中国荷斯坦牛FADS2基因3′端SNP突变对乳中脂肪酸组成的影响
徐晨希,王梦琦,朱小瑞,张玉锋,夏海磊,刘贤慧,王小龙,张慧敏,杨章平,毛永江
(扬州大学动物科学与技术学院,江苏扬州 225009)
中国荷斯坦牛FADS2基因3′端SNP突变对乳中脂肪酸组成的影响
徐晨希,王梦琦,朱小瑞,张玉锋,夏海磊,刘贤慧,王小龙,张慧敏,杨章平,毛永江
(扬州大学动物科学与技术学院,江苏扬州 225009)
摘要:【目的】FADS2是多不饱和脂肪酸合成中关键的限速酶之一,可催化食物中亚油酸(LNA, C18:2n6)合成γ-亚麻酸(GLA, C18:3n6)、二十碳五烯酸(EPA, C20:5n3)、二十二碳六烯酸(DHA, C22:6n3)等长链脂肪酸。试验旨在探讨中国荷斯坦牛FADS2基因3′端非编码区SNP突变对乳中脂肪酸含量的影响。【方法】随机选择20头无亲缘关系的中国荷斯坦牛样本,用直接测序列法检测FADS2基因3′端非编码区SNP突变位点。再以江苏某大型奶牛场551头中国荷斯坦牛为材料,用飞行时间质谱法对前期发现的2个SNP位点进行检测,同时利用最小二乘模型分析了SNP突变及其单倍型对乳中脂肪酸含量及其不饱和指数的影响。【结果】 中国荷斯坦牛FADS2基因3′端非编码区存在3个SNP突变位点:c.1571 A>G、c.2743 A>G、c.2776 A>G。c.1571 A>G位点GG型为优势基因型,基因型频率为0.800,G为优势等位基因,基因频率为0.887。c.2776 A>G位点AA为优势基因型,基因型频率为 0.673,A为优势等位基因,基因频率为 0.819。Χ2检验表明:c.2776 A>G位点基因型分布均符合Hardy-Weinberg 平衡(P>0.05) ,而c.1571 A>G位点基因型分布均偏离Hardy-Weinberg 平衡(P<0.05)。FADS2基因c.1571 A>G位点与c.2776 A>G位点间连锁不平衡系数r2为0.028,未达到显著水平(P>0.05)。c.1571 A>G位点与c.2776 A>G位点有3种单倍型,GA、GG和AA频率分别为0.705、0.181和0.114。多因素方差分析表明:FADS2-1571对C14:1含量、C14和C18不饱和指数的影响达到极显著水平(P<0.01),对C18:0、SFA和MUFA含量的影响达到显著水平(P<0.05)。GG型个体乳中C14:1含量、C14和C18不饱和指数显著高于AG型(P <0.05)。FADS2-2776位点对C16:1含量、 C16和C20不饱和指数的影响达到极显著水平(P<0.01),对C14:1含量的影响达到显著水平(P<0.05),GG型个体乳中C16:1含量、C16和C20不饱和指数显著高于AG型和AA型(P<0.05)。同时,FDAS2-1571-2776单倍型对C16:1含量和C20不饱和指数的影响达到显著水平(P<0.05),单倍型GG型个体乳中C16:1含量和C20不饱和指数显著高于GA型和AA型(P<0.05)。【结论】FADS2基因3′端非编码区SNP突变对中国荷斯坦牛乳中脂肪酸组成有重要影响。在进一步验证其功能情况下,可作为影响中国荷斯坦奶牛乳脂肪酸组成的主效基因加以利用。
关键词:中国荷斯坦牛;FADS2;SNPs;脂肪酸组成
联系方式:徐晨希, E-mail:chouchouxcx@163.com。王梦琦,E-mail:770406499@qq.com。徐晨希和王梦琦为同等贡献作者。通信作者毛永江,E-mail:cattle@yzu.edu.cn
0 引言
【研究意义】乳脂是牛奶重要的组成部分,乳脂的营养价值主要决定于乳中脂肪酸(fatty acid,FA)的构成。脂肪酸按其饱和程度可分为饱和脂肪酸(saturated fatty acid, SFA)、单不饱和脂肪酸(mono unsaturated fatty acid,MUFA)和多不饱和脂肪酸(poly unsaturated fatty acid,PUFA),按其链长度可分为长链脂肪酸(long chain fatty acids, LCFA, 13碳以上)、中链脂肪酸(middle chain fatty acids, MCFA,含6-12碳)和短链脂肪酸(short chain fatty acids,SCFA, 5碳以下)[1]。研究发现普通牛奶中 SFA、MUFA和PUFA的比值通常在70:25:5[2]。牛奶中不饱和脂肪酸以十八碳系列为主,主要有油酸(C18:1),亚油酸(C18:2),亚麻酸(C18:3)。