Prevalence and Antimicrobial Susceptibility of Mycobacterium abscessus in a General Hospital, China*
LI Yan Ming, TONG Xun Liang, XU Hong Tao, JU Yang, CAI Meng, and WANG Chen
1. Department of Respiratory and Critical Care Medicine, Beijing Hospital, Beijing 100730, China; 2. Department of Geriatrics, Beijing Hospital, Beijing 100730, China; 3. Department of Laboratory Medicine, Beijing Hospital,Beijing 100730, China; 4. Department of Hospital Infection Control and Management, Beijing Hospital, Beijing 100730, China; 5. Department of Respiratory Medicine, Chinese-Japanese Friendship Hospital, Beijing 100029,China
Original Article
Prevalence and Antimicrobial Susceptibility of Mycobacterium abscessus in a General Hospital, China*
LI Yan Ming1,4,&, TONG Xun Liang2,&, XU Hong Tao3, JU Yang1, CAI Meng4, and WANG Chen5,#
1. Department of Respiratory and Critical Care Medicine, Beijing Hospital, Beijing 100730, China; 2. Department of Geriatrics, Beijing Hospital, Beijing 100730, China; 3. Department of Laboratory Medicine, Beijing Hospital,Beijing 100730, China; 4. Department of Hospital Infection Control and Management, Beijing Hospital, Beijing 100730, China; 5. Department of Respiratory Medicine, Chinese-Japanese Friendship Hospital, Beijing 100029,China
Abstract
Objective To gain greater insight into the prevalence drug resistant profiles of M. abscessus from a general hospital in Beijing, China.
Methods Partial gene sequencing of 16S, hsp65, and rpoB were used to distinguish the species of NTM isolates. All strains identified as M. abscessus were further enrolled in the drug susceptibility testing by using broth microdilution method.
Results We found that M. avium complex was the most frequent NTM organism, accounting for 54.1% (33/61) of all isolates. Behind MAC, the second most common organisms were M. abscessus (22 out of 61, 36.1%). Average rates of resistance were 4.5% for AMK, 9.1% for LZD, and 13.6% for CLA,respectively. In contrast, resistance to LEV (17/22, 77.3%), IMI (9/22, 40.9%), and SMX (10/22, 45.5%)was noted in more than 40% of M. abscessus isolates. DNA sequencing revealed that all the CLA-resistant isolates harbored nucleotide substitutions in position 2058 (1/3, 33.3%) or 2059 (2/3,66.7%) of 23S rRNA.
Conclusion In conclusion, our data demonstrated that M. intracellulare and M. abscessus were the most common NTM species in the general hospital of Beijing. CLA, AMK, LZD showed promising activity,where as LEV, IMI, and SMX exhibited poor activity against M. abscessus in vitro.
Nontuberculous mycobacteria; Mycobacterium abscessus; China
www.besjournal.com (full text) CN: 11-2816/Q Copyright ©2016 by China CDC
INTRODUCTION
A lthough Mycobacterium tuberculosis is one of the most important mycobacterial species threatening the public health all over the world, other species, known as nontuberculous mycobacteria (NTM), are being responsible for increasing emergency of NTM disease in human[1-3]. NTM are commonly isolated from different environmental sources, including soil,treated and untreated water, animals and food,divided into slow growing mycobacteria (SGM) and rapid growing mycobacteria (RGM) according to their behavior in the culture[2].
Of the rapid growing mycobacteria relevant to human disease (including M. fortuitum, M. abscessus,and M. chelonae), M. abscessus is the most common type to cause lung disease, and is also the most difficult to treat in the clinical practice[4-5], the mobility rate of which have be nearly 20% in susceptible individuals in the past decades[5]. The major problem during treatment is that M. abscessus is not responsive to ‘standard’ antituberculosis agents, but susceptible to other common antibiotics,such as macrolides, beta-lactams or tetracyclines[6]. The drug susceptibility of the isolates is variable,which make it essential to obtain in vitro drug susceptibility profile of individual M. abscessus strains to generate an effective therapeutic regimen for the patients[7].
