后萌发率维持在95%左右(图3和图4-A); 在75 mmol L-1NaCl的培养基中, 在24 h WT不萌发, SiNF-YA5转基因拟南芥少量萌发, 在 48 h, 两者的萌发率相差最大, WT为26.3%, SiNF-YA5转基因拟南芥为84.5%, 72 h以后SiNF-YA5转基因拟南芥萌发率接近100%,两者差异逐渐减小(图3和图4-A); 在100 mmol L-1NaCl和125 mmol L-1NaCl的培养基中, 分别在36 h和48 h, SiNF-YA5转基因拟南芥和WT的萌发率才开始出现差异, SiNF-YA5转基因拟南芥的萌发率始终显著高于WT (图3和图4-A)。以上结果表明, 高盐处理条件下, SiNF-YA5转基因拟南芥的萌发率明显高于WT, 随着NaCl浓度的增加, WT和SiNF-YA5转基因拟南芥的萌发速率都减慢, 但SiNF-YA5转基因拟南芥的萌发率始终显著高于WT。另外, 在萌发第5天统计, 在100 mmol L-1和125 mmol L-1NaCl处理条件下, SiNF-YA5转基因拟南芥绿苗数高于WT, 并达到极显著水平(图 4-B)。说明在拟南芥中过表达SiNF-YA5基因提高了拟南芥萌发期对高盐胁迫的耐受性, SiNF-YA5正向调节植物对高盐胁迫的耐性。

图2 16318hGFP-SiNF-YA5蛋白的亚细胞定位分析结果Fig. 2 Subcellular localization of 16318hGFPSiNF-YA5 protein

图3 高盐处理下SiNF-YA5转基因拟南芥和WT种子萌发情况Fig. 3 Seed germination situation of SiNF-YA5 transgenic Arabidopsis and WT under high salt stress condition
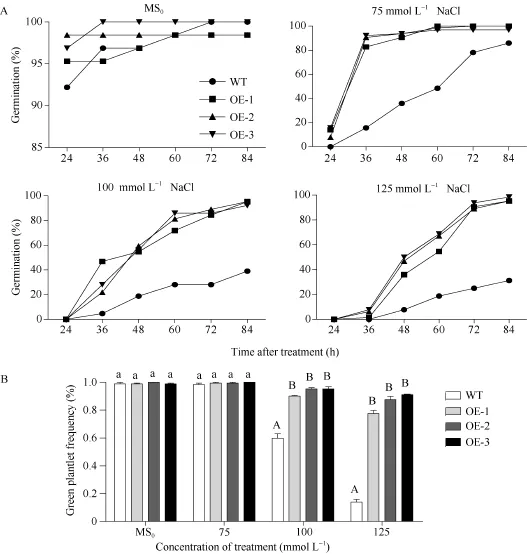
图4 高盐处理下SiNF-YA5转基因拟南芥和WT种子的萌发率和绿苗率Fig. 4 Seed germination rates and green plantlet rates of SiNF-YA5 transgenic Arabidopsis and WT under high salt stressA: 高盐处理下的种子萌发率; B: 高盐处理下的绿苗率; 采用单因素方差分析法对数据进行统计分析, 柱上不同的小写字母代表柱值在0.05水平上差异显著, 不同大写字母代表柱值在0.01水平上差异显著。A: seed germination rate under high salt treatment; B: green plantlet frequency under high salt treatment; Data statically analysis was made by the means of one-way ANOVA. The values marked with different lowercase letters on the columns are significantly different at the 0.05 level; the values marked with different capital letters on the columns are significantly different at the 0.01 level.