当然,多不饱和脂肪酸中还含有对人体很有益的花生四烯酸(C20:4)和二十二碳六烯酸(C22:6,简称DHA)[2]。随着对乳脂的深入研究, 乳中PUFA的营养保健作用及其对乳制品组织结构和风味的影响作用开始受到人们普遍关注。【前人研究进展】最近的研究表明:PUFA具有促进动物生长发育、调控编码脂类代谢相关基因的表达、抑制炎症反应及促进免疫机能、提高动物繁殖性能等功能[3]。牛奶中FA的组成不仅受到环境因素的影响,更多的受内在因素影响,如物种、品种、基因、泌乳阶段、胎次、挤奶频率等[4-7]。脂肪酸脱氢酶(fatty acid desaturases,FADS)是一类能催化脂肪酸生成PUFAs的关键酶。其中FADS2是FADS基因家族中最重要的成员之一,是多不饱和脂肪酸合成代谢途径中关键的限速酶,可催化 2种必需脂肪酸亚油酸((LA, C18:2)和α-亚麻酸(ALA, C18:3)转化为γ-亚麻酸(GLA, C18:3)和十八碳四烯酸(18:4),并进一步脱氢形成长链多不饱和脂肪酸(long chain- poly unsaturated fatty acids, LCPUFAS),包括花生四烯酸(ARA,20:4n-6)、二十碳五烯酸(EPA, 20:5n-3) 和二十二碳六烯酸(DHA, 22:6n-3)等[8-11]。NWANKWO等[12]、NAKAYAMA等[13]的研究表明:人 FADS2基因SNP突变影响人体脂肪组织及血浆中 PUFA的含量,从而影响人类健康。最近人类全基因组关联分析(genome-wide association studies,GWAS)也证实了FADS2基因对脂类代谢疾病的作用[14-16]。另外,FADS2基因启动子区的甲基化对人类肝脏FADS2基因的表达活性也有较大影响[17]。在家畜方面也有部分有关 FADS2基因与生产性能,特别是脂肪产量和脂肪酸组成有关的报道。RENAVILLE 等[18]研究表明:FADS2基因多态性显著影响意大利猪肉中花生四烯酸和亚油酸含量及 γ-亚麻酸与亚油酸的比值;BOSCHETTI 等[19]发现慢速和中等速度生长的肉鸡胸肌中 FADS2基因表达量较高,也具有较高含量的长链多不饱和脂肪酸(LCPUFA),同时FADS2基因SNP突变对胸肌中FADS2基因表达量和LCPUFA也有显著影响;da COSTA等[20]发现肉牛FADS2基因表达量对其肝脏中脂肪酸组成有显著影响。奶牛FADS2基因研究方面,IBEAGHA-AWEMU等[21]对加拿大荷斯坦牛FADS2基因SNP多态性及其与乳中FA含量进行了关联分析。结果发现FADS2基因共24个SNP突变,位于内含子、编码区和3′非编码区(3′un-transcript region, 3′UTR)的 SNP数量分别为16、1和7个。其中FADS2基因3'UTR 1571 A/G和2776 A/G突变对乳中PUFA有显著影响。生物信息学分析表明,FADS2基因1571 A/G为miRNA-744结合位点。故推测miRNA-744可通过与FADS2基因结合,从而调控FADS2基因的表达,进而调节乳中 PUFA含量。【本研究切入点】在国内,梅秀丽[22]比较了不同饲养方式下优质鸡 FADS2基因的表达及其对脂肪酸组成的影响,朱世康等[23-26]对鸡FADS2基因5′端和3′端SNP多态及其与部分经济性状的相关性进行了研究,但未见其他畜种(包括奶牛)FADS2基因突变及其乳中FA组成方面的研究。【拟解决的关键问题】检测中国荷斯坦牛FADS2基因3′UTR SNP突变,同时分析SNP突变对乳中脂肪酸组成的影响,以期为中国荷斯坦牛乳中脂肪酸含量的分子育种提供理论依据。
1 材料与方法
1.1 试验材料
2011年3—4月于江苏省扬州市某大型奶牛场采集中国荷斯坦牛血样共 551头。采用尾静脉采血 10 mL/头,ACD抗凝剂,-20℃冷冻保存备用。采样时,同时选择体况相近(BCS=3.0±0.5)、无临床乳房炎、胎次在2—3胎、处于泌乳中后期(产奶100—300 d)的中国荷斯坦牛共300头,在进行DHI测定时,取全天混合奶样(早∶中∶晚=4∶3∶3)100 mL,其中50 mL送上海光明 DHI中心进行乳中体细胞数测定,另外50 mL用于实验室脂肪酸测定。牧场基本情况如下:全场共有奶牛1 000余头,其中成年泌乳奶牛近500头,散栏饲养,鱼骨式挤奶大厅进行挤奶,日挤奶 3次,采用全混合日粮(total mixed ration, TMR)饲喂。
1.2 DNA提取及FADS2基因3′端SNP突变检测