In China, M. abscessus is one of the most common nontuberculous mycobacetria causing lung disease[8]. To gain greater insight into the prevalence drug resistant profiles of M. abscessus, we firstly identified the in vitro susceptibility of M. abscessus isolates from patient’s respiratory specimens from a general hospital in Beijing, China. Thirteen antibiotics,which were extensively used in the clinical practice for the treatment of NTM infections, were selected to perform the minimum inhibitory concentration (MIC) of M. abscessus.
MATERIALS AND METHODS
Bacterial Strains
Data were obtained from patients at Beijing Hospital during the 36-month period between April 1,2012, and March 31, 2015. During this period,sputum samples from all TB suspect patients were collected, and the sputum was digested with NALC-NaOH (4%) for 15 min, and inoculated onto Löwenstein-Jensen (L-J) medium according to the previous report[9]. The NTM isolates were firstly distinguished from M. tuberculosis isolates with PNB and TCH modified L-J medium. Partial gene sequencing of 16S, hsp65, and rpoB were used to distinguish the species of NTM isolates[8]. All strains identified as M. abscessus were further enrolled in the drug susceptibility testing. In addition to clinical isolates, one reference strain of M. abscessus,ATCC35761 were obtained from National Tuberculosis Reference Laboratory of China. The protocols applied in this study were approved by the Ethics Committee of Beijing Hospital, and informed consent was obtained from all patients whose sputum specimens were used in scientific studies.
Species Identification
All the clones growing on the L-J medium were scraped and genomic DNA was extracted by a rapid-boiling method[10]. The genomic DNA was used for the sequencing of multiple genes, including 16S rRNA, hsp65, and 16S-23S rRNA internal transcribed spacer (ITS) sequence, to perform molecular species identification[8]. The 50 μL PCR mixtures were prepared as follows: 5 μL 10×PCR buffer, 200 μmol/L of each dNTP, 0.2 μmol/L of each primer set, 5 μL crude genomic DNA, 1 U HotStar Taq polymerase (Qiagen). The amplification was performed in athemocycler (Bioer, Hangzhou, China) as follows: 5 min at 94 °C for initial denaturation, and then 35 cycles of denaturation at 94 °C for 1 min, annealing at 58 °C for 1 min, and extension at 72 °C for 2 min,followed by a final extension at 72 °C for 10 min. The PCR products were sent to Sango Company (Beijing,China) for sequencing. All the sequencing results were aligned to the GenBank database over the Internet by using the NCBI BLAST server (www.ncbi.nlm.nih.gov).
Minimal Inhibition Concentration (MIC)
Antimicrobial susceptibility testing was performed using broth microdilution method according to the guidelines by the National Committee for Clinical Laboratory Standards (NCCLS)[11]. Briefly, organisms scraped from the culture media was transferred to saline with 0.02% Tween 80. The suspension was mixed vigorously on a votex for 1 min until the bacterial colonies were dispersed homogeneously. Then the suspension was diluted to the density of a 0.5 McFarland standard. Followed by further dilution two hundred times with cation-adjusted Mueller-Hinton broth media (CAMHB), the diluted bacterial suspension was inoculated at a final concentration of 3.75×105CFU/mL. MICs were determined for 13 antimicrobial agents, which were used in the treatment regimen against M. abscessus infection, were selected for drug susceptibility profile analysis, including clarithromycin (CLA), azithromycin (AZM), amikacin (AMK), cefoxitin (CFX), imipenem (IMI), linezolid (LZD), moxifloxacin (MOX), levofloxacin (LEV),tigecycline (TGC), capreomycin (CAP), tobramycin (TOB), sulfamethoxazole (SMX), and clofazimine (CLO). All agents mentioned above were purchased from Sigma-Aldrich. The breakpoints used todetermine the resistance of M. abscessus were referenced from the the guidelines by NCCLS, which were shown in Table 1.
DNA Sequencing
The fragments of 23S rRNA and 16S rRNA were amplified by PCR, respectively. The primer pairs were synthesized as previously reported. PCR products were sent to Qingke Company (Beijing,China) for sequencing service. The resulting sequences were aligned to the homologous sequences of the reference M. abscessus strain (ATCC35761) using BLASTn in the National Center for Biotechnology Information website (www.ncbi.nlm. nih.gov/BLAST).