2.5 SiNF-YA5转基因拟南芥苗期耐盐性鉴定
垂直培养 7 d后显示, 在正常 MS0培养基上, SiNF-YA5转基因拟南芥根表面积和植株鲜重与WT比较没有明显差别(图 5和图 6-A), 而在 100 mmol L-1和 125 mmol L-1NaCl胁迫处理下, 与WT相比, SiNF-YA5转基因拟南芥的根表面积及植株鲜重增加, 在125 mmol L-1NaCl处理下的根表面积差异达到极显著水平(图 6-A), 转基因株系的鲜重与WT相比差异达到显著水平(图6-B)。以上结果表明在植物中过表达SiNF-YA5基因可以显著提高转基因拟南芥苗期耐盐性。在 150 mmol L-1NaCl处理条件下, 转基因拟南芥及 WT都趋向于死亡, 差异不显著(图5)。
2.6 高盐胁迫响应相关基因的表达分析
图 7显示, 在盐胁迫下, 盐胁迫响应相关基因Na+/H+转运蛋白基因(NHX1)和种子胚胎发育后期富集的脱水保护蛋白基因(LEA7)在SiNF-YA5转基因拟南芥中的表达明显高于WT, 说明SiNF-YA5可能通过控制拟南芥中盐胁迫相关基因NHX1和LEA7的表达来提高植物耐盐性。
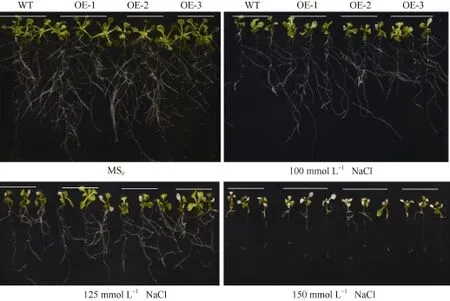
图5 高盐处理下SiNF-YA5转基因拟南芥和WT苗期表型Fig. 5 Phenotype of SiNF-YA5 transgenic Arabidopsis and WT seedlings under high salt stress

图6 高盐处理下SiNF-YA5转基因拟南芥和WT苗期根表面积和鲜重Fig. 6 Root surfaces and fresh weights of SiNF-YA5 transgenic Arabidopsis and WT seedlings under high salt stressA: 高盐处理下的根表面积; B: 高盐处理下植株鲜重。采用单因素方差分析法对数据进行统计分析, 柱上不同的小写字母代表柱值在0.05水平上差异显著, 不同大写字母代表柱值在0.01水平上差异显著。A: root surface under high salt treatment; B: fresh weight under high salt treatment. Data statically analyzed by using method of one-way ANOVA. The values marked with different lowercase letters on the columns are significantly different at the 0.05 level; the values marked with different capital letters on the columns are significantly different at the 0.01 level.

图7 NaCl处理下SiNF-YA5转基因拟南芥中盐胁迫相关基因表达水平Fig. 7 Expression levels of two stress-tolerant genes in SiNF-YA5 transgenic Arabidopsis under NaCl treatment
2.7 SiNF-YA5转基因植株对ABA敏感性分析
在ABA处理下, WT和SiNF-YA5转基因株系萌发率无差异(图8-A和图9)。取上述MS0培养基上正常生长7 d的WT和SiNF-YA5转基因拟南芥幼苗, 分别转接到正常MS0和含有30 μmol L-1和40 μmol L-1ABA的MS0培养基上照光, 垂直培养7 d, 结果显示, WT和转基因拟南芥在地上部分和地下部分无显著差异(图8-B)。说明无论在萌发期还是苗期, SiNF-YA5转基因拟南芥与 WT相比对ABA 敏感性没有差异, 证明 SiNF-YA5不参与ABA信号途径, 它通过ABA非依赖途径调控植物的耐盐性。
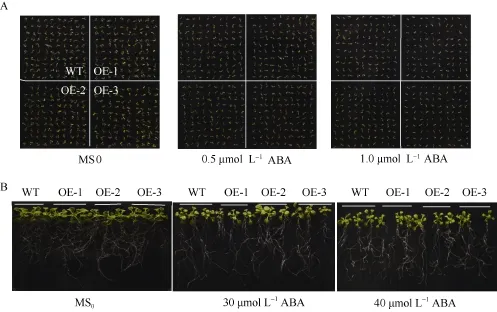
图8 SiNF-YA5转基因拟南芥和WT对ABA敏感性Fig. 8 Sensitivity analysis of SiNF-YA5 transgenic Arabidopsis and WT under ABA treatmentA: SiNF-YA5转基因拟南芥和WT种子在含有0.5 μmol L-1和1.0 μmol L-1ABA的培养基上萌发情况; B: SiNF-YA5转基因拟南芥幼苗和WT幼苗在含有30 μmol L-1和40 μmol L-1ABA的培养基上对ABA敏感性。A: seed germination situation of SiNF-YA5 transgenic Arabidopsis and WT under 0.5 μmol L-1and 1.0 μmol L-1ABA treatment; B: sensitivity analysis of SiNF-YA5 transgenic Arabidopsis seedlings under 30 μmol L-1and 40 μmol L-1ABA treatment.