奶牛血液基因组DNA采用常规酚氯仿提取法,TE溶解,取部分DNA样品稀释至100 ng·µL-1,-20℃保存备用。
根据GenBank公布的牛的FADS2基因DNA序列(BC123735.1), 参考IBEAGHA-AWEMU等[21]对加拿大荷斯坦牛FADS2基因3′检测到的SNP位点信息(c.1571 A>G、c.2743 A>G和c.2776 A>G),用Primer5.0 软件设计2对引物,用于扩增3′端包括上述3个突变位点在内的DNA片段。PCR引物信息及扩增条件见表1。

表1 FADS2基因3'端PCR引物序列及扩增条件Table 1 The primers and PCR condition for amplification of 3' UTR of FADS2 gene
从所采551个样本中随机选择20个DNA样品,经PCR 扩增、测序,所得序列用DNAMAN(Ver 5.2)与GenBank中FADS2参考序列(BC123735.1)进行比对,确认中国荷斯坦牛FADS2基因3′端SNP突变位点。经对 20个样本的初步分析,c.2743 A>G和c.2776 A>G由于距离较近,连锁程度较高,连锁不平衡系数r2达0.7以上。故在接下来的SNP分型过程中,只对c.1571 A>G和c.2776 A>G进行分析。对大样本 FADS2 SNP突变检测采用飞行时间质谱法(MassARRAY® MALDI-TOF System,Sequenom,Inc.,USA)。同时,为保证SNP分析结果可靠性,其中40个样本重复测定2次(测试员并不知道这个40个是重复样本)。结果表明该方法SNP分型结果准确性为100%。
1.3 脂肪酸测定
脂肪酸测定在采样后3个月内完成。根据食品安全国家标准-婴幼儿食品和乳品中脂肪酸的测定(GB 5413.27-2010)并加以改进(食品安全国家标准)[27],用气相色谱仪进行测定(美国Agilent7890A,色谱柱为DB-23, 30 m×0.25 mm×0.25 μm),气相色谱仪使用程序升温进行检测[28]。使用Sigma公司的37种脂肪酸甲酯的混标作为标准品,用气相色谱仪对样品中所有脂肪酸进行测定。详细方法介绍见文献[29]。
1.4 统计分析
1.4.1 遗传学分析 用遗传学软件 Shesis进行常规群体遗传学统计分析(包括基因频率、基因型频率、Hardy-Weinberg平衡检测等),同时进行连锁不平衡和单倍型分析[30]。
1.4.2 关联分析 利用多因素方差分析法分析FADS2基因多态性及其单倍型对乳中脂肪酸组成的影响,模型如下:
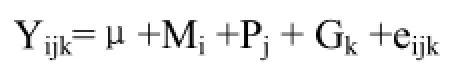
式中:Yijk为原料乳脂肪酸的测定值;μ为总体均值;Mi为泌乳阶段的固定效应(i =1,2),泌乳阶段按100 d间隔划为一个阶段,即泌乳后101—200 d为泌乳中期,201—300 d为泌乳后期;Pj为胎次固定效应(j=1,2);Gk为FADS2基因SNP位点基因型或单倍型的固定效应,eijk为随机残差。不同基因型或单倍型间的多重比较用Duncan法。分析单倍型对乳中FA组成的影响时,只选择在2个位点均是纯合子的个体进行分析。另外,由于部分牛只乳中体细胞数大于5.0×105·mL-1,初步认定患有隐性乳房炎,在最后进行关联分析时删除了这部分样品的数据。最后用于FADS2基因 SNP多态与脂肪酸含量分析的样品量为275头。
以上统计分析由 SPSS软件(Ver 16.0)GLM (General Linear Model)过程完成。
2 结果
2.1 FADS2基因多态性检测
对FADS2基因3′端2对引物扩增的PCR产物进行测序和比对分析,发现中国荷斯坦牛FADS2基因3′端SNP存在3个SNP突变位点:c.1571 A>G、c.2743 A>G、c.2776 A>G。结果与IBEAGHA-AWEMU等[21]对加拿大荷斯坦牛FADS2基因3′检测到的SNP位点信息完全一致,没有发现新的突变位点。另外,由于c.2743 A>G和c.2776 A>G由于距离较近,连锁程度较高,连锁不平衡系数r2达0.7以上, 因此在随后的飞行时间质谱法中,只选择c.2776 A>G进行检测。
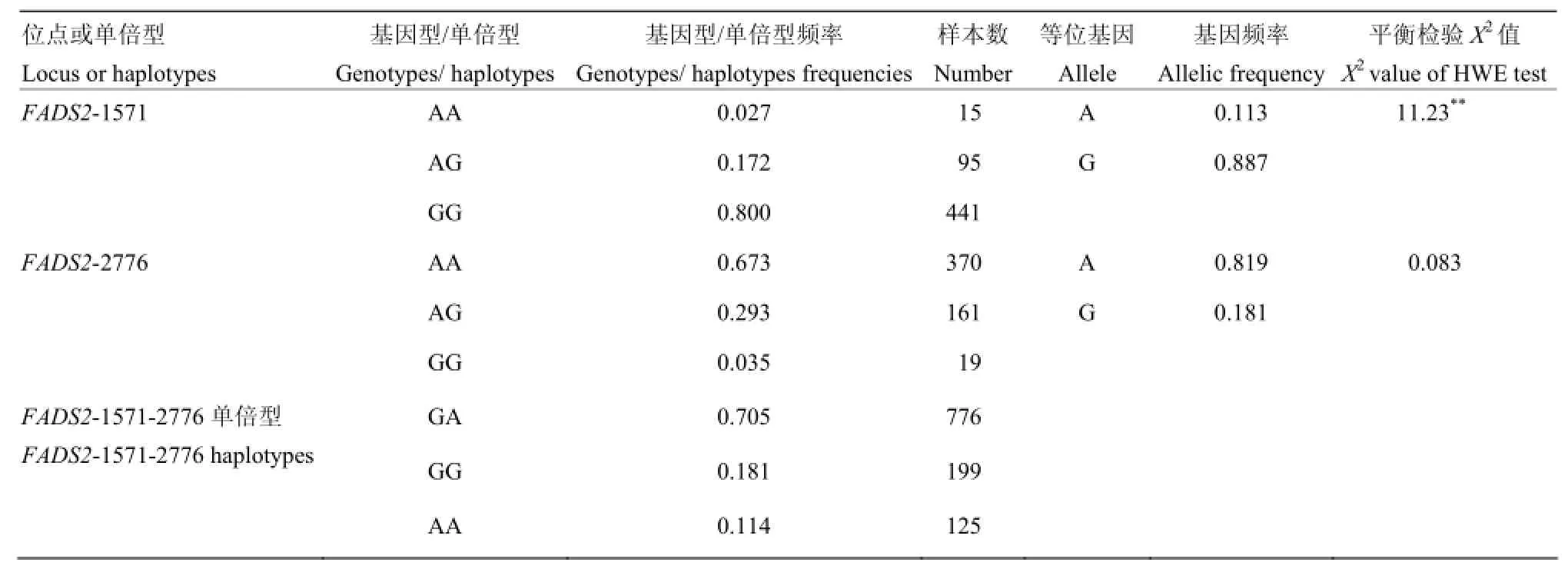
表2 FADS2基因 3′UTR等位基因、基因型频率、单倍型频率及H-W平衡检验Table 2 The frequencies of genotypic, allelic and haplotypes, value of X2test significance for 3′UTR of FADS2 gene