RESULTS
Identification of NTM Species
A total of 61 NTM isolates identified by conventional biochemical method were enrolled in further molecular identification. By the use of multilocus sequence analysis, the NTM strains could be divided into the species level. As shown in Table 1,M. avium complex was the most frequent NTM organism, accounting for 54.1% (33/61) of all isolates. Of 33 isolates classified as M. avium complex, there were 25 (36.1%) M. intracellulare and 8 (13.1%) M. avium isolates, respectively. Behind MAC, the second most common organisms were M. abscessus (22 out of 61, 36.1%); other organisms (6 out of 61,9.8%) included M. Kansasii (n=3), M. fortuitum (n=2),and M. chelonae (n=1).

Table 1. Breakpoint Values of Different Antimicrobial Agents
Drug Susceptibility Profiles
The distribution of MICs of each antimicrobial agent for M. abscessus isolates was shown in Table 2. Overall, CLA was highly active against M. abscessus strains, with MIC50of 0.06 μg/mL and MIC90of 8 μg/mL, respectively. Similarly, AMK, LZD, MOX,TGC, and CLO also showed activity against M. abscessus, the MIC50s and MIC90s of which were lower than 2 and 16 μg/mL, respectively. When using the breakpoint listed in Table 1, resistance to AMK (4.5%, 1/22) was rare, and only one AMK-resistant isolate was encountered in this study. Average rates of resistance were 9.1% for LZD and 13.6% for CLA, respectively. For fluoroquinolones,MOX and LEV exhibited different drug resistant profiles, and statistical analysis revealed that the percentages of MOX-resistance (6/22, 27.3%)isolates were significantly lower than LEV-resistant (17/22, 77.3%) among M. abscessus (P<0.01). In addition to LEV, resistance to IMI (9/22, 40.9%) and SMX (10/22, 45.5%) was noted in more than 40% of M. abscessus isolates (Table 3).
Mutations Conferring CLA and AMK Resistance
We further analyzed the genomic mutation conferring antimicrobial agent resistance. Mutations in 23S rRNA, which are associated with CLA resistance in mycobacteria, were firstly detected by DNA sequencing in all CLA-resistant M. abscessus isolates. As shown in Table 4, all the CLA-resistant isolates harbored nucleotide substitutions in position 2058 (1/3, 33.3%) or 2059 (2/3, 66.7%) of 23S rRNA. For AMK resistance, we found no genetic mutation within rrs gene, indicating that other drug-resistant mechanism may play an essential role in the AMK resistance in M. abscessus.
DISCUSSION
Lung diseases caused by NTM have been increasing worldwide, while the distribution of mycobacteria species varies significantly by geographic region[8]. In United States, M. avium complex is the most frequent pathogen associated with NTM lung diseases, followed by M. kansasii[12]. In England and Welsh, M. kansasii was the most common[13]. According to previous literatures from different regions of China, M. avium complex and M. abscessus accounted for the majority of isolated NTM species[8]. In line with previous findings, our data demonstrated that these two species were alsothe most predominant NTM species in the general hospital of Beijing. In China, most of patients with TB suspects primarily seek health care in the general hospital rather than TB specialized hospital or TB dispensary[14]. Different from pulmonary TB patients,patients caused by NTM infection can receive treatment in the general hospitals, which may avoid the nosocomial infection with tuberculosis during in-patient period. Hence, the high prevalence of NTM in China highlights the urgent need to perform rapid species identification among TB suspects in the general hospitals.