图9 ABA处理下SiNF-YA5转基因拟南芥和WT种子的萌发率Fig. 9 Seed germination rates of SiNF-YA5 transgenic Arabidopsis and WT under ABA treatment
3 讨论
NF-Y转录因子是一类重要的逆境调控因子, 它由3类亚基构成, NF-YA亚基可进一步被分为7个亚族。本研究从谷子中克隆 1个 NF-YA类基因SiNF-YA5, 根据已经发表的谷子NF-YA类转录因子进化树分析结果显示 SiNF-YA5属于第 III亚族[37],与水稻NF-YA蛋白(OsHAP2E)进化关系最近, 而与拟南芥NF-YA蛋白进化关系较远。目前, 已经报道许多 NF-Y类转录因子依赖 ABA信号途径参与干旱、耐盐胁迫反应。大豆GmNF-YA3受ABA和NaCl胁迫诱导表达, 在拟南芥中过表达 GmNF-YA3能够提高植物的抗旱性。在正常条件下, 在 GmNF-YA3过表达拟南芥中, ABA合成及信号传导相关基因和胁迫相关基因转录水平提高[20]。拟南芥 AtNF-YA1基因依赖 ABA信号途径负向调控植物盐胁迫耐性,抑制幼苗的生长。在苗期, 过表达AtNF-YA1基因的转基因拟南芥提高了植物对盐和ABA的敏感性, 当ABA抑制剂存在时, 过表达 AtNF-YA1拟南芥对盐敏感的表型恢复[38]。谷子 SiNF-YA1 (Si037045m)和SiNF-YB8 (Si032469m)基因通过ABA信号途径激活胁迫相关基因表达, 改善植物生理特性从而正向调节植物耐盐性和耐旱性[37]。同时发现SiNF-YA5与拟南芥AtNF-YA1、谷子SiNF-YA1和SiNF-YB8均不在同一亚族, 推测SiNF-YA5可能通过与上述基因不同的其他途径调控耐盐和抗旱性。本文研究结果表明,在高盐处理条件下, SiNF-YA5转基因拟南芥在萌发期的萌发率显著高于WT (图4); 在苗期, SiNF-YA5转基因拟南芥的根系比WT发达, 鲜重显著大于WT (图 6)。然而, 与 SiNF-YA1和 SiNF-YB8基因不同, SiNF-YA5转基因拟南芥在萌发期和苗期对 ABA均不敏感(图8), 所以推测SiNF-YA5通过ABA非依赖途径提高植物对 NaCl胁迫的耐性。Chamindika创建拟南芥NF-YA类转录因子的10个过表达材料, 观察ABA调节的种子萌发和植物生长发育的情况, 结果发现所有的材料生长发育受到抑制, 但萌发期对ABA敏感性存在差异。在ABA不敏感的过表达材料中进行基因表达检测发现 ABA信号途径相关基因下调[39]。同样, NF-YC类转录因子在种子萌发期对ABA的反应也不完全相同, 种子萌发期NF-YC4突变体对ABA敏感, 而NF-YC3 和NF-YC9突变体则不敏感[40]。这些研究结果都表明 NF-Y类转录因子调控耐盐的途径存在差异, 同时存在着ABA依赖型和ABA非依赖型的耐盐信号调控途径。
在SiNF-YA5转基因株系中, 我们检测到参与盐胁迫响应基因NHX1和LEA7的表达量较WT都显著提高(图7)。同时, 分别分析NHX1和LEA7基因的启动子区域, 发现两者均有4个CAATT-box结构域(NF-Y类转录因子结合元件), 因此推测SiNF-YA5可能主要通过 ABA非依赖途径直接激活下游基因NHX1和 LEA7基因表达完成植物的耐盐调控。NHX1是第1个在拟南芥中发现的Na+/H+转运蛋白,能够促进钠离子在液泡中的积累[41]。盐胁迫环境下构建及分析 SOS 转录调控网络时发现拟南芥 bZIP类转录因子At5g65210首先通过ABA非依赖途径接收细胞膜上感受器传递的外界 Na+信号, 然后调控液泡膜上的 Na+/H+转运基因 NHX1[42]。NHX1主要利用液泡膜的H+-ATPase和液泡膜H+-PPase产生的跨膜质子梯度将胞质中的 Na+逆浓度梯度运入液泡中降低 Na+对植物细胞的毒害作用[43]。LEA蛋白是一类参与细胞抗逆保护的蛋白质, 广泛存在于植物的种子中, 能够在干旱胁迫时保护膜系统以及生物大分子免受脱水伤害。植物遭受干旱、盐渍及低温等胁迫时, 体内LEA基因的表达会增加。已有研究报道LEA基因可以增加转基因水稻耐盐和抗旱性[44]。关于SiNF-YA5提高转基因植物对高盐胁迫耐性的信号途径还需要进一步分析, 本研究初步阐明了谷子SiNF-YA5在调节高盐胁迫响应中的 ABA非依赖型信号途径, 为进一步了解谷子抗逆机制提供了新的依据。
4 结论
从谷子中分离出NF-Y类转录因子基因SiNF-YA5。SiNF-YA5被定位于细胞核及细胞膜。SiNF-YA5受低氮、干旱、高盐等胁迫的诱导表达。在植物中过表达SiNF-YA5可以显著提高植物在萌发期及苗期的耐盐性。SiNF-YA5转基因植物对ABA的敏感性与WT差异不显著, 证明SiNF-YA5通过ABA非依赖途径调控植物的耐盐性。
[1] Boyer J S. Plant productivity and environment. Science, 1982, 218: 443-448
[2] Xiong L M, Schumaker K S, Zhu J K. Cell signaling during cold, drought, and salt stress. Plant Cell, 2002, 14: S165-S183
[3] Zhu J K. Salt and drought stress signal transduction in plants. Annu Rev Plant Biol, 2002, 53: 247-273
[4] The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 2000, 408: 796-815
[5] Riechmann J L, Ratcliffe O J. A genomic perspective on plant transcription factors. Curr Opin Plant Biol, 2000, 3: 423-434
[6] Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol, 1999, 17: 287-291
[7] Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol, 2006, 57: 781-803
[8] Jiang C, Iu B, Singh J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol, 1996, 30: 679-684
[9] Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA, 2000, 97: 11632-11637
[10] Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB)function as transcriptional activators in abscisic acid signaling. Plant Cell, 2003, 15: 63-78
[11] Mantovani R, Li X Y, Pessara U, Hooft van Huisjduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem, 1994, 32: 20340-20346