2.2 等位基因、基因型频率分布及连锁不平衡分析
如表2表示,c.1571 A>G位点GG型为优势基因型,基因型频率为0.800,G为优势等位基因,基因频率为0.887。c.2776 A>G位点AA为优势基因型,基因型频率为0.673,A为优势等位基因,基因频率为0.819。经X2检验,c.2776 A>G位点基因型分布符合Hardy-Weinberg平衡,而c.1571 A>G位点基因型分布偏离Hardy-Weinberg平衡。
连锁不平衡分析表明:c.1571 A>G位点与c.2776 A>G位点间连锁不平衡系数r2为0.028。经检验,该连锁不平衡系数未达到显著水平(P>0.05),说明两位点间不连锁。单倍型分析表明:c.1571 A>G位点与c.2776 A>G位点有3种单倍型,GA、GG和AA频率分别为0.705、0.181和0.114,AG型在群体中未出现。
2.3 脂肪酸测定
在本研究中,用气相色谱仪在原料乳中共检出23种脂肪酸(表3)。其中棕榈酸(C16:0)所占比例最大(29.56%),油酸(C18:1)次之(24.32%),豆蔻酸(C14:0)、硬脂酸(C18:0)和亚油酸(C18:2)都占有较高比例,而长链多不饱和脂肪酸(C20:3、C20:4和C22:6等)含量相对较少。就不饱和指数而言,C20的不饱和指数最高(88.18±1.06),其次为C18(70.13±0.41),最低为C16(4.66±0.21);SFA、MUFA和PUFA的比例分别为66.68∶27.43∶5.89,SCFA∶MCFA∶LCFA的比例分别为3.30∶17.12∶79.58。
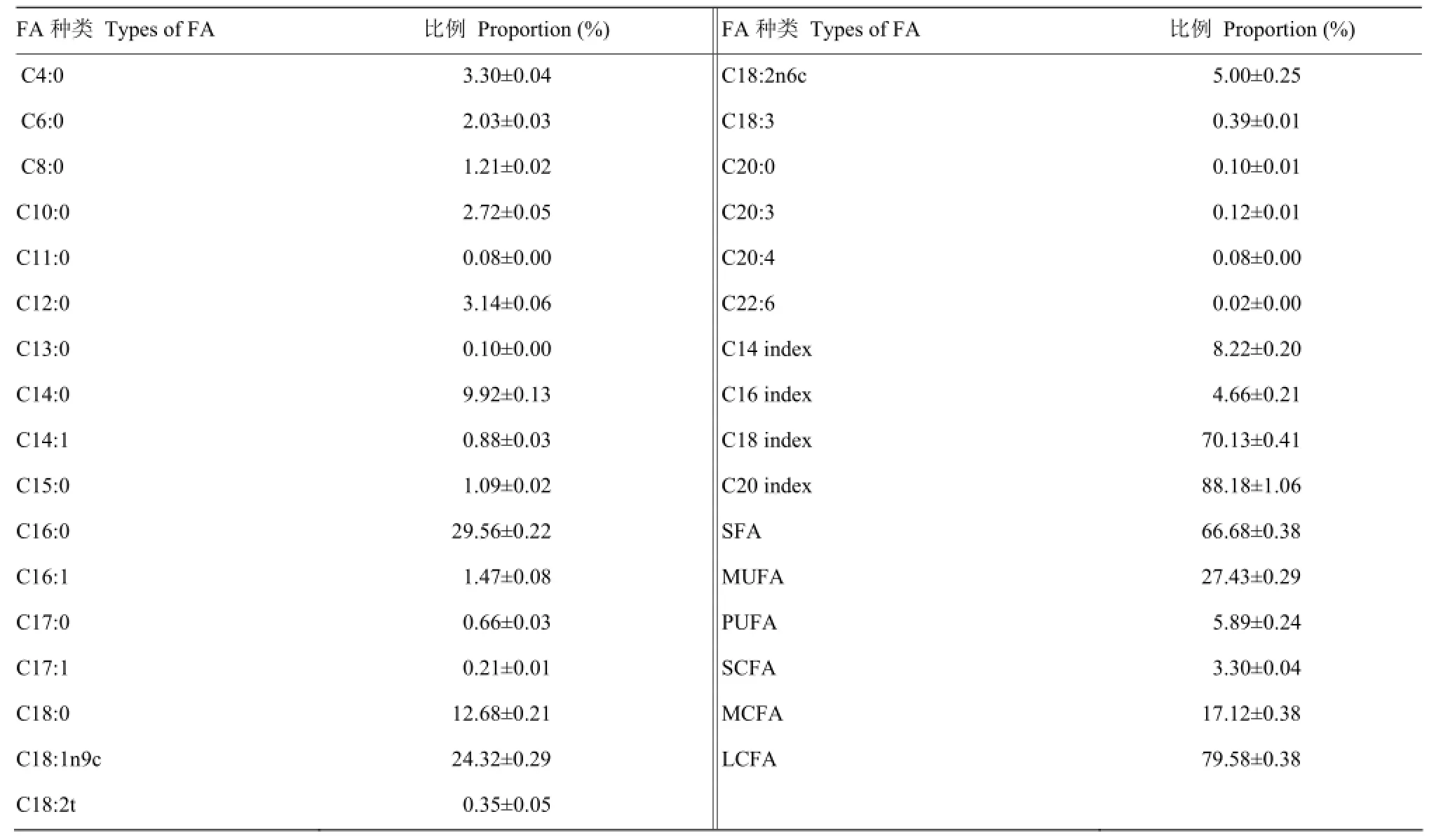
表3 原料乳脂肪酸所占比例Table 3 The proportion of each milk fatty acids in raw milk (M±SD)
2.4 FADS2基因对乳中脂肪酸类型及脂肪酸饱和指数的影响
FADS2基因1571位点不同基因型乳中不同类型脂肪酸含量及方差分析见表4。由表4可知:FADS2 1571位点对C14:1含量、C14和C18不饱和指数的影响达到极显著水平(P<0.01),对 C18:0、SFA和MUFA含量的影响达到显著水平(P<0.05),对C4:0含量和短链FA的影响接近显著水平(0.1>P>0.05),对其它不同类型脂肪酸含量及FA不饱和指数无显著影响(P>0.05)。多重比较表明:GG型个体乳中C14:1含量、C14和C18不饱和指数显著高于AG型(P<0.05),C18:0和SFA含量显著低于AG型(P<0.05);AA型个体乳中 MUFA含量显著高于 AG型(P<0.05)。
FADS2基因2776位点不同基因型乳中不同类型脂肪酸含量及方差分析见表5。由表5可知:FADS2 2776位点对C16:1含量、 C16和C20不饱和指数的影响达到极显著水平(P<0.01),对C14:1含量的影响达到显著水平(P<0.05),对C14:0 和C20:0含量的影响接近显著水平(0.1>P>0.05),对其他不同类型脂肪酸含量及 FA不饱和指数无显著影响(P>0.05)。多重比较表明:GG型个体乳中C16:1含量、C16和C20不饱和指数显著高于AG型和AA型(P <0.05),AG型个体乳中C14:1含量显著高于AA型和GG型(P<0.05)。
FDAS2-1571-2776不同单倍型对乳中不同类型脂肪酸含量及方差分析见表 6。由表 6可知:FDAS2-1571-2776单倍型对C16:1含量和C20不饱和指数的影响达到显著水平(P<0.05),对C4:0 含量和C16不饱和指数的影响接近显著水平(0.1>P>0.05),对其他不同类型脂肪酸含量及FA不饱和指数无显著影响(P>0.05)。多重比较表明:单倍型GG型个体乳中C16:1含量和C20不饱和指数显著高于GA型和AA型(P<0.05)。
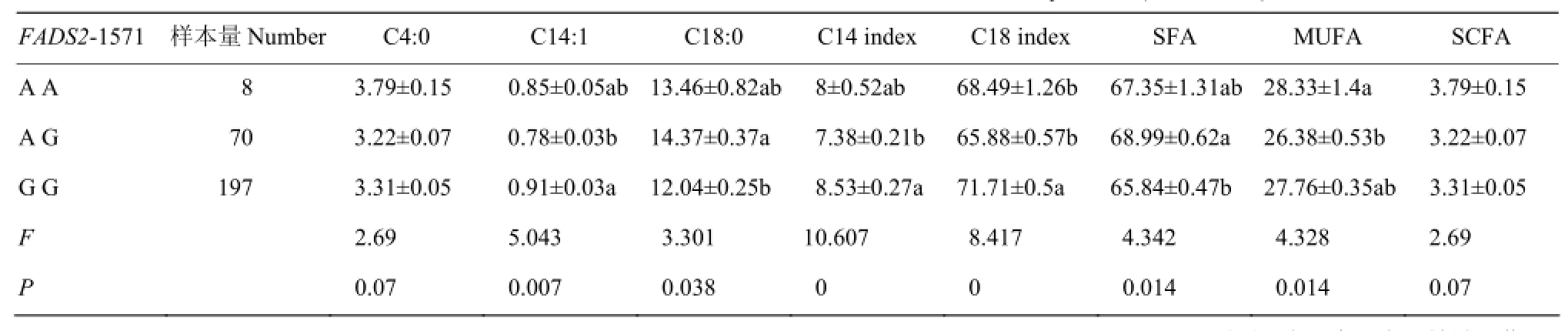
表4 FADS2基因1571位点对不同类型脂肪酸含量的影响Table 4 The effect of FADS2-1571 on the relative content of different kinds of milk fatty acids (%, M±SE)
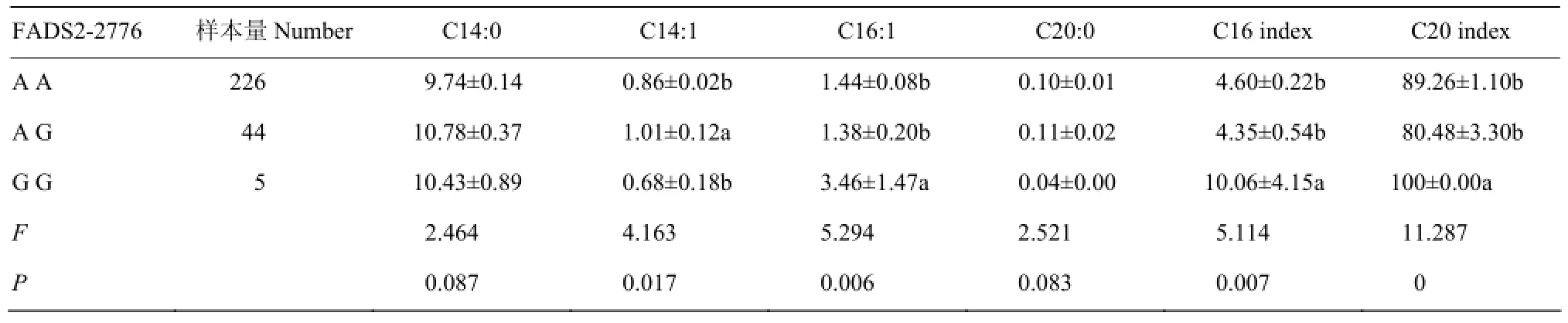