Table 2. In vitro Susceptibility of 22 M. abscessus Strains
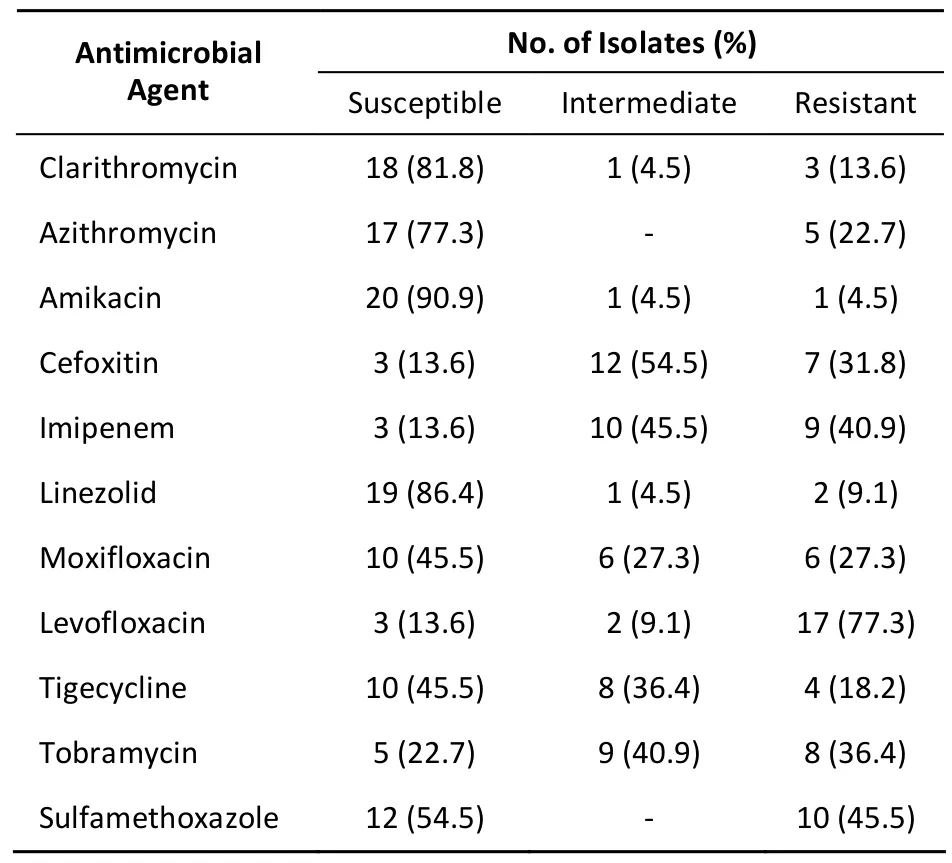
Table 3. Percentage of M. abscessus Isolates Against Different Antimicrobial Agents

Table 4. Mutations Conferring CLA and AZM Resistance in 3 Clinical M. abscessus Isolates
Due to the broad spectrum of drug resistance against many antibiotics, M. abscessus has been considered as the most resistant organisms to chemotherapeutic agents[6]. Before anti-infection treatment, in vitro susceptibilities to antimicrobial agents for clinical M. abscessus isolates are recommended to generate the effective chemotherapy regimens. According to the ATS/IDSA guidelines, CLA, CFX, and AMK are recommended for the treatment of patients infected with M. abscessus[1]. In the present study, we observed that CLA and AMK were in vitro active against M. abscessus, which was in consistent to previous report[15]. In contrast, 31.8% of M. abscessus in the present study showed resistant against CFX,indicating the addition of CFX in the treatment regimen might not improve the prognosis among one third of M. abscessus diseases. Compared withCFX, LZD, a member of the oxazolidinone class of antibiotics[16], showed better in vitro activity against M. abscessus isolated from China. The frequency of LZD-resistant M. abscessusisolates observed in this study (9.1%) is similar to that in India (7%)[17],although it is lower than that in Britain (63%)[18]and that in South Korea (20%)[19]. Several explanations may be attributed to this difference. On one hand, LZD is a new antimicrobial agent introduced in developing countries, such as China and India. The relatively short period used for the antimicrobial treatment in these regions may be the most important reason for the low proportion of LZD-resistance among M. abscessus isolates. On the other hand, the M. abscessus strains from Britain were mainly isolated from individuals with cystic fibrosis (CF), an indicator associated with poor clinical outcome[18]. The higher resistant prevalence aganist LZD may be due to their constant exposure to antibiotics. Our findings have demonstrated that LZD serves as an alternative for clinical treatment of M. abscessus,whereas the high price of LZD poses the biggest obstacle to its routine use against M. abscessus diseases in China.