[12] Frontini M, Imbriano C, Manni I, Mantovani R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle, 2004, 3:217-222
[13] Kahle J, Baake M, Doenecke D, Albig W. Subunits of the heterotrimeric transcription factor NF-Y are imported into the nucleus by distinct pathways involving importin beta and importin 13. Mol Cell Biol, 2005, 25: 5339-5354
[14] Steidl S, Tuncher A, Goda H, Guder C, Papadopoulou N, Kobayashi T, Tsukagoshi N, Kato M, Brakhage A. A single subunit of a heterotrimeric CCAAT-binding complex carries a nuclear localization signal: piggy back transport of the pre-assembled complex to the nucleus. J Mol Biol, 2004, 342: 515-524
[15] Ceribelli M, Dolfini, D, Merico D, Gatta R, Vigano A M., Pavesi G, Mantovani R. The histone-like NF-Y is a bifunctional transcription factor. Mol Cell Biol, 2008, 28: 2047-2058
[16] Maity S N, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci, 1998, 23:174-178
[17] Siefers N, Dang K K, Kumimoto R W, Bynum W E, Tayrose G, Holt B F. Tissue-specific expression patterns of Arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol, 2009, 149: 625-641
[18] Thirumurugan T, Ito Y, Kubo T, Serizawa A, Kurata N. Identification, characterization and interaction of HAP family genes in rice. Mol Genet Genomics, 2008, 279: 279-289
[19] Nelson D E, Repetti P P, Adams T R, Creelman R A, Wu J, Warner D C, Anstrom D C, Bensen R J, Castiglioni P P, Donnarummo M G, Hinchey B S, Kumimoto R W, Maszle D R, Canales R D, Krolikowski K A, Dotson S B, Gutterson N, Ratcliffe O J, Heard J E. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc Natl Acad Sci USA, 2007, 104: 16450-16455
[20] Ni, Z Y, Hu Z, Jiang Q Y, Zhang H. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol Biol, 2013, 82: 113-129
[21] Qu B, He X, Wang J, Zhao Y, Teng W, Shao A, Zhao X, Ma W, Li B, Li Z, Tong Y. A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol, 2014; 167: 411-423
[22] Stephenson T J, McIntyre C L, Collet C, Xue G P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in Triticum aestivum. Plant Mol Biol, 2007, 65: 77-92
[23] Sun X C, Ling S, Lu Z H, Ouyang Y D, Liu S S, Yao J. OsNF-YB1, a rice endosperm-specific gene, is essential for cell proliferation in endosperm development. Gene, 2014, 551: 214-221
[24] Miyoshi K, Ito Y, Serizawa A, Kurata N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J, 2003, 36: 532-540
[25] Mu J, Tan H, Hong S, Liang Y, Zuo J. Arabidopsis transcription factor genes NF-YA1, 5, 6, and 9 play redundant roles in male gametogenesis, embryogenesis, and seed development. Mol Plant, 2013, 6: 188-201
[26] Alam M M, Tanaka T, Nakamura H, Ichikawa H, Kobayashi K, Yaeno T, Yamaoka N, Shimomoto K, Takayama K, Nishina H, Nishiguchi M. Overexpression of a rice heme activator protein gene (OsHAP2E) confers resistance to pathogens, salinity and drought, and increases photosynthesis and tiller number. Plant Biotechnol J, 2015, 13: 85-96
[27] Stephenson T J, McIntyre C L, Collet C, Xue G P. TaNF-YB3 is involved in the regulation of photosynthesis genes in Triticum aestivum. Funct Integr Genomics, 2011, 11: 327-340