表5 FADS2基因2776位点对不同类型脂肪酸含量的影响Table 5 The effect of FADS2-2776 on the relative content of different kinds of milk fatty acids (%, M±SE)
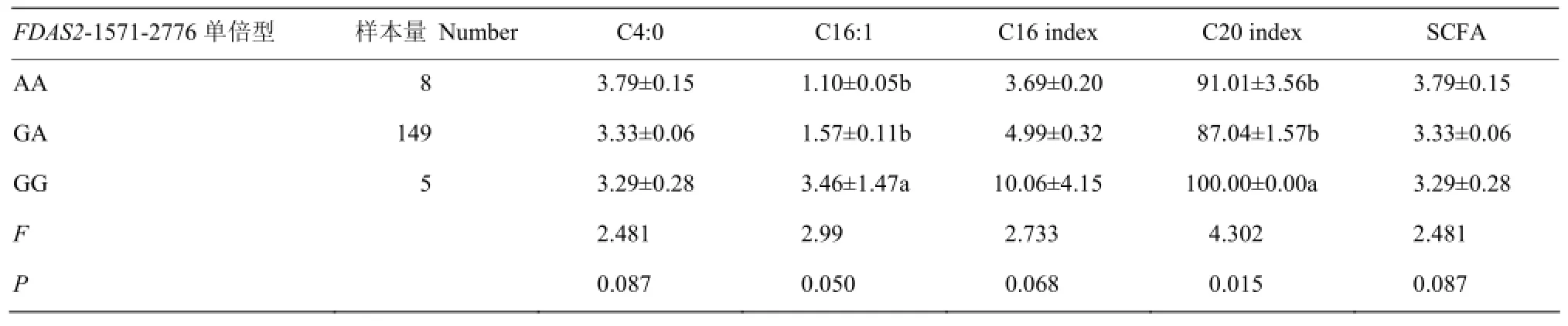
表6 FADS2基因1571-2776不同单倍型对脂肪酸含量的影响Table 6 The effects of haplotypes of FADS2-1571-2776 on the relative content of different kinds of milk fatty acids (%, M±SE)
3 讨论
3.1 FADS2基因遗传多态性分析
本研究首次对中国荷斯坦牛FADS2基因3′UTR区部分序列进行测定,发现 3个 SNP位点,与IBEAGHA-AWEMU等[21]对加拿大荷斯坦牛FADS2基因3′UTR区检测到的SNP位点信息进行对比,没有发现新的SNP,随后对其中2个SNP在大群体中进行了检测。经比较表明:c.1571 A>G 位点A基因频率低于加拿大荷斯坦牛相应基因频率, c.-2776 A>G位点A基因频率与加拿大荷斯坦牛相应基因频率基本一致,这种差异可能是由于所用种公牛不一致造成,同时也说明FADS2基因3′UTR区c.1571 A>G与c.-2776 A>G间连锁程度很低。另外,由于其他牛种或品种 FADS2基因序列没有被报道,今后有必要加强这一领域的研究,特别是乳脂率含量较高的品种(如娟姗牛、更赛牛、牦牛、水牛等)。
3.2 FADS2基因多态与乳脂肪酸组成及不饱和指数的关系
FADS2是PUFA合成代谢途径中关键的限速酶,其基因序列SNP突变对基因表达活性及血液和乳汁等体液中PUFA含量有较大影响,这已得到大量有关人类相关研究的证实[12-13,31-32]。本研究结果表明:FADS2基因 3′ UTR c.1571 A>G对 C14:1含量、MUFA含量、C14和C18不饱和指数的影响均达到显著水平(P<0.05),GG型个体乳中C14:1含量、C14 和 C18不饱和指数显著高于 AG型(P<0.05)。IBEAGHA-AWEMU等[21]对加拿大荷斯坦牛 FADS2基因与乳中FA含量的研究也得出类似结论。这说明FADS2基因3′UTR区c.1571 A>G对乳中UFA的含量有显著影响。生物信息学分析表明(Target Scan 7.0,http://www.targetscan.org/):该位点是bta-miR-744结合位点,可能是该位点的突变改变了microRNA与基因的结合,从而改变相关基因表达量的变化,最终导致乳中UFA含量的改变,但这一假设需要进一步用试验来验证。
在本研究中,FADS2 2776位点对C16:1含量、C16和C20不饱和指数的影响达到极显著水平(P<0.01),对C14:1含量的影响达到显著水平(P<0.05),GG型个体乳中C16:1含量、C16和C20不饱和指数显著高于AG型和AA型(P<0.05)。而IBEAGHAAWEMU等[21]的研究表明:FADS2-2776 G基因显著增加在乳中 omega-6 FAs(C20:3n6 和 C20:4n6)含量,而A基因显著降低乳中omega-6 FAs含量。本研究中虽未发现FADS2 2776位点对C20:3n6和C20:4n6含量的影响,但对C20不饱和指数有显著影响,且G基因对增加乳中C20:3n6和C20:4n6含量也是有利的。
最后,虽然本研究初步发现FADS2基因3′UTR区部分 SNP突变及其单倍型对乳中脂肪酸(特别是PUFA含量)和部分不饱和指数有显著影响。也应看到,由于各方面原因,本研究原料乳中FA测定的样本量相对较少,有必要对大样本奶牛群体进行每月一次的跟踪测定。如能改进乳中测定方法,在进行奶牛群生产性能测定(dairy herd improvement, DHI)时同时分析乳中FA含量,这对准确估计原料乳中FA含量的遗传参数和把乳中FA含量纳入奶牛的育种目标将大有帮助。
4 结论
在中国荷斯坦牛FADS2基因3′UTR区检测到3 个SNP突变,该突变对中国荷斯坦牛乳中脂肪酸组成有重要影响。在进一步验证其功能情况下,可作为影响中国荷斯坦奶牛乳脂肪酸组成的主效基因加以利用。
References
[1] HARVATINE K J, BOISCLAIR Y R, BAUMAN D E. Recent advances in the regulation of milk fat synthesis. Animal, 2009(3):40-54.