CLA resistance in mycobacterium has been proved to be associated with mutations located in the 23S rRNA gene[20-22]. In agreement with previous studies, all CLA-resistant M. abscessus isolates harbored nucleotide substitutions at position 2058 and 2059 of the 23S rRNA gene, suggesting that this gene may used as an promising target for the prediction of CLA resistance in M. abscessus[6,23]. We also found an AMK-resistant M. abscessus isolate in this study. Although numerous literatures have mutations affecting the 16S rRNA gene confer high-level resistance to AMK[6,24], no nucleotide substitution has been detected in this AMK-resistant strain of this study. Further molecular analysis will provide new insight onthe functions of other additional resistance mechanisms conferring AMK resistance in M. abscessus, including efflux pump and cellular permeability.
In conclusion, our data demonstrated that M. intracellulare and M. abscessus were the most common NTM species in the general hospital of Beijing. CLA, AMK, LZD showed promising activity against M. abscessus in vitro, whereas LEV, IMI, and SMX exhibited poor activity against M. abscessus. In addition, mutations located in 23S rRNA could be used as a promising target for the prediction of CLA resistance in M. abscessus.
ACKNOWLEDGEMENTS
We would like to thank all the staff from the Beijing Hospital for their assistance.
Accepted: January 21, 2016
REFERENCES
1. Glassroth J. Pulmonary disease due to nontuberculous mycobacteria. Chest, 2008; 133, 243-51.
2. Winthrop KL. Pulmonary disease due to nontuberculous mycobacteria: an epidemiologist's view. Future Microbiol,2010; 5, 343-5.
3. McShane PJ, Glassroth J. Pulmonary Disease Due to Nontuberculous Mycobacteria: Current State and New Insights. Chest, 2015; 148, 1517-27.
4. Van Ingen J, Boeree MJ, Dekhuijzen PN, et al. Environmental sources of rapid growing nontuberculous mycobacteria causing disease in humans. Clin Microbiol Infect, 2009; 15, 888-93.
5. Jarand J, Levin A, Zhang L, et al. Clinical and microbiologic outcomes in patients receiving treatment for Mycobacterium abscessus pulmonary disease. Clin Infect Dis, 2011; 52, 565-71.
6. Nessar R, Cambau E, Reyrat JM, et al. Mycobacterium abscessus: a new antibiotic nightmare. J Antimicrob Chemother, 2012; 67, 810-18.
7. Daley CL, Glassroth J. Treatment of pulmonary nontuberculous mycobacterial infections: many questions remain. Ann Am Thorac Soc, 2014; 11, 96-7.
8. Zhang Z, Pang Y, Wang Y, et al. Differences in risk factors and drug susceptibility between Mycobacterium avium and Mycobacterium intracellulare lung diseases in China. Int J Antimicrob Agents, 2015; 45, 491-5.
9. Pang Y, Du J, Zhang ZY, et al. The feasibility of sputum transportation system in China: effect of sputum storage on the mycobacterial detection. Biomed Environ Sci, 2014; 27,982-6.
10. Pang Y, Zhou Y, Wang S, et al. A novel method based on high resolution melting (HRM) analysis for MIRU-VNTR genotyping of Mycobacterium tuberculosis. J Microbiol Methods, 2011; 86,291-7.
11. Clinical and Laboratory Standards Institute. Susceptibility testing of mycobacteria, nocardia, and other aerobic actinomycetes; approved standard, 2nd ed; CLSI document M24-A2. Clinical and Laboratory Standards Institute, Wayne,PA. 2011; 19-43.
12. Prevots DR, Shaw PA, Strickland D, et al. Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med, 2010;182, 970-6.
13. Moore JE, Kruijshaar ME, Ormerod LP, et al. Increasing reports of non-tuberculous mycobacteria in England, Wales and Northern Ireland, 1995-2006. BMC Public Health, 2010; 10,612.
14. Du J, Pang Y, Liu Y, et al. Survey of tuberculosis hospitals in China: current status and challenges. PLoS One, 2014; 9,e111945.