[28] Combier J P, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, Niebel A. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev, 2006, 20: 3084-3088
[29] Zanetti M E, Blanco F A, Beker M P, Battaglia M, Aguilar O M. A C subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell, 2010, 22:4142-4157
[30] Hackenberg D, Keetman U, Grimm B. Homologous NF-YC2 subunit from Arabidopsis and tobacco is activated by photooxidative stress and induces flowering. Int J Mol Sci, 2012, 13:3458-3477
[31] Leyva-Gonzalez M A, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One, 2012, 7:e48138
[32] Li W X, Oono Y, Zhu J, He X J, Wu J M, Iida K, Lu X Y, Cui X P, Jin H L, Zhu J K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell, 2008, 20: 2238-2251
[33] Warpeha K M, Upadhyay S, Yeh J, Adamiak J, Hawkins S I, Lapik Y R, Anderson M B, Kaufman L S. The GCR1, GPA1, PRN1, NFY signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol, 2007, 143:1590-1600
[34] 刘敬科, 刁现民. 我国谷子产业现状与加工发展方向. 农业工程技术: 农产品加工业, 2013, (12): 15-17
Liu J K, Diao X M. Foxtail millet processing industry status and development trend in our country. Agric Eng Technol (Agric Prod Process), 2013, (12): 15-17 (in Chinese)
[35] Yoo S D, Cho Y H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc, 2007, 2: 1565-1572
[36] Beehtold N, Ellis J, Pelletier G. In plant Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Life Sci, 1993, 316: 1194-1199
[37] Feng Z J, He G H, Zheng W J, Lu P P, Chen M, Gong Y M, Ma Y Z, Xu Z S. Foxtail millet NF-Y families: genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front Plant Sci, 2015, 6: 1142
[38] Li Y J, Fang Y, Fu Y R, Huang J G, Wu C A, Zheng C C. NFYA1 is involved in regulation of postgermination growth arrest under salt stress in Arabidopsis. PLoS One, 2013, 8(4): e61289
[39] Siriwardana C L, Kumimoto R W, Jones D S, Holt B F. Gene Family Analysis of the Arabidopsis NF-YA transcription factors reveals opposing abscisic acid responses during seed germination. Plant Mol Biol Rep, 2014, 32: 971-986
[40] Kumimoto R W, Siriwardana C L, Gayler K K, Risinger J R, Siefers N, Holt B F. NUCLEAR FACTOR Y transcription factors have both opposing and additive roles in ABA-mediated seed germination. PLoS One, 2013, 8: e59481
[41] Gaxiola R A, Rao R, Sherman A, Grisafi P, Alper S L, Fink G R. The Arabidopsis thaliana proton transporters, AtNHX1 and Avpl, can function in cation detoxification in yeast. Proc Natl Acad Sci USA, 1999, 96: 1480-1485
[42] 谢崇波, 金谷雷, 徐海明, 朱军. 拟南芥在盐胁迫环境下 SOS转录调控网络的构建及分析. 遗传, 2010, 6: 639-646
Xie C B, Jin G L, Xu H M, Zhu J. Construction and analysis of SOS pathway-related transcription a regulatory network underlying salt stress response in Arabidopsis. Hereditas (Beijing), 2010, 6: 639-646 (in Chinese with English abstract)
[43] Wu Y Y, Chen Q J, Chen M, Chen J, Wang X C. Salt-tolerant transgenic perennial ryegrass (Lolium perenne L.) obtained by Agrobacterium tumefaciens-mediated transformation of the vacuolar Na+/H+antiporter gene. Plant Sci, 2005, 169: 65-73
[44] Babu R C, Zhang J X, Blum A, Ho T H D, Wu R, Nguyen H T. HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci, 2004, 166: 855-862