[2] WOODS V B, FEARON A M. Dietary sources of unsaturated fatty acids for animals and their transfer into meat, milk and eggs: a review. Livestock Science, 2009, 126: 1-20.
[3] LEE H, PARK W J. Unsaturated fatty acids, desaturases, and human health. Journal of Medicinal Food, 2014, 17(2): 189-197.
[4] TALPUR F N, BHANGER M I, KHOOHARO A A, MENON G Z. Seasonal variation in fatty acid composition of milk from ruminants reared under the traditional feeding system of Sindh, Pakistan. Livestock Science, 2008, 118: 166-172.
[5] TALPUR F N, BHANGER M I, KHUHAWAR M Y. Comparison of fatty acid and cholesterol content in the milk of Pakistani cow breeds. Journal of Food Composition and Analysis, 2006, 19: 698-703.
[6] HADDAD I, MOZZON M, STRABIOLI R, FREGA N C. Stereospecific analysis of triacylglycerols in camel (Camelus dromedarius) milk fat. International Dairy Journal, 2010, 20:863-867.
[7] WIKING L, NIELSEN J H, BÅVIUS A K, EDVARDSSON A,SVENNERSTEN-SJAUNJA K. Impact of milking frequencies on the level of free fatty acids in milk, fat globule size, and fatty acid composition. Journal of Dairy Science, 2006, 89: 1004-1009.
[8] GUILLOU H, RIOUX V, CATHELINE D, THIBAULT J N,BOURIEL M, JAN S, D'ANDREA S, LEGRAND P. Conversion of hexadecanoicacid to hexadecenoic acid by rat Delta 6-desaturase. The Journal of Lipid Research, 2003, 44(3): 450-454.
[9] DANDREA S, GUILLOU H, JAN S, CATHELINE D, THIBAULT J N, BOURIEL M, RIOUX V, LEGRAND P. The same rat Delta6-desaturase not only acts on 18- but also on 24-carbon fatty acids invery-long-chain polyunsaturated fatty acid biosynthesis. Biochemical Journal, 2002, 361(1): 49-55
[10] MARTINELLI N, GIRELLI D, MALERBA C, GUARINI P, ILLIG T,TRABETTI E, SANDRI M, FRISO S, PIZZOLO F, SCHAEFFER L,HEINRICH J, PIGNATTI P F, CORROCHER R, OLIVIERI O. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. The American Journal of Clinical Nutrition, 2008, 88:941-949.