15. Chu HS, Chang SC, Shen EP, et al. Nontuberculous mycobacterial ocular infections--comparing the clinical and microbiological characteristics between Mycobacterium abscessus and Mycobacterium massiliense. PLoS One, 2015; 10,e0116236.
16. Zhang L, Pang Y, Yu X, et al. Linezolid in the treatment of extensively drug-resistant tuberculosis. Infection, 2014; 42,705-11.
17. Singh S, Bouzinbi N, Chaturvedi V, et al. In vitro evaluation of a new drug combination against clinical isolates belonging to the Mycobacterium abscessus complex. Clin Microbiol Infect, 2014;20, 1124-7.
18. Broda A, Jebbari H, Beaton K, et al. Comparative drug resistance of Mycobacterium abscessus and M. chelonae isolates from patients with and without cystic fibrosis in the United Kingdom. J Clin Microbiol, 2013; 51, 217-23.
19. Lyu J, Jang HJ, Song JW, et al. Outcomes in patients with Mycobacterium abscessus pulmonary disease treated with long-term injectable drugs. Respir Med, 2011; 105, 781-7.
20. Zhao X, Wang Y, Pang Y. Antimicrobial susceptibility and molecular characterization of Mycobacterium intracellulare in China. Infect Genet Evol, 2014; 27, 332-8.
21. Brown BA, Wallace RJ Jr, Onyi GO, et al. Activities of four macrolides, including clarithromycin, against Mycobacterium fortuitum, Mycobacterium chelonae, and M. chelonaelike organisms. Antimicrob Agents Chemother, 1992; 36,180-4.
22. Meier A, Heifets L, Wallace RJ Jr, et al. Molecular mechanisms of clarithromycin resistance in Mycobacterium avium: observation of multiple 23S rDNA mutations in a clonal population. J Infect Dis, 1996; 174, 354-60.
23. Wallace RJ Jr, Meier A, Brown BA, et al. Genetic basis for clarithromycin resistance among isolates of Mycobacterium chelonae and Mycobacterium abscessus. Antimicrob Agents Chemother, 1996; 40, 1676-81.
24. Prammananan T, Sander P, Brown BA, et al. A single 16S ribosomal RNA substitution is responsible for resistance to amikacin and other 2-deoxystreptamine aminoglycosides in Mycobacterium abscessus and Mycobacterium chelonae. J Infect Dis, 1998; 177, 1573-81.
Biomed Environ Sci, 2016; 29(2): 85-90 10.3967/bes2016.009 ISSN: 0895-3988
November 2, 2015;
*This work was supported by the grant 2012BAI05B02 from National Key Technology Research and Development Program; China and grant 81400037 from the National Natural Science Foundation of China.
&These authors contributed equally to this study.
#Correspondence should be addressed to WANG Chen, E-mail: cyh-birm@263.net
Biographical notes of the first authors: LI Yan Ming, female, born in 1971, MD, majoring in respiratory and intensive care; TONG Xun Liang, female, born in 1985, MD&PhD, majoring in allergy and infection disease research.
 Biomedical and Environmental Sciences2016年2期
Biomedical and Environmental Sciences2016年2期
- Biomedical and Environmental Sciences的其它文章
- The Survey of Cronobacter spp. (formerly Enterbacter sakazakii)in lnfant and Follow-up Powdered Formula in China in 2012*
- The Cellular Toxicity of PM2.5Emitted from Coal Combustion in Human Umbilical Vein Endothelial Cells*
- Cadmium Activates Reactive Oxygen Species-dependent AKT/mTOR and Mitochondrial Apoptotic Pathways in Neuronal Cells*
- Bioremediation of Hexavalent Chromium Pollution by Sporosarcina saromensis M52 lsolated from Offshore Sediments in Xiamen, China*
- Gene Knockdown in Human Rhinovirus 1B Using 2′-OMe-modified siRNAs Results in the Reactivation of the Interferon Response
- Protective Effects of Lycopene on Furan-treated Diabetic and Non-diabetic Rat Lung