Transcription Factor SiNF-YA5 from Foxtail Millet (Setaria italica) Conferred Tolerance to High-salt Stress through ABA-independent Pathway in Transgenic Arabidopsis
HUANG Suo1,**, HU Li-Qin1,**, XU Dong-Bei1,2, LI Wei-Wei1,3, XU Zhao-Shi1, LI Lian-Cheng1, ZHOU Yong-Bin1,2, DIAO Xian-Min1, JIA Guan-Qing1, MA You-Zhi1, and CHEN Ming1,*
1Institute of Crop Science, Chinese Academy of Agricultural Sciences / National Key Facility For Crop Gene Resource and Genetic Improvement / Key Laboratory of Biology and Genetic Improvement of Triticeae Crop, Ministry of Agriculture, Beijing 100081, China;2College of Agronomy, Northwest A&F University / State Key Laboratory of Arid Region Crop Adversity Biology, Yangling 712100, China;3College of Life Science and Technology, Harbin Normal University / Key Laboratory of Molecular Cytogenetics and Genetic Breeding of Heilongjiang Province, Harbin 150025, China
Nuclear transcription factor Y (NF-Y) consisting of three subunits, NF-YA, NF-YB, and NF-YC, plays an essential role in many biologic processes, including growth, development, and abiotic stress response. In this study, an NF-Y like transcription factor gene SiNF-YA5 was isolated from foxtail millet variety Longgu 25. The full-length sequence of SiNF-YA5 gene is 924 bp, encoding 307 amino acids. Molecular weight and isoelectric point of SiNF-YA5 protein are 33.76 kD and 9.19, respectively. There is a conserved CBF domain from the 149th to the 210th amino acids of SiNF-YA5. According to the subcellular localization analysis, SiNF-YA5 was mainly localized and expressed on the plasma membrane and nucleus in plant cell. Gene functionalanalysis showed that under different NaCl concentration treatments, the germination rate of SiNF-YA5 transgenic Arabidopsis was significantly higher than that of wild-type (WT) Arabidopsis during seed germination stage; root surface area and fresh weight of SiNF-YA5 transgenic Arabidopsis remarkably increased compared with WT during seedling stage. Those results indicated that the overexpression of SiNF-YA5 in transgenic plants could enhance tolerance to high salt. Gene expression analysis showed that the expressions of two salt stress related genes, namely NHX1 and LEA7, increased significantly in SiNF-YA5 transgenic plants. On the other hand, there was no obvious difference in sensitivity to ABA between SiNF-YA5 transgenic Arabidopsis and WT showed during seed germination and seedling stages indicating that SiNF-YA5 could enhance salt tolerance through ABA-independent pathway in transgenic plants.
Foxtail millet (Setaria italic); NF-Y like transcription factor; High salt stress; ABA independent signaling pathway
10.3724/SP.J.1006.2016.01787
本研究由国家转基因新品种生物培育科技重大专项(2016ZX08002-002)和中国农业科学院创新工程资助。
This work was funded by the National Majar Project for Developing New GM Crops (2014ZX08002-002) and the Innovation Project of Chinese Academy of Agricultural Sciences.
*通讯作者(Corresponding author): 陈明, E-mail: chenming02@caas.cn, Tel: 13683360891**同等贡献(Contributed equally to this work)
联系方式: E-mail: hnndhs@126.com, Tel: 17701300735
稿日期): 2016-03-06; Accepted(接受日期): 2016-06-20; Published online(
日期): 2016-07-04.
URL: http://www.cnki.net/kcms/detail/11.1809.S.20160704.0826.014.html