[11] KOHAMA T, OLIVERA A, EDSALL L, NAGIEC M M, DICKSON R, SPIEGEL S. Molecular cloning and functional characterization of marine sphingosine kinase. The Journal of Biological Chemistry, 1998,273(37): 23722-23728.
[12] NWANKWO J O, SPECTOR A A, DOMANN F E. A nucleotide insertion in the transcriptional regulatory region of FADS2 gives rise to human fatty acid delta-6-desaturase deficiency. The Journal of Lipid Research, 2003, 44(12): 2311-2319.
[13] NAKAYAMA K, BAYASGALAN T, TAZOE F, YANAGISAWA Y,GOTOH T, YAMANAKA K, OGAWA A, MUNKHTULGA L,CHIMEDREGZE U, KAGAWA Y, ISHIBASHI S, IWAMOTO S. A single nucleotide polymorphism in the FADS1/FADS2 gene is associated with plasma lipid profiles in two genetically similar Asian ethnic groups with distinctive differences in lifestyle. Human Genetics,2010, 127: 685-690.
[14] ILLIG T, GIEGER C, ZHAI G, ROMISCH-MARGL W,WANG-SATTLER R, PREHN C, ALTMAIER E, KASTENMULLER G, KATO B S, MEWES H W, MEITINGER T, DE ANGELIS M H,KRONENBERG F, SORANZO N, WICHMANN H E, SPECTOR T D, ADAMSKI J, SUHRE K. A genome-wide perspective of genetic variation in human metabolism. Nature Genetics, 2010, 42(2):137-141.
[15] KATHIRESAN S, MELANDER O, GUIDUCCI C, SURTI A,BURTT N P, RIEDER M J, COOPER G M, ROOS C, VOIGHT B F,HAVULINNA A S, WAHLSTRAND B, HEDNER T, CORELLA D,TAI E S, ORDOVAS J M, BERGLUND G, VARTIAINEN E,JOUSILAHTI P, HEDBLAD B, TASKINEN M R, CHEH C N,SALOMAA V, PELTONEN L, GROOP L, ALTSHULER D M,ORHO-MELANDER M. Six new loci associated with blood lowdensity lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nature Genetics, 2008, 40(2): 189-197.
[16] TANAKA T, SHEN J, ABECASIS G R, KISIALIOU A, ORDOVAS J M, GURALNIK J M, SINGLETON A, BANDINELLI S, CHERUBINI A, ARNETT D, TSAI M Y, FERRUCCI L. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI study. PLoS Genetics, 2009, 5(1): e1000338.
[17] HOWARD T D, MATHIAS R A, SEEDS M C, HERRINGTON D M,HIXSON J E, SHIMMIN L C, HAWKINS G A, SELLERS M,AINSWORTH H C, SERGEANT S, MILLER L R, CHILTON F H. DNA methylation in an enhancer region of the FADS cluster is associated with FADS activity in human liver. PLoS ONE, 2014, 9(5):e97510.
[18] RENAVILLE B, PRANDI A, FANB B, SEPULCRI A, ROTHSCHILD M F , PIASENTIER E. Candidate gene marker associations with fatty acid profiles in heavy pigs. Meat Science , 2013, 93: 495-500.
[19] BOSCHETTI E, BORDON A, MELUZZI A, CASTELLINI C, DAL BOSCO A, SIRRI F. Fatty acid composition of chicken breast meat is dependent on genotype-related variation of FADS1 and FADS2 gene expression and desaturating activity. Animal, 2015, 16: 1-9.
[20] DA COSTA A S H, BESSA R J B, PIRES V M R, ROLO E A,PINTO R M A, FONTES C M G A, PRATES J A M. Is hepatic lipid metabolism of beef cattle influenced by breed and dietary silage level?. BMC Veterinary Research, 2014, 10: 65-78.
[21] IBEAGHA-AWEMU E M, AKWANJI K A, BEAUDOIN F, ZHAO X. Associations between variants of FADS genes and omega-3 and omega-6 milk fatty acids of Canadian Holstein cows. BMC Genetics,2014, 15: 25
[22] 梅秀丽. 优质鸡不同饲养方式下 FADS1-FADS2基因的表达及其对脂肪酸组成的影响[D]. 雅安: 四川农业大学, 2012. MEI X L. Fatty acid desaturase (FADS1) and fatty acid desaturase 2(FADS2) gene expression and the effects on fatty acids content of high-quality chicken in different housing systems[D]. Ya'an: Sichuan Agricultural University, 2012. (in Chinese)
[23] 朱世康. 鸡FADS2基因5'_端SNP鉴定及启动子功能分析[D]. 郑州: 河南农业大学, 2013. ZHU S K. Polymorphisms in the 5' region of chicken FADS2 gene and the promoter function analysis[D]. Zhengzhou: Henan Agricultural University, 2013. (in Chinese)
[24] 洪雪莹. 鸡FADS2基因3_UTR区多态性检测及其与经济性状关联性分析[D]. 郑州: 河南农业大学, 2012. HONG X Y. Polymorphisns in the 3'UTR region of chicken FADS2 Gene and their association with economic traits[D]. Zhengzhou:Henan Agricultural University, 2012. (in Chinese)
[25] 卢冉. 鸡Δ6脂肪酸脱氢酶基因启动子区域多态性及基因时空表达的研究[D]. 郑州: 河南农业大学, 2011. LU R. Study of chickenΔ6 fatty acid desaturase gene's promotor region polymorphism and gene expression[D]. Zhengzhou: HenanAgricultural University, 2011. (in Chinese)
[26] ZHU S K, TIAN Y D, ZHANG S, CHEN Q X, WANG Q Y, HAN R L,KANG X T. Adjacent SNPs in the transcriptional regulatory region of the FADS2 gene associated with fatty acid and growth traits in chickens. Genetics Molecular Research, 2014, 13(2): 3329-3336.
[27] 中华人民共和国卫生部. 食品安全国家标准-婴幼儿食品和乳品中脂肪酸的测定: GB 5413.27—2010[S/OL]. 北京: 中国标准出版社,2010. National Health and Family Planning Commission of the People's Republic of China. National Food Safety Standard-Determination of Fatty Acids in Foods for Infants and Young Children, Milk and Milk Products: GB 5413.27—2010[S/OL]. Beijing: Standards Press of China, 2010. (in Chinese)
[28] 林秋萍, 李瑾, 冯书惠. 气相色谱法快速测定牛奶中脂肪酸. 食品科学, 2005, 26(8): 346-348. LIN L P, LI J, FENG S H. Fast quantitative determination of fatty acid in milk. Food Science. 2005, 26(8): 346-348. (in Chinese)
[29] 毛永江, 常玲玲, 杨章平, 吴海涛, 陈莹, 施雪奎, 李云龙, 梁祥焕,尹召华. 中国荷斯坦牛乳中体细胞评分与脂肪酸含量和组成的相关分析. 中国农业科学. 2011, 44(24): 5073-5082. MAO Y J, CHANG L L, YANG Z P, WU H T, CHEN Y, SHI Y K, LI Y L, LIANG X H, YIN Z H. Correlation between the SCS and the amount and composition of fatty acids in milk of Chinese Holstein. Scientia Agricultura Sinica, 2011, 44(24): 5073-5082. (in Chinese)
[30] SHI Y Y, HE L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Research, 2005, 15(2): 97-98.
[31] SOLAKIVI T, KUNNAS T, JAAKKOLA O, RENKO J, LEHTIMÄKI T,NIKKARI S T. Delta-6-desaturase gene polymorphism is associated with lipoprotein oxidation in vitro. Lipids in Health and Disease, 2013,12: 80.
[32] SONG Z K, CAO H Y, QIN L, JIANG Y F. A case-control study between gene polymorphisms of polyunsaturated fatty acid metabolic rate-limiting enzymes and acute coronary syndrome in Chinese Han Population. BioMed Research International, 2013, ID: 928178. http:ldx.doi.org/10.1155/2013/928178.
(责任编辑 林鉴非)
Effects of SNPs in the 3' Untranslated Regions of FADS2 on the Composition of Fatty Acids in Milk of Chinese Holstein
XU Chen-xi, WANG Meng-qi, ZHU Xiao-rui, ZHANG Yu-feng, XIA Hai-lei, LIU Xian-hui, WANG Xiao-long,ZHANG Hui-min, YANG Zhang-ping, MAO Yong-jiang
(College of Animal Science and Technology, Yangzhou University, Yangzhou 225009, Jiangsu)
Abstract:【Objective】D-6-fatty acid desaturase 2 (FADS2) is one of the key limiting enzymes in the conversion of dietaryessential 18 carbon PUFAs (18C-PUFAs) such as linoleic acid (LNA,C18:2n-6) to γ-linolenic acid (GLA, 18:3n6), eicosapentaenoic acid (EPA, C20:5n3) and docosahexaenoic acid (DHA, C22:6n3). The objective of this study was to investigate the effects of SNPs in the 3' untranslated regions of FADS2 gene on the compositions of fatty acids in milk of Chinese Holstein.【Method】In this study,20 Chinese Holstein cows were selected randomly for PCR amplification and sequencing of the 3' untranslated regions of FADS2 gene used for SNP discovery. Then the Chinese Holstein cows (n =551) were genotyped using Sequenom MassARRAY (Sequenom Inc., San Diego, CA) based on the previous SNP information in this study, and the associations between SNPs or haplotypes and compositions of fatty acids, unsaturated indexes of fatty acids in milk were analyzed by the least squares method in the GLM procedure of SPSS.【Result】Three SNPs (c.1571 A>G, c.2743 A>G and c.2776 A>G) were identified in the 3' untranslated regions of FADS2 gene. The genotype GG was the dominant genotype, which the frequencies were 0.800 for c.1571 A>G, allele G was dominant allele with the frequency of 0.887. The genotype AA was the dominant genotype, which the frequencies were 0.673 for c. 2776 A>G. The allele A was dominant allele with the frequency of 0.819. The coefficient of disequilibrium was 0.028 between c.1571 A>G and c.2776 A>G, which was not significant at P<0.05. There were 3 haplotypes for FADS2 c.1571 A>G and c.2776 A>G, which the frequencies were 0.705, 0.181 and 0.114 for GA, GG and AA, respectively. Χ2test indicated that the c.2776 A>G fitted with Hardy-Weinberg equilibrium in the population (P>0.05), and the c.1571 A>G deviated from Hardy-Weinberg equilibrium (P<0.05). The SNP c.1571 A>G showed a very significant association with C14:1, unsaturated indexes of C14 and C18 (P<0.01), and significant association with C18:0, SFA and MUFA (P<0.05). The individuals with genotype GG had higher C14:1, unsaturated indexes of C14 and C18 than genotypes AG (P<0.05). The SNP c.2776 A>G showed a very significant association with C16:1, unsaturated indexes of C16 and C20 (P<0.01), and significant association with C14:1 (P<0.05). The individuals with genotype GG had higher C16:1, unsaturated indexes of C16 and C20 than genotypes AG and AA (P<0.05). In the meantime, the haplotype of FADS2 1571-2776 showed a significant association with C16:1, unsaturated indexes of C20 (P<0.05). The individuals with genotype GG had higher C16:1, unsaturated indexes of C20 than genotypes GA and AA (P<0.05). 【Conclusion】The SNPs in the 3' untranslated regions of FADS2 gene have significant genetic effects on composition of fatty acids in milk, but further investigation will be required to elucidate the biological and practical relevance of these SNPs.
Key words:Chinese Holstein; FADS2; SNPs; composition of fatty acids
收稿日期:2015-10-28;接受日期:2016-03-18
基金项目:国家自然科学基金(31272407, 31372286,31472067)、江苏省优势学科(PAPD)、国家级大学生创新训练计划(201411117031)、江苏省大学生实践创新训练计划(201411117031Z)

